Abstract
Single-seeded fruit of the sacred lotus Nelumbo nucifera Gaertn var. China Antique from NE China have viability as long as ~1300 years determined by direct radiocarbon-dating, having a germination rate of 84%. The pericarp, a fruit tissue that encloses the single seeds of Nelumbo, is considered one of the major factors that contribute to fruit longevity. Proteins that are heat stable and have protective function may be equally important to seed viability. We show proteins of Nelumbo fruit that are able to withstand heating, 31% of which remained soluble in the 110°C-treated embryo-axis of a 549-yr-old fruit and 76% retained fluidity in its cotyledons. Genome of Nelumbo is published. The amino-acid sequences of 11 “thermal proteins” (soluble at 100°C) of modern Nelumbo embryo-axes and cotyledons, identified by mass spectrometry, Western blot and bioassay, are assembled and aligned with those of an archaeal-hyperthermophile Methancaldococcus jannaschii (Mj; an anaerobic methanogen having a growth optimum of 85°C) and with five mesophile angiosperms. These thermal proteins have roles in protection and repair under stress. More than half of the Nelumbo thermal proteins (55%) are present in the archaean Mj, indicating their long-term durability and history. One Nelumbo protein-repair enzyme exhibits activity at 100°C, having a higher heat-tolerance than that of Arabidopsis. A list of 30 sequenced but unassembled thermal proteins of Nelumbo is supplemented.
Keywords: Nelumbo nucifera China Antique, Eleven 100°C-Heat-Soluble Proteins, Hyperthermophile-Mesophile Protein-Alignments, Stress-and-Repair Proteins, 30 Additional Thermal-Proteins
Introduction
Fruit of Nelumbo nucifera Gaertn var. China Antique hundreds of years in age, discovered in a dry lakebed in NE China at Xipaozi Village, Pulandian, Liaoning Province (Chen et al., 1965), were collected in 1952 by botanists of Academia Sinica, Beijing Institute of Botany (Chang, 1978). The oldest germinated fruit was directly radiocarbon dated as being 1300 yr-old (Shen-Miller et al. 1995). At present, a total of 16 old fruit of China Antique collected from the same lakebed have been shown to germinate within ~3 days (84% germination). Remarkably, this rate of germination of the old fruit is comparable not only to the modern controls (93%) but also to those of other crops 2–3 yr in age (Priestley, 1986).
The Nelumbo pericarp (the fruit coat that encloses single seed) is the first line of defense of Nelumbo that protects the seed and preserves its viability. Such pericarps are robust and impermeable to water as documented by Shen-Miller et al. (submitted). The coat of an old fruit has been shown to have elastic stiffness and hardness equivalent to that of the antlers of elk (Shen-Miller et al. submitted). These Nelumbo fruit coats are wax-covered and reinforced with layers of cells covered with water-insoluble suberin (Shaw, 1929; Shen-Miller et al. submitted). It is further infused with latex (Esau & Kosakai, 1975; Tele Images-Nature, 2003; Shen-Miller et al. submitted). The robust protective Nelumbo pericarp is superbly ‘architectured’ – both structurally and chemically to protect the seed within.
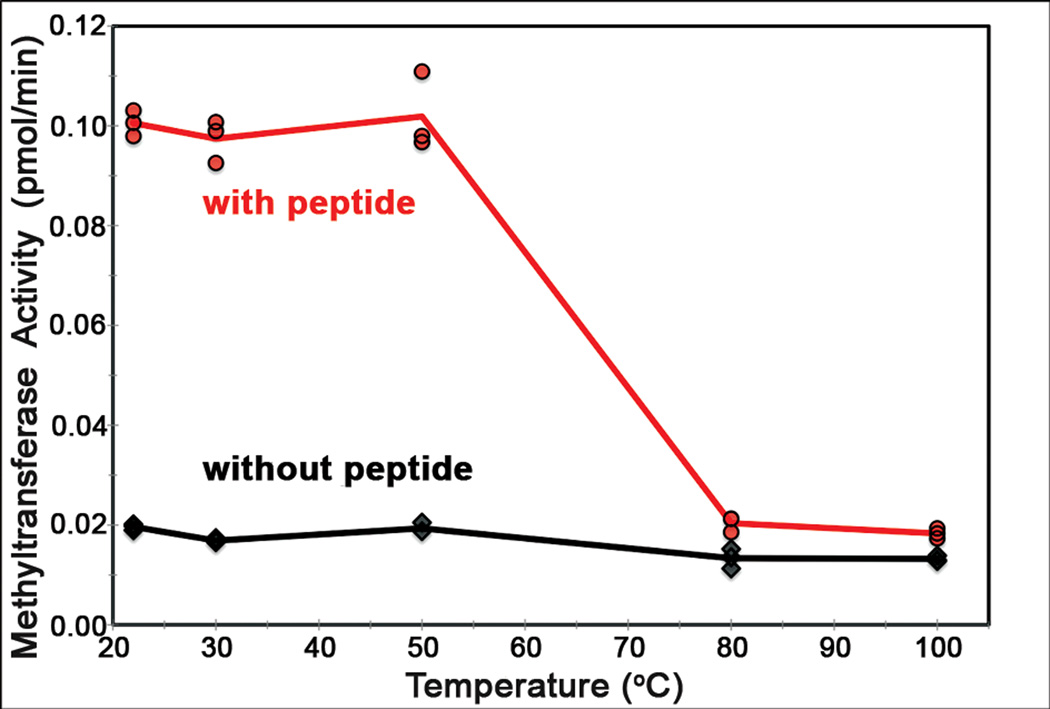
The genome of Nelumbo nucifera var. China Antique has recently been annotated (Ming et al. 2013), having a size of ~1 Gb and coding for ~27,000 proteins. In this paper we report the presence of proteins in embryo axes and cotyledons of Nelumbo seeds that remain soluble after having been heated to as high as 110°C. The fluidity exhibited by Nelumbo seed-proteins under stress may certainly play a role in long-term maintenance of seed viability. All of the 11 “thermal” proteins (heat soluble at 100°C) here identified and aligned have roles in cellular protection and restoration of plant functions under stress. Additionally, a list of 30 thermal proteins of Nelumbo assembled in Supplement 2 (http:_______) were identified by mass spectrometry. One thermal protein, PIMT (protein L-isoaspartyl methyltransferase), exhibits protein-repair activity after heating to 100°C. The optimal activity of enzymes of most mesophile plants occurs in temperature range of 25°–50°C (Vieille and Zeihus, 2001). Interestingly, two of the Nelumbo thermal enzymes, PIMT and CuZn-SOD (Supp. 1), tested here and elsewhere (Fig. 6; Chen et al. 2011), show to maintain higher activity following heat treatment than, respectively, those of Arabidopsis (Villa et al. 2006) and pearl millet (Mahanty et al. 2012).
Fig. 6.
PIMT bioassay of heat stability of the activity of protein L-isoaspartyl methyltransferase in soluble extracts of embryo axis of modern Nelumbo nucifera Gaertn var. China Antique, heated for 10 min at ~22°, 30°, 50°, 80°, 100°C. The enzyme activity is shown in the presence (red curve) and absence (black curve) of a synthetic substrate L-isoaspartyl-containing peptide in triplicate assays.
Ohga (1926) was the first to report discovery of old Nelumbo fruits at Xipaozi Village, NE China (Chen et al. 1965), the source of the seeds studied here. He reports germination of these fruits after 120°C steam treatment (Ohga, 1927), and Ding et al (2008) using 100°C air for 15 min result in 0% germination in maize, whereas, 24 hr in such heat Nelumbo still has ~14% germination. Huang et al. (2000) also show Nelumbo fruit germination after 100°C heating, other crop seeds, such as bean, corn and peanut did not. These data suggest that Nelumbo proteins active during seed germination are highly heat stable.
Stability of proteins at high temperatures is due to many factors (Unsworth et al. 2007), among others the co-occurrence of metabolites and sugars and the presence of polar amino acids. Specific polar residues (viz., Asp, Glu, Lys, Arg, Tyr: D-E-K-R-Y) can enhance the occurrence of intra-subunit ion-pair formation that confers thermal stability to the proteins. Like thermal proteins known from hyperthermophiles and used for bioengineering (Vieille and Zeihus, 2002; Unsworth et al. 2007), the thermal proteins here identified in seeds of Nelumbo could prove useful in protein engineering, their presence in a mesophile expanding the temperature range of enzymatic activity.
Multiple shared sequence alignments are documented for 11 Nelumbo thermal proteins and the archaeal hyperthermophile Methancaldococcus jannaschii (an anaerobic autotrophic methanogen having a growth optimum of 85°C; Stetter, 1996; Vieille and Zeikus, 2001; old name Methanococcus jannaschii) as well as with five mesophile angiosperms (Arabidopsis, corn, grape, poplar and soybean) (Supp. 1, http: _______).
M. jannaschii the first archaeal hyperthermophile sequenced and having ~1738 predicted protein-coding genes (Bult et al. 1996), is a marine single-celled methanogen occurring at abyssal depth (~2600 m below the sea level) in the vicinity of the “white smokers” of hydrothermal vents in boiling water and extreme hydrodstatic pressure (DeWeerdt, 2002). The oceans of the early Earth were likely hot (Kasting and Howard, 2006; Robert and Chaussidon, 2006), populated at least in part by archaeal hyperthermophiles. Carbon isotopic data indicate that methanogens, all members of the Archaea, were extant at ≥2.8 billion years ago (Schopf, 1994). The proteins of Mj used in the alignment presented here have been evolutionarily highly conserved, their coding sequences occurring also in the genome of Nelumbo and angiosperms. More than half (55%) of the 11 Nelumbo thermal proteins identified are present in Mj, documenting their existence over billions of years.
About Sacred Lotus Nelumbo nucifera
Nelumbo nucifera Gaertn belongs to the smallest family of angiosperms, the Nelumbonaceae that includes only a single genus Nelumbo and its two species, N. nucifera (Asia) and N. lutea (N. America). These taxa are related neither to the water lily (Nymphaeaceae; e.g., Egyptian lily-of-the-Nile, Nymphpaea caerulea), to Lotus japonica (Leguminosae), nor to the “lotos-fruits” of Ziziphus lotus (Rhamnaceae). Ancient Greeks used “lotus” (“lotos”) to refer to many unrelated plants including Nelumbo (Shen-Miller et al. submitted). To avoid such confusion, “lotus” will not be used in the following (and hopefully not in future literature) pertaining to Nelumbo. Instead, we will refer to the plant for which data are reported here by its genus name, “Nelumbo,” not encumbered by italics.
Materials and Methods
Proteins Isolation and Heat Treatment
After pre-testing Nelumbo fruit viability using the sink/float water test (“floaters” invariably being not germinable), those deemed viable were selected for experimentation. Whole embryo axes and cotyledon tissues of equivalent dry weight (~25 mg) of a “sinker” fruit of 459-yr-old China Antique OL96-60 (Table 1, Shen-Miller et al. submitted) and a modern control were powdered and extracted with Tris buffer (50 mM Tris-HCl, 5 mM MgCl2, 0.5 mM DTT). The extracts were spun and the supernatants estimated for protein content by absorbance measured at 595 nm and by use of the standards of bovine serum albumin (Bradford, 1976). Aliquots of protein extracts in pressure–capped microfuge tubes were heated for 10 min at 30°, 50°, 80°, 100° and 110°C in temperature controlled water-baths (30° to 100°C) and glycerol-baths (110°C). After chilling on ice, the tubes were spun, the protein quantity of the resultant supernatants (constituting the “crude proteins;” Table 1) were estimated, and the aliquots were frozen and stored for SDS gel-electrophoresis (Fig. 1). Similar procedures were used for samples of trypsin digestion of in-gel protein for tandem mass-spectrometry (Fig. 2).
Table 1.
Thermostable proteins in embryo axes and cotyledons of a modern and a 549-yr-old fruit of Nelumbo nucifera Gaertn var. China Antique, heated for 10 min at room temperature (Rt, ~22°), 30°, 50°, 80°, 100° and 110°C; ± (standard deviation; n=5).
| Temperatures (°C) | Rt | 30 | 50 | 80 | 100 | 110 |
|---|---|---|---|---|---|---|
| Embryo Axis – Modern | ||||||
| µg·mg−1 | 70.3±12.3 | 63.5±1.0 | 66.2±14.8 | 32.6±16.2 | 9.1±2.0 | 7.1±5.3 |
| Recovery, % | 100 | 90±2 | 94±7 | 46±13 | 13±3 | 10±5 |
| Embryo Axis - 549-yr-old | ||||||
| µg·mg−1 | 73.0±3.3 | - | 100±34 | 31.8±0.3 | - | 22.6±3.6 |
| Recovery, % | 100 | - | 137±34 | 44±2 | - | 31±5 |
| Cotyledons – Modern | ||||||
| µg·mg−1 | 62.6±3.4 | - | 57.0±15.8 | 32.9±5.1 | 17.0±3.4 | 21.9±3.5 |
| Recovery, % | 100 | - | 91±9 | 53±10 | 28±6 | 36±6 |
| Cotyledons - 549-yr-old | ||||||
| µg·mg−1 | 14.7±5.9 | - | 17.7±1.2 | 17.5±5.4 | - | 11.1±0.2 |
| Recovery, % | 100 | - | 120±40 | 119±18 | - | 76±1.0 |
Fig. 1.
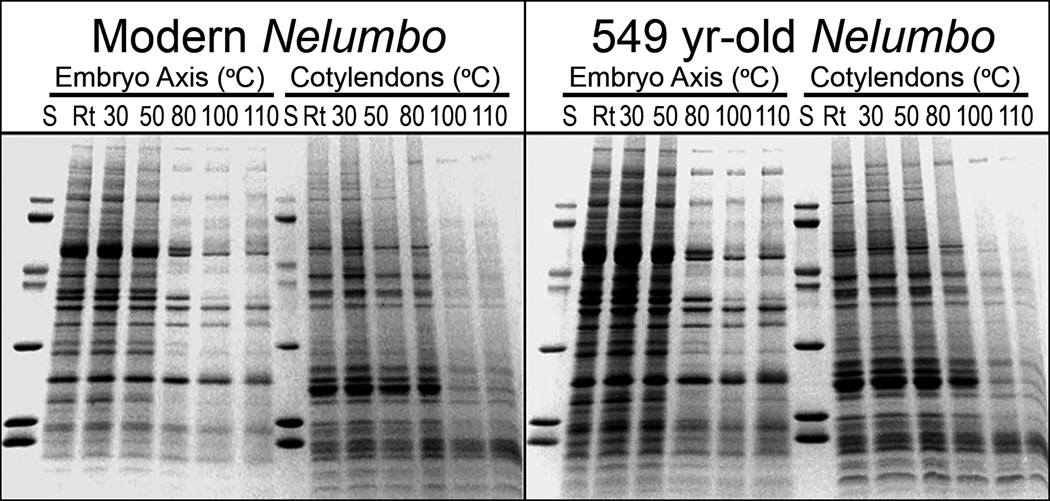
1D-SDS gels of heat-soluble proteins in fruit of Nelumbo nucifera Gaertn var. China Antique: Embryo axes and cotyledons of a modern and a 549-yr-old fruit heated for 10 min at room temperature (Rt, ~22°), 30°, 50°, 80°, 100°, and 110°C; S, protein standards from top to bottom: 78, 67, 45, 39, 27, 18 and 12 kDa.
Fig. 2.
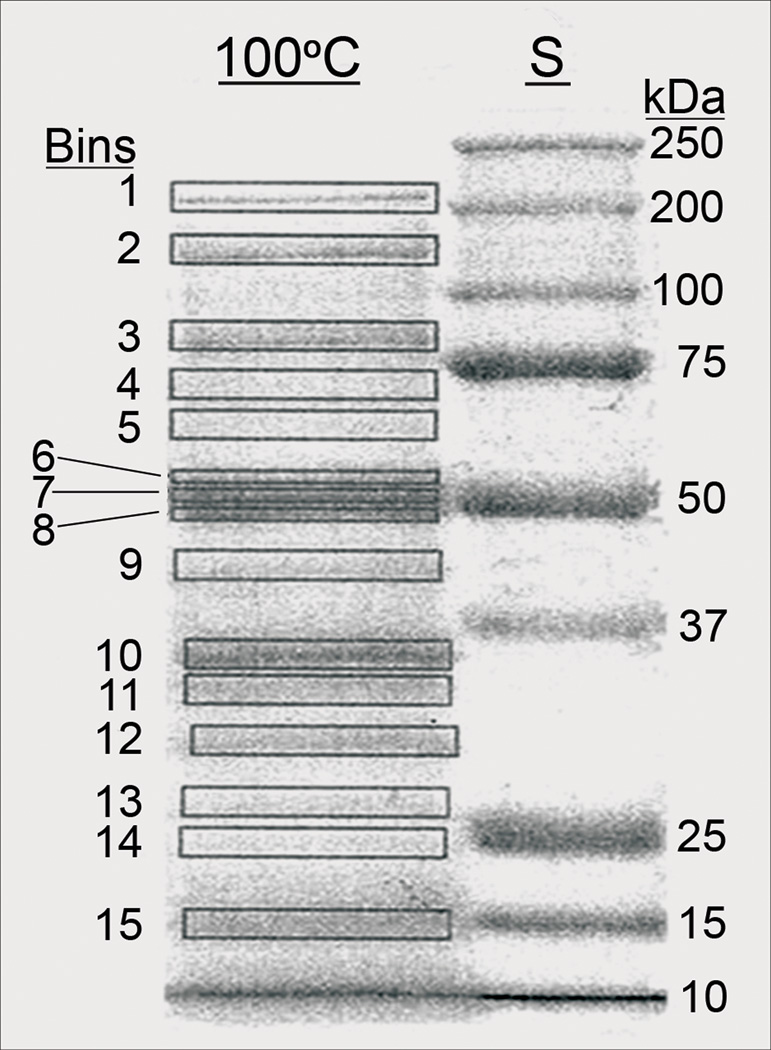
Embryo-axis proteins of a modern fruit of Nelumbo nucifera Gaertn var. China Antique, heated for 10 min at 100°C. S, protein standards: 250 to 10 kDa. Protein bands are boxed into Bins 1 through 15. Proteins eluted from these Bins have been identified by MS (see Supplements 1 [aligned], 2).
Gel Electrophoresis
The method of Laemmli (1970) was employed for 1D SDS-PAGE gel-electrophoresis of the heat-treated proteins, using linear gradient gels of 5 to 25% w/v polyacrylamide. Equal aliquots of heat-treated proteins from embryo axes and cotyledons and corresponding unheated controls were electrophoresed and compared with protein standards having masses ranging from 78 to 12.4 kDa. The resultant gels were stained with 0.1% Coomassie Blue w/v in 30% methanol with 10% HAc v/v, developed at 50°C, and photographed (Fig. 1). For MS identification of protein, 1D-SDS gels (BioRad) of 100°C treated extracts of embryo axes and cotyledons of modern China-Antique were prepared (Fig. 2).
Western Blots
Western blotting of graded 1D SDS-PAGE gels of the heat-treated crude proteins of modern embryo axes and cotyledons of China Antique followed the procedures of Towbin et al. (1979) and Burnett (1981). The blotted Millipore membranes (PVDF, BioRad) were stained with 0.2% Ponceau-S to mark the protein standards. The membrane was destained with water and finally with PBS-T (4 mM KH2PO4, 16 mM Na2HPO4, 115 mM NaCl, in 0.05% Tween-20, w/v). For protein identification, the destained membrane containing the blotted Nelumbo protein was incubated at room temperature together with dilutions of the appropriate antiserum for 1–12 hr in PBS-T. The primary antibodies used included rabbit anti-spinach chloroplast chaperonin CPN20, anti-pea chloroplast CPN60 (Bertsch et al. 1992; Fig. 5), anti-barley dehydrin (Close, 1997; Fig. 3), and rabbit anti-Arabidopsis PIMT1 (Xu et al. 2004; Fig. 4).
Fig. 5.
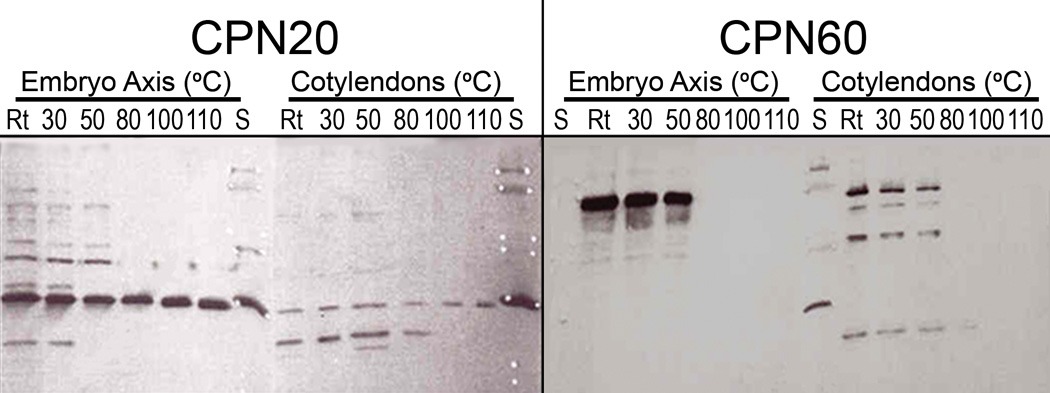
Western blot of CPN20 (~27 kDa) and CPN60 (~60 kDa) of embryo axis and cotyledons of modern Nelumbo nucifera China Antique heated for 10 min at different temperatures; S, protein standards from top to bottom: 78, 67, 45, 39, 27, 18 and 12 kDa (cf. under CPN20 cotyledon, S, “white-dots”).
Fig. 3.
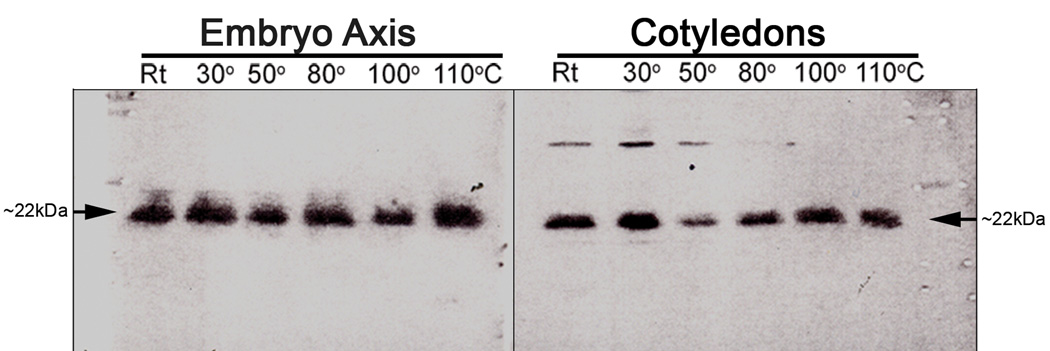
Western blots of dehydrin from embryo axis and cotyledons of modern Nelumbo nucifera Gaertn var. China Antique. Crude protein heated for 10 min at room temperature (RT, ~22°), 30°, 50°, 80°, 100°, 110°C; arrows indicate location of dehydrin, ~22 kDa.
Fig. 4.
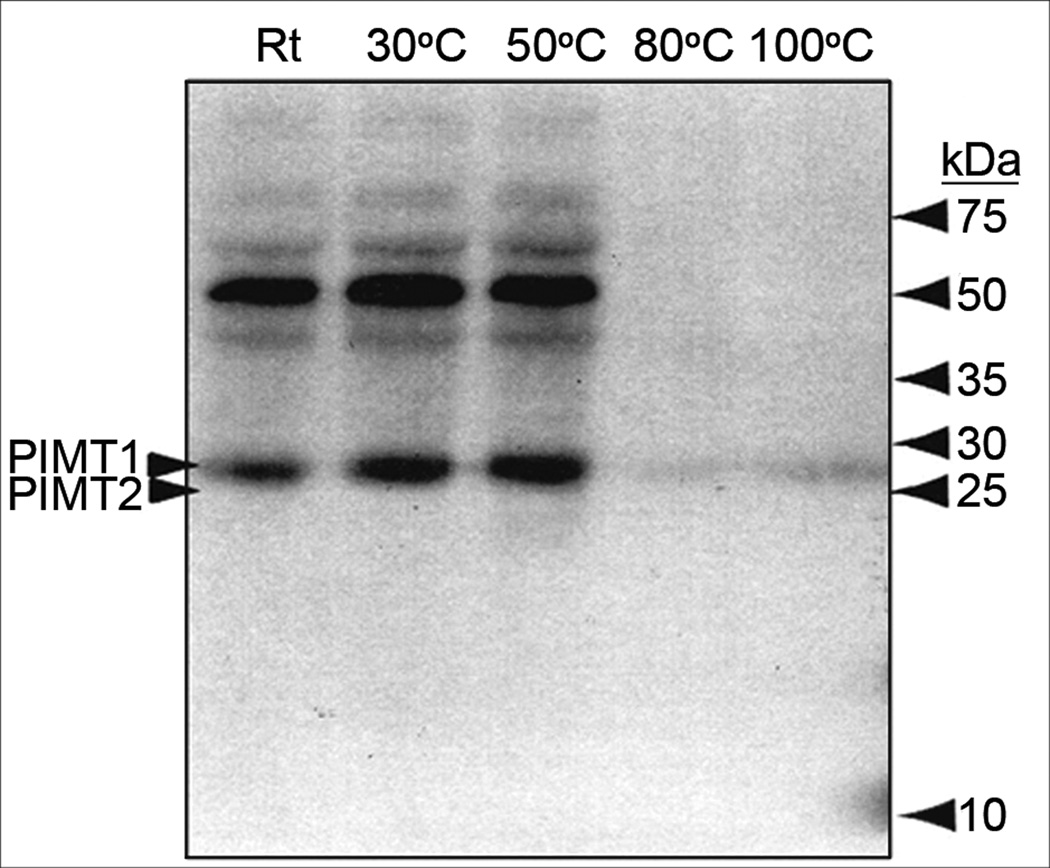
Western blot of PIMT (protein L-isospartyl methyltransferase) of modern Nelumbo nucifera China Antique embryo-axis extracts heated for 10 min at different temperatures; PIMT1 and PIMT2 arrows at ~25 kDa denote PIMTs of Arabidopsis (Xu et al. 2004).
After incubation, the membranes were washed 3X with TBS-T to remove unbound primary antisera and incubated for 45 min to 2 h in a 1:5000 dilution of a secondary antibody of anti-rabbit sheep IgG linked to alkaline phosphatase or horseradish peroxidase. After incubation, the membranes were washed with PBS-T to remove unbound secondary antibodies, and were then washed 3X in PBS without Tween-20. Shielded from light, color development of these washed blots (e.g., alkaline phosphatase) was carried out without shaking in a 15-ml solution of 10 mM Tris-HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2 that contained 45 µl 5-bromo-4-chloro-3-indolyl phosphate (50 mg/ml, w/v) and 25 µl 4-nitro blue tetrazolium chloride (100 mg/ml, v/v) dissolved sequentially in 40 and 70% dimethylformamide v/v. Membranes containing the stained proteins were photographed (Figs. 3–5).
Protein Digestion and Mass Spectrometry
Coomassie-blue stained gel-bands numbered by bins (Fig. 2) were individually excised and placed in microfuge tubes to prepare proteins for identification. The gel-embedded proteins were reduced, iodoacetaminde-alkylated and trypsin digested using Promega, a sequencing grade modified trypsin (Shevchenko et al. 1996). Product peptides were extracted with 50% acetonitrile-0.1% trifluoroacetic acid and the resulting extract was dried by vacuum centrifugation. Peptides were then dissolved in 10 µl of a 0.1% formic acid (FA) solution and analyzed by liquid-chromatography tandem-mass spectrometry (LC-MS/MS) using electrospray ionization (ESI) on an Applied Biosystems OSTAR® Pulsar XL (OqTOF) mass spectrometer equipped with a nanoelectrospray interface (Protana), a Proxen (Odense) nano-bore stainless steel emitter (30µm i.d), and using an LC packings nano-LC system. The nano-LC was equipped with a specially constructed precolumn (150 µm×5 mm) and an analytical column (75 µm×150 mm) packed with Jupiter Proteo C12.4-µm resin (Phenomenex). Typically, 6 µl samples were loaded onto the precolumn, washed with loading solvent (0.1% FA) for 4 min and injected into the LC column. The eluents used were 0.1% FA, aqueous (Solvent A) and 95% CH3CN containing 0.1% FA (Solvent B). A column solvent-flow of 200-nL per-min was applied to the gradient: 3% B to 6% B in 6 sec, 6% B to 24% B in 18 min, etc., to a final step of 36% B to 80% B in 2 min, where it was maintained at 80% B for 7.9 min. At the conclusion of the run, the column was equilibrated with 3% B for at least 15 min before the next run.
Peptide ion spectra were recorded automatically during LC-MS/MS by information-dependent analysis (IDA) software on the mass spectrometer. Argon was used as the collision gas, and collision energies for maximum fragmentation efficiencies were calculated by the instrumental software using empirical parameters based on the charge and mass-to-charge ratio of the peptide precursor ion.
Proteins were identified by database search utilizing the Mascot database search engine (Matrix Science). Searches were performed against a database of proteins predicted from the genome, supplemented with keratin and trypsin sequences. These protein sequence searches allowed a maximum of two missed cleavages and a mass size of 0.3 Da for precursor and product ions. Protein hits were accepted based on >2 ascribed peptides, with at least one peptide possessing an ion score >32, such a score being defined as -10*Log (P), where P is the probability that the observed match is a random event. For the database of predicted Nelumbo proteins, ion scores >32 corresponded to <5% probability that the peptide match was random. Correspondences between MS/MS spectra and ascribed sequences were also verified manually.
PIMT Bioassay
The powdered embryo axis prepared as described above was extracted on ice by mixing for 10 min with 100 mM pH 7.5 HEPES composed of 10 mM β-mercaptoethanol, 10 mM Na-hydrosulfite, 10 mM sodium-metabisulfite, 1 µM Leupeptin, 25 µM PMSF and 10% glycerol (Villa et al. 2006). After centrifugation at 14,000×g for 10 min at 4°C the extracts were collected. Aliquots of the extracts were then heated for 10 min in water bath at room temperature (~22°C), 30°C, 50°C, 80°C and 100°C, and re-centrifuged. The protein concentrations of the heat-treated supernatants were assayed by the Lowry method after precipitation with 10% trichloroacetic acid and use of the standards of bovine serum albumin.
The heat-treated protein samples were used for Western blotting with antibodies to Arabidopsis PIMT1 (above) and for determination of methyltransferase activity as described below. Twelve µl (13 – 46 µg protein) of each thermally treated supernatant was incubated for 60 min at 40°C in a reaction mixture of substrate consisting 10µM [14C]AdoMet (60 mCi/mmol-1), 625 µM Val-Tyr-Pro-(L-isoAsp)-His-Ala [VYP-(L-isoAsp)-HA], and 200 mM phosphate pH 6.7 citrate buffer and having a final volume of 40 µl. The assay tubes were immediately frozen on dry ice, subsequently thawed on wet ice for 5–10 min, and quenched with 40 µl 0.2N NaOH in 1% NA-dodecylsulfate for hydrolysis of methyl esters to methanol. Sixty µl aliquots of the quenched reaction mixture of each treatment were spotted onto a triple-pleated 1.5×8 cm filter paper (No. 1650962, BioRad Lab) and placed in the neck of a 20-ml scintillation vial that contained 5 ml of the counting fluor (Safety Solve High Flashpoint Cocktail; Res Prod Intl). The vials were then capped and kept at room temperature for 2 hr during which volatile [14C]-methanol diffused into the fluor and unreacted [14C]AdoMet remained on the filter paper. The reaction vials, after removal of filter paper, were counted by liquid scintillation. Reactions were assayed in triplicate (Fig. 6).
Protein Identification and Sequence Alignment
Nelumbo thermal proteins are identified from sequencing by liquid-chromatography tandem mass-spectrometry (Pennington and Dunn, 2000). The proteins were identified from the newly annotated Nelumbo genome (Ming et al. 2013) using standard proteomics experimental strategies. In addition, the deduced protein sequences were further verified by EST identification (expressed sequence tag; Gouet et al. 1999), BLASTed (Basic Local Alignment Tool, by NCBI; Camacho et al. 2009) and aligned. Protein sequences were aligned using MUSCLE (Edgar, 2004) and visualized using ESPript (Gouet et al. 1999) (Supp. 1, http:_________).
Results and Discussions
Thermal Proteins
Figure 1 shows 1-D SDS gels of Nelumbo fruit heat-soluble proteins at room temperature (Rt, ~22°), 30°, 50°, 80°, 100° and 110°C isolated from embryo axis and cotyledon tissues of a modern and a 549-yr-old fruit of China Antique, compared with protein standards (S) having masses from 78 to 12 kDa. On the whole these Nelumbo proteins of axes withstand heat without diminishing solubility through 50°C and those of cotyledons through 80°C (Fig. 1). Proteins in the axes are evidently somewhat less heat stable than those of cotyledons. After five-and-a-half centuries of aging, proteins in the old fruit have remained remarkably intact (Fig. 1).
After 110°C of heating, ~31% of the proteins of the embryo axis of this old fruit and ~76% of its cotyledons remained soluble (Table 1), a further example of the greater heat-stability of the proteins of Nelumbo cotyledons. Surprisingly, the percentages of soluble protein remaining in the old axis and cotyledons after 110°C heating were more than twice those of its modern counterpart. This difference in thermal stability (Table 1), also visible in the SDS gels (Fig. 1), may reflect fruit maturity. That the modern fruits require more than twice as long (~6–9 days) to germinate than the old fruits (Shen-Miller et al. submitted) may evidence their relative lack of maturity, accommodating lesser amount of heat-stress proteins (Table 1). The modern Nelumbo fruit is from a 3-yr-old crop, harvested at the Kenilworth Aquatic Gardens, Washington DC. This fruit is an offspring of fruit originated from the dry lakebed in Xipaozi Village, China (Shen-Miller et al. 1995) where all of the old fruit used in such studies have been collected (e.g., Ohga, 1926; Shen-Miller et al. 2002; Shen-Miller et al. submitted).
In Table 1 the total amount of embryo axis proteins in room temperature (Rt, ~22°C) of the modern control is essentially identical to that after 549 yr of aging (70 µg·mg−1 modern vs. 73 µg·mg−1 old). Nevertheless, the cotyledon proteins of the old fruit (15 µg·mg−1) are markedly less than those of the modern (63 µg·mg−1). This difference may be a result of long-term metabolism of the cotyledons in the old fruit over hundreds of years. Notably, the proteins remaining in the old cotyledons are evidently more thermal stable than those of the modern control (Table 1; respectively, 76% vs. 36%, at 110°C), a further example that relates protein heat stability to fruit maturity (cf. Table 1, old vs. modern embryo axis at 110°C, respectively, 31% vs. 10%).
Thermal Protein Identification
Modern Nelumbo proteins of China Antique from Xipaozi Village, NE China were treated for 10 min at 100°C. Supernatants of the crude protein of embryo axes and cotyledons were electrophoresed using the 1-D SDS gels (Fig. 2; embryo axis). Stained protein bands were boxed into 15 bins (Bins 1–15 vs. protein standards ~200-15 kDa). The proteins from individual bins were processed for trypsin digestion and analyzed by liquid chromatography and tandem MS. Product ion spectra were matched to those calculated for putative Nelumbo proteins predicted from its genome (Perkins et al. 1999).
Eight Nelumbo thermal proteins of embryo axes and cotyledons were identified by sequence and aligned with those of the archaeal hyperthermophile Methancaldococcus jannaschii (Mj) and five mesophile angiosperms (Arabidopsis, corn, grape, poplar and soybean). Molecular masses of the identified Nelumbo proteins match well the protein standards. The identified thermal proteins are listed below in the order of size, from small to large (Fig. 2, Bin-15 to Bin-3); their aligned sequences are shown in Supplement 1 (http:_______, Figs. S-1a through S-1h):
CuZn-SOD, NNU_001676 (Bin-15, ~15kDa)
1-CysPRX, NNU_016172 (Bin-14, ~25 kDa)
CPN20, NNU_010559 (Bin-14, ~25 kDa)
Vicilin, NNU_007171 (Bin-7, ~50 kDa)
ENO1, NNU_002362, _020386, _021001; (Bin-7, ~50 kDa)
EF-1α, NNU_024576, _024577, (Bin-7, ~50 kDa)
HSP80, NNU_010290 (Bin-3, ~75 kDa)
Met-Synthase, NNU_013651 (Bin-3, ~75 kDa)
Three additional thermal proteins from embryo axes and cotyledons of Nelumbo verified by Western blots (Figs. 3–5) and listed below in the order of increasing size, are aligned with those of the Mj and angiosperms (Supplement 1, Figs. S-1i to S-1k):
-
(i)
Dehydrin, NNU_006332, NNU_013652, NNU_013851 (~22 kDa)
-
(j)
PIMT, NNU_002938, NNU_004234 (~25 kD)
-
(k)
CPN60, NNU_011934, NNU_023642 (~60 kDa)
Sequence Alignment of Thermal Proteins of Nelumbo Embryo Axes and Cotyledons (Supp. 1, http: ________, Figs S-1a to S-1k):
(a) CuZn-SOD (cotyledons; NNU_001676; Fig. 2 Bin15, ~17 kDa; Fig. S-1a): As a result of the mid-Precambrian Great Oxidation Event ~2.4 billion years ago (Holland, 2002), when free oxygen first became a significant component of Earth’s atmosphere, aqueous Fe2+ became scarce, soluble Cu2+ became the dominant cofactor at the active site of dismutases (Alscher et al. 2002). CuZn-SOD, a superoxide dismutase of aerobic bacteria and eukaryotes (Alscher et al. 2002) thus appears to be a relatively late evolutionary innovation and has no evident sequence-similarity with earlier-evolved Fe-SOD and Mn-SOD of other organisms, neither with that of archaeal hyperthermophile Mj. Hence, the antioxidative plant enzyme CuZn-SOD, not present in Mj (Fig. S-1a) appears to have become prevalent at ~2.4-b-yr when it first played an important role in catalyzing the dismutation of superoxide (O2−, a reactive oxygen species, ROS) and thereby protecting cellular integrity.
The CuZn-SOD of Nelumbo cotyledons, NNU_001676, is highly conserved among such angiosperms as Arabidopsis, corn, grape, soybean and poplar (Fig. S-1a), composed of 152 residues (~16.7 kDa) and having two cysteines, Cys56 and Cys145, that form a disulfide bridge identical to that of the CuZn-SOD in the halophyte Polygonum sibirium Laxm (Qu et al. 2010) and harsh-arid monocot pearl-millet Pennisetum glaucum (Mahanty et al. 2012). In P. sibirium, the two CuZn-SOD catalytic sites (GFHVHALGD, the first site at amino acid position 43–51, and GNAGGRI*ACGII at 137–148) are essentially identical both in their composition and sequence location to those of Nelumbo and other plants (Fig. S-1a). The first active site in Nelumbo is totally conserved, and the second differs only by having a “V” in place of “I*” (Fig. S-1a). NNU_001676 is composed of ~18% polar residues that form thermally stable intramolecular ion-pairs (D-E-K-R-Y, Unsworth et al. 2007), with Asp, Lys and Glu being the most abundant.
(b) 1-CysPRX (NNU_016172; Fig. 2 Bin14, ~25 kDa; Fig. S-1b): In animals, 1-Cys-peroxiredoxin is bifunctional, exhibiting both glutathione peroxidase and phospholipase A2 activities (Chen et al. 2000). It has the simultaneous catalytic role of regulating phospholipid turnover and of providing oxidative protection by detoxifying reactive oxygen ROSs. The two active sites performing these functions have, respectively, the consensus sequences of GDS32WG and PVC47TTE (Chen et al. 2000). In plants as well as Mj, the active site of peroxisdase PVC46TTE is highly conserved (Fig. S-1b), but the phospholipase sequence (in which Ser32 is the catalytic nucleophile) is not present in the Mj-angiosperm alignments (Fig. S-1b). Nelumbo has two 1-CysPRXs: NNU_016172 is predominantly conserved among the plants as well as in the archaean Mj (Fig. S-1b), consisting of ~213 residues (~23.4 kDa).
(c) CPN20 (NNU_010559; Fig. 2 Bin14, ~27 kDa; Fig. S-1c): Chaperonin20 is a molecular chaperone that together with its companion chaperonin CPN60 (Fig. S-1k), forms protein-folding machinery powered by ATP serves to restore nascent or misfolded proteins to functional conformation (Thirumalia and Lorimer, 2001). It is a highly heat-stable protein through 110°C in both Nelumbo embryo axis and cotyledons (Fig. 5). Of the three dimers identified in Nelumbo, Cpn20 NNU_010559 is one. A second dimer, NNU_010102, has sequence similarity more close to a dimer of Homo sapiens than to that either of grape or rice, whereas other two Nelumbo dimers are similar to those of Arabidopsis. CPN20 (NNU_010559, ~27.1 kDa) is composed of ~246 residues. With the exception of the first ~40 residues at the N-terminus, the remaining ~200 amino acids are highly conserved among the aligned angiosperms (Fig. S-1c). The “MASI” motif at the N-terminus of NNU_010559 reflects its localization in chloroplasts as is evidenced in an Arabidopsis chloroplastic CPN10 (Hill and Hemmingsen, 2001). In Nelumbo amino acids 54–63 (SIKPLGDRVL), 76–80 (GILLP) plus 3 shorter segments up to amino acid 96 show similarity or identity (shown by pink or red type) to the Arabidopsis CPN10. CPN20 per se is not found in the archaean Mj, however, the larger companion chaperonin CPN60 is present (Fig. S-1k).
(d) Vicilin (NNU_007171, Fig. 2 Bin7, ~54 kDa; Fig. S-1d): Vicilin, a component of the plant seed-storage protein 7S-globulin, has the dual function of providing nutritional reserve and microbial defense (De Souza et al. 2011). The vicilin NNU_007171 of Nelumbo, similar to that of Macademia integrifolia, contains a short hydrophobic N-terminus (Marcus et al., 1999). During seed germination, two similar motifs of Macademia vicilin (Cys-X-X-X-Cys) have been identified as promoting the release of exudates that exhibit antimicrobial activity. Nelumbo vicilin NNU_007171 contains three Cys residues, at positions 10, 150, and 348, with long stretches of Xs between. Vicilin is not found in the archaean Mj. Further, the aligned vicilin proteins show similarity but less identity among the aligned plants (Fig. S-1d), having much differentiation at either the N- or C-termini. In all of the aligned plant sequences compared with Nelumbo vicilin, most identities occur in two sets of amino acid pairs, PY and AG, between positions 60 and 70, 420 and 430. In these alignments, the longest stretch of sequence similarity to Nelumbo is EQIKAMS at position 243–249. Vicilin NNU_007171 (~53.5 kDa) is composed of ~486 residues, ~30% of which are the thermal-stable polar amino acids (Unsworth et al. 2007) from the N-terminus to the mid-region of the protein.
(e) ENO1 (NNU_002362, NNU_020386, NNU_021001; Fig. 2-Bin7, ~48–53 kDa; Fig. S-1e): Three Nelumbo enolase1s have been aligned (Fig. S-1e). All of which show high conservation with the ENO of Mj, and with those of the other angiosperms (Fig. S-1e), as well as with that of corn (Lal et al. 1998), and that exhibit similarity to the corn sequences at the N-terminus of the protein (viz., MAVTITWVKARQ, in which the amino acid differences are underlined, and identity in bold). This same terminus sequence occurs also in the maize enolase isozymes ANP45A and ANP45B (Lal et al. 1998). Activity of ENO1 in corn roots is induced 10-fold under anaerobic conditions, and the two isozymes appear to remain functional during long-term anoxia when the activity of other enolases declines to a nondetectable level (Lal et al. 1998). NNU_002362 (~53.4 kDa), the longest, is composed of ~485 amino acids. In Nelumbo, the ENO1 protein has a Tyr53 residue (similar to the Tyr46 in vertebrate ENO1) that has been shown to be phosphorylated (Lal et al. 1998). The regions adjacent to the Tyr(Y) residue in Nelumbo are highly conserved among the aligned species except Mj (Fig. S-1e). In animals, dephosphorylation at this site is reported to occur under anoxia that increases activity of the enzyme (Lal et al. 1998).
Enolase1 is a highly conserved polypeptide. The consensus sequence of its anaerobic responsive element, ARE at the 5’-terminus, is present in all organisms examined (Van Der Straeten et al. 1991; identity with Nelumbo in bold type) including Nelumbo and the others shown in Figure S-1e, in which in plants, the E is replaced by Q at position 18. Six active sites that are homologous in the enolases of plants and vertebrates (E-170, D-248, E-289, DD-314-5, K-339 and K-402; Van Der Straeten et al. 1991) are fully conserved among Nelumbo, the archaean Mj, and the aligned angiosperms (red columns in Fig. S-1e).
(f) EF-1α (NNU_024576, NNU_024577, NNU_026673; Fig. 2, Bin7, ~42–49 kDa; Fig. S-1f): Three Nelumbo elongation-factor EF-1α proteins have been identified, two of which, NNU_024576 and NNU_024577 are in tandem and exhibit 100 % sequence identity (Fig. S-1f). The third protein, NNU_026673, shows sequence similarity with this tandem pair except for having fewer residues toward the C-terminus and contains a total of 377 amino acids (~41.5 kDa) rather than 443 (48.7 kDa) (Fig. S-1f). All three Nelumbo EF-1αs exhibit sequence conservation with the other aligned plant sequences and the archaean Mj (Fig. S-1f). The Nelumbo EF-1α proteins have sequence similarities also with those of carrots, wheat, and barley in the regions from position 6–30: (VHINIVVIGHVDSGKSTTTGHLIYK), 167–174 (EVS.SYLK), 274–294 (SVEMHHETLQEALPGDNVGFN), and 297–304 (NVAVKDLK) (bold type indicates identity; Durso and Cyr, 1994). The lack of alignment (black type) at the C-termini of these proteins in Figure S-1f is due to the absence of sequences in the shorter Nelumbo protein NNU_026673; otherwise their C-terminus sequences are of high similarity among the alignments (Fig. S-1f).
(g) HSP80 (NNU_010290, Fig. 2 Bin3, ~77 kDa; Fig. S-1g): Heat-shock protein 80 falls in the HSP70-90 group of the evolutionarily conserved hsps. The Nelumbo HSP80, NNU_010290 (~76.9 kDa), composed of 699 residues, shows a very high degree of conservation in comparison with those of the other aligned angiosperms (Fig. S-1g). Of these, the Nelumbo N-terminus exhibits the longest sequences identical to those of the other species, beginning from amino acid 5 followed by 34 residues and flanked by an identical amino acid A at each end: (AETFAFQAEINQLLSLIINTFYSNKEIFLRELISNA; Fig. S-1g). This long sequence and many more other identical residues are present also in the HSP80 both of tomato (Koning et al. 1992) and Triticum aestivum (GeneBank CAA67191.1). Interestingly, the HSP80 protein is absent in the hyperthermophile Mj. The Nelumbo HSP80 contains a large number (264) of the heat-stable polar residues D-E-K-R-Y that comprise ~37.8% of this protein (Unsworth et al. 2007). These polar amino acids are most abundant in the 170–290 and 410–585 regions (Fig. S-1g).
(h) Met-Synthase (NNU_013651; Fig. 2 Bin3, ~84 kDa; Fig. S-1h): The methionine synthase of Nelumbo, comprised of 765 residues (~84.2 kDa), is another highly conserved protein among the angiosperms aligned (Fig. S-1h). The Arabidopsis protein present in the alignment (Fig. S-1h) is a cytosolic cobalamin-independent Met-synthase (GenBank U97200). The archaean Mj has a small Met-Synthase composed of ~308 amino acids, fewer than half those of the plant homologues (Mj alignment data not included). However, the first ~70 residues at the N-terminus of the plant proteins (viz., MAS.IVG.YPRM..KRE…..ESF.D…….LQKV etc. to ~70 residues) are identical or similar to that of the Mj. In Mj, beginning from amino acid position 83, the rest of the residues are highly conserved with the last third of the plant sequences through the C-terminus.
(i) Dehydrin (NNU_006332, NNU_013652, NNU_013851; Fig. 3, ~22 kDa; Fig. S-1i): Thermal stability of Nelumbo dehydrin is shown by the Western blot (Fig. 3). The dehydrins of plants analyzed does not evidence strong similarity in the alignment, due to missing sequence-regions in several plants (Fig. S-1i), including grape, corn1, Arabidopsis4, 5, soy1 and Nelumbo. All plants, including those for which data are presented in Supplement 1 Figure S-1i, have been shown to typically exhibit conserved and repetitive sequences, in which the “k-segments” (EKKGIMDKIKEKLPG) are of common occurrence (Close, 1997).
Figure S-1i compares the alignments of three Nelumbo dehydrins (NNU_006332, _013652, _013851) with other angiosperms. Nelumbo dehydrin NNU_013851 (the longest of its three) has a K-like segment beginning at amino acid 85 (EKKGILEKIKEKLPG; bold-type infers identity, the underlined residues being similar to those of the above K-segment), and have nearby His-residues that can be protonated (Eriksson et al. 2011), at either end, but not immediately adjacent to the segment. The dehydrins of Arabidopsis1 and 2 have identical K-like segments at this same location and His residues situated similarly to those in Nelumbo dehydrin NNU_013851. This Nelumbo dehydrin is composed of ~197 amino acids and has an average molecular mass of ~21.7 kDa, similar mass to that of the thermal proteins shown on the Western blot (Fig. 3; arrows, ~22 kDa). Nelumbo dehydrin has the richest concentration of the D-E-K-R-Y polar amino acids of all Nelumbo thermal proteins so far assembled, 54.6%.
A Nelumbo medium-length dehydrin protein, NNU_006332, has the K-segment of EKKGMMDKIKEKLPG which differs at the three underlined residues from NNU_013851 but is identical to that of Arabidopsis5 having only one His-flanking residue close to its lead-end. The third and shortest Nelumbo dehydrin (NNU_013652) is composed of a hefty ~38% of thermostable amino acids and exhibits slightly different K-segment (KKKGEH…….G) at the same position of the K-segments of the other two dehydrins of Nelumbo and the other plants. This dehydrin has an additional His-residue within the segment. The plant dehydrin proteins have no equivalent in the archaean Mj.
(j) PIMT (NNU_002938, NNU_004234; Fig. 4, ~25–33 kDa; Fig. S-1j): Protein repair L-isoaspartyl methyltransferase (PIMT) is another Nelumbo protein whose thermal stability is verified by the Western blot (Fig. 4). PIMT recognizes L-isoaspartyl residues spontaneously produced from L-asparaginyl and L-aspartyl residues during aging and stress (Clarke, 2003). These isomerized residues are methyl esterified by PIMT at the α-carboxyl in the S-adenosyl-L-methionine-dependent reaction, followed by non-enzymatic reactions that lead to conversion of the normal L-aspartyl residues. Similar to Arabidopsis (Xu et al. 2004), Nelumbo has two PIMTs in its genome, one long (NNU_002938, 301 residues) and one short (NNU_004234, 231 amino acids). The amino acid sequences of these two Nelumbo proteins are 89% identical to each other over 229 residues. The longer one has an additional 72 residues at its N-terminus not present in the shorter form (Fig. S-1j).
The longer Nelumbo protein (NNU_002938) shows 63% identity over 286 residues to the PIMT2 of Arabidopsis (Villa et al. 2006), including some identities in the N-terminal 72 sequences of Nelumbo. In Arabidopsis, PIMT2 is located in the nucleus, suggesting that the Nelumbo 002938 species may also be nuclear. The shorter form of the Nelumbo PIMT shows 69% identity with PIMT1 of Arabidopsis over 230 residues.
The alignment of PIMTs in higher plants with that of the archaen Mj demonstrates a very high degree of conservation among these enzymes, indicating their ancient origin. For example, the L-isoasparate substrate-binding motifs at residues 59–67 (TISAPHMHA) and 202–209 (VRYVPLTS) are similar or identical in all plants sequences studied (Griffith et al. 2001) as well as the archaean enzyme (Fig. S-1j). The nuclear location signal “NLS” (KIIKKRKKKMR) of lead sequences of Arabidopsis PIMT2 (Xu et al. 2004) is present in the lead sequences of Soy2 PIMT as KRKSEKKKKMR (with both signals having been confirmed by the NLStradamus NLS prediction program; Nguyen et al. 2009). The nuclear location of the forms of PIMT in corn2, Nelumbo NNU_002938, and poplar2 have not been shown, nor have the Arabidopsis PIMT2 and Soy2 PIMT shown to be plastid-, mitochondrial- or ER-localized (Predotar Program; Small et al. 2004). The PIMTs of corn1 and poplar2 could be situated in plastids/mitochondria and ER, respectively, but no clear Predotar-based data are available to evidence the location of the long Nelumbo PIMT NNU_002938 in these cellular components.
(k) CPN60 (NNU_011934, NNU_023642; Fig. 5, ~60 kDa; Fig. S-1k): Chaperonin 60 is the third Nelumbo protein whose heat stability is evidenced by the Western blot (Fig. 5). CPN60 (cpn60β) is a homologue of E coli GroEL (Viitanen, et al. 1995). There are two CPN60s represented in the Nelumbo genome. Both are conserved among the aligned plants and the archean Mj, and both are similar in size being ~96% identical, composed of ~542 amino acids, and having a molecular mass ~59.6 kDa.
High sequence similarity to the other taxa analyzed is exhibited by Nelumbo CPN60 in which all aligned sequences in various regions are mostly similar, some identical (Fig. S-1k). The Mj sequences at position 75 to 107 and 152 to 170 (Fig. S-1k), the so-called equatorial domains, exhibit high conservation with the ATP-dependent thermosome Mj protein MJ0999 (~60 kDa) that has been shown to be up-regulated at 95°C (Kowalski et al. 1998; Boonyaratanakornkit et al. 2005). The highly conserved motif of Nelumbo GDGTT beginning at position 93 (Fig. S-1k) has been shown to form the ATP-binding site for GroEL (Kowalski et al. 1998). This conserved motif is found in all of the aligned angiosperms, including Mj (Fig. S-1k), plus those of the not aligned methanogen Methanopyrus kandleri (98°C growth optimum), Pyrococcus sp. (100°C growth optimum) and yeast (Kowalski et al. 1998).
In addition to the 11 Nelumbo thermal proteins presented above, 30 other thermal proteins of Nelumbo China Antique are identified by mass spectrometry. They are not aligned, presented in Supplement 2.
Thermal-Stable Polar Amino-Acids in Nelumbo Proteins
Table 2 summarizes the distribution of thermostable, polar amino-acids (D-E-K-R-Y) present in the 11 Nelumbo thermal proteins, listed in the descending order of their abundance. Of these, amino acids capable of forming intramolecular thermally stable ion-pairs (Unsworth et al. 2007) are dominant Lys, Glu and Asp (Table 2). The dehydrin of Nelumbo stands out particularly, being composed of 55% of such heat-stable residues.
Table 2.
Heat-stable protein polar amino-acid residues (D-E-K-R-Y; Unsworth et al. 2007) in fruit of Nelumbo nucifera Gaertn var. China Antique, and ± protein sequence alignments of Nelumbo with those of Mj, Methancaldococcus jannaschii (85°C optimal growth), aa, amino acid.
| Thermoprotein | Most abundant residues | % Total aa | ± Mj |
|---|---|---|---|
| Dehydrin | Glu, Lys | 55 | − |
| HSP80 | Lys/Glu, Asp | 38 | − |
| CPN60 | Lys, Glu | 33 | + |
| Vicilin | Glu, Lys, Arg/Asp | 30 | − |
| EF-1α | Lys, Asp, Glu | 29 | + |
| Met-Synthase | Glu, Lys, Asp | 26 | + |
| 1-CysPRX | Lys, Asp | 26 | + |
| Enolase-1 | Lys, Glu | 24 | + |
| CPN20 | Lys, Glu, Asp | 22 | − |
| PIMT | Glu, Lys, Asp | 21 | + |
| CuZn-SOD | Asp, Lys, Glu | 18 | − |
PIMT has 21% thermostable polar amino-acid (Table 2). This protein-repair enzyme has 100% solubility and activity through 50°C (Figs. 4 and 6). The solubility of CPN60 (Fig. 5) is also observed through 50°C yet this protein contains a much higher percentage of the polar residues than PIMT (Table 2). On the other hand, the abundance of such polar amino acids of CPN20 and PIMT is essentially equal, ~22% and 21%, respectively (Table 2), but the two enzymes differ substantially in thermal solubility (Figs. 4, 5). Protein thermal stability is not determined by the presence of such polar residues alone. Vieille and Zuckus (2001) and Unsworth et al. (2007) have reported that, in addition to the occurrence of these amino acids, the presence of other substances (e.g., salts, polyamines, soluble sugars) and posttranscriptional modifications (e.g., glycosylation) can equally contribute to heat stability.
Notably, 55% of the Nelumbo thermal proteins are present in the archaean hyperthermophile Methancaldococcus jannaschii (85°C growth optima; Table 2), indicating the high thermal stability and antiquity of these proteins.
Western Blots of Thermal Proteins
Western blot verifications of thermal stability of three Nelumbo proteins dehydrin, PIMT and CPN60 (Figs. 3–5) are presented, in the ascending order of size.
Dehydrin (~22 kDa, Figs. 3, S-1i): Dehydrin is a LEA (late embryogenesis abundant) protein that responds to freezing temperature and draught (Close, 1996). The ultra-high thermal stability of Nelumbo dehydrin is illustrated in the Western blot shown in Figure 3, documenting the results for an embryo axis and cotyledon proteins after heating for 10 min at room temperature (Rt, ~22°C) and from 30° to 110°C. The Nelumbo proteins of both axis and cotyledons remained intact throughout the heating series to 110°C with no diminished intensity (noted by the arrows at ~22 kDa in Fig. 3). Such heat stability of dehydrin is reflected in the very high amount of its thermostable polar-residues D-E-K-R-Y (Table 2; 55%). The Nelumbo embryo axis evidently contains a higher concentration of dehydrin than the cotyledons.
PIMT (~25 kDa, Figs. 4, S-1j): Two PIMT proteins were annotated from the Nelumbo genome, NNU_004234 and NNU_002938 (Fig. S-1j). The former has an expected polypeptide size of 24.9 kDa and the latter, 33.1 kDa. The Western blot in Figure 4 shows a clear band of reactivity at ~25 kDa that corresponds closely to the position noted for the Arabidopsis recombinant PIMT1 and PIMT2αω proteins (Xu et al., 2004) and to the expected size of the NNU_004234 Nelumbo enzyme. The 25 kDa immunoreactive species persist in the supernatant of heated fractions to 50°C and a small amount of the enzyme is visible in extracts heated to 80°C and 100°C (Fig. 4). However, we detected no immunoreactive species corresponding to the expected size ~33 kDa (Fig. 4), suggesting that it may be synthesized in lower amounts than the shorter form 004234. Strong staining bands of an unidentified 50 kDa species are noted, occurring to the 50°C heated-fractions. This may be an unrelated Nelumbo protein that cross-reacted with the antibody to the Arabidopsis protein.
CPN20 and CPN60 (~27 kDa and ~60 kDa, respectively; Figs. 5, S-1c, S-1k): In the restorative folding of nascent or damaged proteins, a “lid-and-basket” structure is formed between chaperonins CPN20 and CPN60 (Thirumalia and Lorimer, 2001). Nelumbo CPN20 (~27 kDa; the lid) remains soluble in the embryo axis and cotyledons through heat treatments to 110°C (Fig. 5). In contrast, the CPN60 (~60 kDa; the basket) is heat stable only through 50°C for both tissues (Fig. 5) Judging from the stain intensities of proteins in the axis and cotyledons of Nelumbo, the axis appears to contain more of both these chaperonins than the cotyledons (Fig. 5).
PIMT Bioassay
PIMT is the only Nelumbo thermal protein for which enzymatic activity has been tested at different temperatures. The methyltransferase activity of such supernatant fractions in the presence and absence of a peptide substrate is shown (Fig. 6). The data presented in Figure 6 (n=3) illustrate that the heated extracts of Nelumbo are able to catalyze the methylation of a synthetic substrate L-isoaspartyl-containing peptide. A robust peptide-dependent activity (red curve) is shown in samples at room temperature (~22°C) as well as those heated to 30° and 50°C -- compared with a smaller but readily detectable peptide-dependent activity in these samples heated at 80° and 100°C (~9 and ~6%, respectively). PIMT enzymes heated at 80° and 100°C remain active but in lower enzyme concentrations (Fig. 6). Significantly, these data show that although the concentration of PIMT decreases with heat treatments (Fig. 4), the proteins remaining in the high temperature-treated samples are biologically functional (Fig. 6).
Concluding Discussion
The guanine-cytosine (G-C) content of RNA is evidently correlated directly with the optimal growth temperature of the organisms (Unsworth et al. 2007). Nelumbo, a basal eudicot, has a G-C content of 38% (Ming et al. 2013), higher than that of such core-eudicot angiosperms as Arabidopsis, poplar and grape, but lower than that of corn. The optimum and/or maximum growth temperature of plants may be related to the maintenance-temperature of their protein solubility or optimal protein activity. In hyperthermophiles, the optimal activities of most enzymes coincide with optimal temperatures of organismal growth (cf. Vieille and Zuckus, 2001). Several proteins of the anaerobic methanogen hyperthermophile Methancaldococcus jannaschii (85°C growth optimum) are highly conserved in Nelumbo (Table 2, Supp. 1). Under anaerobic conditions, such as that within the pericarp of Nelumbo, such proteins can maintain solubility and may be enzymatically active.
The roles of the Nelumbo fruit thermoproteins are numerous. They have the function in antioxidation, chaperonine and chaperon, anaerobic glycolysis, microbial defense, membrane maintenance, nutritional provision, repair and abating stresses. The collaborative effort of such group would certainly contribute to extend viability of Nelumbo fruit. The functional roles of Nelumbo thermal proteins are:
• Antioxidant and Membrane Maintenance (CuZn-SOD, 1-CysPRX, dehydrin):
(1) CuZn-SOD: Nelumbo is a widespread crop, particularly throughout Asia. In China its habitat extends from the near-tropics to the frigid northern reaches of Heilongjiang Province, bordering Siberia (Huang, 1987). Gill et al. (2010) found versatility in the CuZn superoxide dismutases of alpine plants collected at high altitudes in the Himalayas, having “an autoclavable property and a reaction maximum at 0°C.” CuZn-SOD is one of four SODs identified in plants that not only have antioxidative properties but are regarded as important anti-aging and heat tolerant enzymes (Qu et al. 2010; Chen et al. 2011). Ninety percent of the CuZn-SOD activity of Nelumbo remains intact at 65°C, and 20% after 95°C for 30 min (Chen et al. 2011). Such enzyme heat stability in the Nelumbo is much higher than that of the C4 pearl-millet (Mahanty, 2012).
Plant CuZn-SOD functions mainly in the cytoplasm and chloroplasts, and can initiate activity under mild water-stress (Alscher et al. 2002) and salt-stress (Qu et al. 2002) with increased expression of SOD activity being correlated with an increased tolerance of membrane damage (Kwon et al. 2002). Analyses of the cotyledons of a viable 14C-dated 466-yr-old Nelumbo China Antique fruit and two viable undated fruit from the same Holocene lakebed at Xipaozi Village showed high concentrations of polyunsaturated lipids, indicating the maintenance of membrane fluidity in these old fruits likely due to a lack of extensive auto-oxidation (Priestley and Posthumus, 1982).
(2) 1-CysPRX (1-Cys-peroxiredoxin) is regarded to have evolved during the early history of life (Dietz, 2011), serving as a detoxifier of ROS released by early-evolved oxygenic photosynthesizers and the nonbiogenic photodissociation of water and CO2 (Schopf, 2011). Workers on circadian rhythms go so far as to suggest that the redox homeostatic mechanisms of PRX and associated cellular time-keeping biology may have co-evolved after the Great Oxidation Event (GOE) 2.4 billion-yr ago (Edgar, et al. 2012). That this Nelumbo protein is present in Methancaldococcus jannaschii may suggest an even earlier evolution, as methanogens seem certain that have been extant ~2.8 billion years ago and probably earlier (Schopf, 1994). The gene coding for 1-CysPRX is strongly expressed during seed germination (rice; Dietz, 2011). 1-CysPRX in the hermetically enclosed Nelumbo embryo axis could provide antioxidative protection during centuries of seed aging. The notable expansion of redoxin genes of the Nelumbo genome (Ming et al. 2013), coding for enzymes involved in antioxidation and cellular O2-damage repair, may have played a crucial role enhancing long-term Nelumbo seed viability.
(3) Dehydrin, a late embryogenesis abundant protein (LEA), is up-regulated during seed maturation when it functions in combating freezing-, drought- and salinity-stress (Close, 1997). It is reasonable that the archaean Mj, having an 85°C growth optimum and inhabiting in the vicinity of submarine thermal vents, has no need for such a protein (Fig. S-1i). Given the functions of dehydrin for detoxification and protection from cold stress, this protein presumably evolved later than the early Precambrian origin of oxygenic photosynthesis but before the first major glacial epoch ~2.4 billion years ago.
All three Nelumbo proteins contain the so-called K-segment common to all dehydrins (see above). Protonation of the His residues flanking such K-segments induces membrane binding by stabilizing membrane topology may thereby modulate lipid fluidity (Eriksson, et al. 2011). Such potential of membrane fluidity has been documented by Priestley and Posthumus (1982) in old China Antique Nelumbo fruit, in which dehydrin may play a role in the viability of the centuries-old Nelumbo fruit. Several more Nelumbo thermal-stable LEA proteins (Dc3, Dc8, D34) are identified by mass spectronmetry presented in Supplement 2.
• Chaperonins, Chaperone, and Stress Proteins (Cpn20, Cpn60, HSP80, EF-1α):
(1 & 2) CPN20 & CPN60 are molecular chaperones, responsible for such cellular processes as protein folding, assembly, translocation and degradation. CPN20 is a dimer of CPN10, an equivalent of the bacterial GroES of E. coli (Viitanen et al. 1995). With coordinated ATP hydrolyses between CPN60 and the co-chaperonin CPN20, mal-folded proteins can be rearranged to functional conformation states (Thirumali and Lorimer, 2001). The genome of Nelumbo is coded for at least seven CPN10s and five CPN20s (Ming et al., 2013). It would be interesting to know whether the greater heat stability of Nelumbo CPN20 (the “lid;” Fig. 5) serves at relatively higher temperatures could stabilize the companion component that forms the underlying “basket” (CPN60) after conjugation for protein folding (Fig. 5).
(3) The HSP80 of Nelumbo, a stress protein, is highly conserved with those of the other angiosperms here aligned (Fig. S-1g). This hsp is absent from the hyperthermophile Mj (Fig. S-1g). It seems to be a later evolved heat-shock protein. Mj has small hsps that are induced under stress, and the Mj HSP16.5 protein forms a large homomeric complex of 24 subunits having a molecular mass of ~400 kDa (Kim et al. 2003). This huge complex of small hsps functions to prevent protein aggregation at elevated temperature, and may thereby serve the same purpose as the chaperones composed of the larger hsps in plants. In Supplement 2 of Nelumbo thermal proteins is listed two more heat-shock species, HSP18.2 and HSP70.
(4) EF-1α elongation factors are regarded to be among the slowest evolving enzymes; they exhibit highly conserved sequences (Gaucher et al. 2001) and have diverse functions in protein biosynthesis, chaperone activity (cf. HSP45), disulfide bond formation, protection from abiotic and biotic stresses and protein degradation (Fu et al. 2012). The occurrence of a tandem pair of EF-1α proteins in Nelumbo reinforces their functional effectiveness. The protein elongation factors in plant cytoplasm are homologs of the EF-Tu in plastids, mitochondria, and bacteria (Fu et al. 2012). Transgenic crop plants containing EF-Tu developed heat stress tolerance, and the induced expression of EF-1α has been shown to prolong the lifespan of Drosophila (Tatsuka et al. 1992).
• Anaerobic Glycolytic Protein (Enolase1):
ENO1 is an ancient glycolytic enzyme originating early in Earth history when the environment was anoxic. In the process of glycolysis, ENO1 converts 2-phosophoglycerate to phosphoenopyruvate, PEP (Van Der Straeten et al. 1991). For survival under anaerobic conditions, most plant cells shift metabolism from the oxidative to the fermentative mode and ENO1 comes into action, exhibiting intense activity particularly in non-green tissues. The glycolytic pathway of anaerobic respiration evidently provided the cellular energy required to sustain viability of the hermetically sealed old Nelumbo fruit, particularly the non-green tissues in cotyledons and the embryonic radicle. This may explain the apparent diminution of protein content in the cotyledons of the 549-yr-old fruit in comparison with its modern counterparts (Table 1).
• Defense, Food and Repair (vicilin, Met-Synthase, PIMT):
(1) Vicilins and cruciferins (legumins) are major members of 7S-globulin seed-storage proteins (Tiedemann et al. 1999; De Souza et al. 2011). In addition to seven vicilins, Nelumbo has 19 cruciferins and three cruciferin-like proteins (network map not shown; Ming et al. 2013). In addition to storage, these proteins have antimicrobial functions (discussed above). The heaviest staining band on the SDS-PAGE gels in Figures 1 and 2, corresponding to a mass of ~50 kDa, shows the location of these most abundant Nelumbo seed-storage proteins. Although not present in the archaean Mj, the vicilin protein has an ancestral chimeric genome derived both from a gram-negative bacterium and most probably an archaean (Dunwell et al. 2000). The vicilins are desiccation resistant, and by their storage function provide energy for germinating embryos. The vicilin of peanut, Ara h1 is highly resistant to harsh environments (e.g., that of highly acidic mammalian intestines and factory food-processing; Dunwell et al. 2000). A Nelumbo Ara h1 is identified as thermally stable (Supp. 2). Also identified are 13S-globulin and legumin proteins LEGA and LEGB presented in Supplement 2.
(2) Met-Synthase catalyzes the biosynthesis of methionine, via the cobalamine independent pathway present in plants and some prokaryotes including archaeans. In addition to providing monomeric blocks for protein building, Met is the precursor of AdoMet (S-adenosyl-L-methionine), a major methyl donor in numerous cellular reactions (cf. PIMT protein repair). In plants, Met plays a central role in the cellular metabolism of protein synthesis, methyl-group transfer, and the biosynthesis of ethylene and polyamines (Ravanel et al. 1998) all processes of relevance to aging-repair and stress protection. AdoMet is an immediate intermediate in the biosynthesis of the plant stress-hormone ethylene (Ravenel et al. 1998). The Met-mediated production of ethylene is typically promoted by such abiotic stresses as heat, water, cold, salinity, hypoxia and water deprivation (Morgan and Drew, 1997). The Met-Synthase in plants appears to have undergone major changes following its early origination in archaeans such as Methancaldococcus jannaschii (Fig. S-1h and discussion above).
(3) PIMT We had tested the capability of supernatants of heated extracts of the Nelumbo PIMT enzyme to catalyze methylation of a synthetic L-isoaspartyl-containing polypeptide. Our results show PIMT activity at 100°C (Fig. 6) indicating that the Nelumbo protein exhibits greater thermostability than the recombinant PIMT enzymes of humans and Arabidopsis heated to ~65°C (Villa et al. 2006). Interestingly, all of these PIMT enzymes exhibit enzymatic activity at elevated temperatures (Thapar et al. 2001), suggesting that the repair of isomerization damage is likely to be significant in resistance to heat stress and an enzyme-repair factor important to the maintenance of viability of the centuries-old Nelumbo fruit. In Supplement 2 is listed another Nelumbo thermostable methyltransferase (METE) identified by MS.
Summary and Future Research
All the assembled 11 thermostable proteins identified in Nelumbo fruit function in the protection and repair of cellular processes, as reflected in the higher germination rated and enzyme activities, can promote healthy aging and long-term seed viability. Fifty-five percent of these proteins occur also in archaean Methancaldococcus jannaschii, a modern microbe of a lineage dating from ≥ 3.0 billion years ago. Thus, these are exceedingly ancient and evolutionarily well-tested proteins that have changed slowly over geologic time and that today serve to maintain the viability of Nelumbo fruit for as long as 1300 years (Shen-Miller et al. 1995). The remarkable long-term existence of these proteins provides firm evidence of the crucial role they have played in the history of life.
The Nelumbo protein-repair enzyme PIMT was the only species tested for biologic activity after being heated at 100°C (Fig. 6). Clearly, bioassay of rest of the thermal proteins here studied needs to be examined. Heat stability of purified CuZn-SOD protein in Nelumbo has been shown to have heat tolerance and maintaining activity at high temperatures (Chen et al. 2011; Ding et al. 2011). Another Nelumbo heat-tolerant protein, annexin, is upregulated after 90°C (Chu et al. 2012). Interestingly, several such 90°C-upregulated proteins are also listed in Supplement 2 of Nelumbo thermal proteins, aldolase reductase, LEA (Dc3, Dc8, D34) and phosphoglycerate kinase. Plant annexin is a multifunctional stress protein. Its binding to lipids may have a membrane protective role as evidenced by membrane fluidity in old Nelumbo seeds (Priestley and Posthumus, 1982). With Nelumbo genome now available for broad-range research we are heartened that information will accrue toward understanding seed aging and longevity, and the betterment of agricultural crops.
From the data presented above, we suggest below some “fruit-for-thought” questions for future studies of the Nelumbo fruit:
How long a maturation period is required for accumulation of heat-stabled proteins?
How does protein heat-solubility relate to protein activity?
What factors govern protein heat-stability?
In addition to the 11 enzymes discussed above and the 30 thermostable proteins identified in Supplement 2, what other heat resistant proteins occur and how might they be related to stress and the exceptional long-term viability of the Nelumbo fruit?
Supplementary Material
Acknowledgements
We thank C. Haas-Blaby for assistance on bioinformatics and protein alignments, J.W. Schopf for constructive comments on the manuscript, J. Lowenson for helpful discussions, and H. Nguyen for assistance in the tandem massspectrometry data. For antisera, we are grateful to P. Viitanen for CPN20 and CPN60; T. Close for dehydrin; and B. Downie and D. Martin for PIMT1. Work in SGC laboratory is supported by NIH grant GM206020. We also thank K.O. Stetter whose seminal research on hyperthermophiles stimulated this ‘hot protein’ study of Nelumbo enzymes.
Abbreviations
- AdoMet
S-adenosyl-L-methionine
- CPN20/60
chaperonine20/60
- CuZn-SOD
copper-zinc superoxide dismutase
- 1-CysPRX
1-Cys peroxiredoxin
- ENO1
enolase1
- EF-1α
elongation factor-1α
- EST
expressed sequence tag
- HSP80
heat-shock protein 80
- LC-MS-MS
liquid-chromatography tandem mass-spectrometry
- Mj
Methancaldococcus jannaschii
- PIMT
L-isoaspartyl methyltransferase
- ROS
reactive oxygen-species
Footnotes
J. Shen-Miller for manuscript preparation, overall experimentation and coordination; P. Lindner for initiation of thermal-protein experiments; Y. Xie, K. Wooding, R.R.O. Loo, and J. A. Loo for protein mass-spectrometry; and S. Villa and S. G. Clarke for PIMT experiments.
Contributor Information
J. Shen-Miller, IGPP Center for the Study of Evolution and Origin of Life, Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Geology Building, Room 5676, 595 Charles E. Young Drive East, Los Angeles, CA 90095-1567, USA, Telephone: (310) 825-2891, Fax: (310) 825-0097, shenmiller@lifesci.ucla.edu
Petra Lindner, Lehrstuhl Mikrobiologie Regensburg University Universitat Str. 31 93053 Regensburg, Germany.
Yongming Xie, Department of Chemistry and Biochemistry University of California, Los Angeles 402 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1569, USA.
Sarah Villa, Department of Chemistry and Biochemistry University of California, Los Angeles 640 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1570, USA.
Kerry Wooding, Department of Chemistry and Biochemistry University of California, Los Angeles 402 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1569, USA.
Steven G. Clarke, Department of Chemistry and Biochemistry University of California, Los Angeles 640 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1570, USA
Rachel R. O. Loo, Department of Chemistry and Biochemistry University of California, Los Angeles 402 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1569, USA
Joseph A. Loo, Department of Chemistry and Biochemistry University of California, Los Angeles 402 Boyer Hall, Hilgard Avenue Los Angeles, CA 90095-1569, USA
References
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot. 2002;53(327):1331–1341. [PubMed] [Google Scholar]
- Bersch U, Soll J, Seetharam R, Viitanen P. Identification, characteristic and DNA sequencing of a functional “double” GroES-like chaperonine from chloroplasts of higher plants. Proc Natl Acad Sci (USA) 1992;89:8696–8670. doi: 10.1073/pnas.89.18.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit BB, Simpson AJ, Whitehead TA, et al. Transcriptional profiling of the hyperthermophilic methanoarchaeon Methanococcus jannaschii in response to lethal heat and non-lethal cold shock. Environ Microbiol. 2005;7(6):789–797. doi: 10.1111/j.1462-2920.2005.00751.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;27:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, et al. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Burnett WN. “Western blotting” electrophoretic transfer of proteins from sodium dodecylsulfate-polyacrylamide gels to unmodified nitrocellulose, and radiographic detection with antibody and radio-iodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BNC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ. Thousand-year-old Nelumbo has awakened. Fossil. 1978;1:22–23. (in Chinese) [Google Scholar]
- Chen CH, Chen SM, Zhou KS. Palynological analysis of the Holocene Nymphaea seed-bearing deposits at the vincinity in Liaoning Peninsula. Quaternaria Sinica. 1965;4:167–173. (in Chinese) [Google Scholar]
- Chen D, Zheng X, Li G, et al. Molecular cloning and expression of two cytosolic copper-zinc superoxide dismutases genes from Nelumbo nucifera. Appl Biochem Biotechnol. 2011;163:679–691. doi: 10.1007/s12010-010-9074-1. [DOI] [PubMed] [Google Scholar]
- Chen JW, Dodia C, Feinstein SI, et al. 1-Cys peroxiredoxin, a Bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000;275(37):28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- Chu P, Chen H, Zhou Y, et al. Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta. 2012;235:1271–1288. doi: 10.1007/s00425-011-1573-y. [DOI] [PubMed] [Google Scholar]
- Clarke S. Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damage proteins for repair. Ageing Res Rev. 2003;2:263–285. doi: 10.1016/s1568-1637(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Close TJ. Dehydrin, a commonality in response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- De Souza CE, Grossi-De-Sa MF, Lima TB, et al. Plant storage proteins with antimibrobial avtivity novel insights into plant defense mechanisms. FASEB. 2011;25(10):3290–3305. doi: 10.1096/fj.11-184291. [DOI] [PubMed] [Google Scholar]
- DeWeerdt SE. The first sequenced extremophile, what scientists have learned from the M. jannaschii genome. Genome News Network, Feb 1, 2002. 2002 www.genomenewsnetwor.org/article/02_03/extremo.shtml. [Google Scholar]
- Dietz KJ. Peroxiredoxins in plants and cyanobacteria. Antioxidants and redox signaling. 2011;15(4):1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YF, Cheng HY, Song SQ. Changes in extreme high-temperature tolerance and activities of antioxidant enzymes of sacred lotus seeds. Science in China Series C: Life Sci. 2008;51(9):824–853. doi: 10.1007/s11427-008-0107-8. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Kuri S, Gane PJ. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev. 2000;64(1):153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso NA, Cyr RJ. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor 1-α. The Plant Cell. 1994;6:893–905. doi: 10.1105/tpc.6.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–467. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson SK, Kutzer M, Procek J, et al. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold induced plant stress protein. Plant Cell. 2011;23:2391–2404. doi: 10.1105/tpc.111.085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K, Kosakai H. Laticifers in Nelumbo nucifera Gaertn.: Distribution and structure. Am Bot. 1975;39:713–719. [Google Scholar]
- Fu J, Momcilovic I, Prasad Roles of protein synthesis elongation factor EF-Tu in heat tolerance in plants. J Bot ID 835836. 2012 [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA. Function-structure analysis of proteins using covarion-based evolutionary approaches elongation factors. USA PNAS. 2001;98:548–552. doi: 10.1073/pnas.98.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Kumar A, Ahuja PS, Sreenivasulu Over-expression of Potentilla superoxide dismutase improve salt stress tolerance during germination and growth in Arabidopsis thaliana. J Plant Genet & Trangenics. 2010;1:1–10. [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: Multisequence alignments in Postscript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Griffith SC, Sawaya MR, Boytz DR, et al. Crystal structures of a repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate. J Mol Biol. 2001;313(5):1103–1116. doi: 10.1006/jmbi.2001.5095. [DOI] [PubMed] [Google Scholar]
- Hill JE, Hemmingsen SM. Arabidopsis thaliana type I and II chaperonins. Cell Stress & Chaperones. 2001;6(3):190–200. doi: 10.1379/1466-1268(2001)006<0190:attiai>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland HD. Volcanic gases, black smokers, and the Great Oxidation Event. Geochim Cosmochim Acta. 2002;66(21):3811–3826. [Google Scholar]
- Huang SZ, Tang XJ, Lu CB, et al. Characteristic of superoxide dismutase in lotus seeds. Acta Physiol Sinica. 2000;26(6):492–496. (English abstract) [Google Scholar]
- Huang GH. China Nelumbo. Academia Sinica Wuhan Bot Inst, Science Publ (in Chinese); 1987. Systematic and distribution of Nelumbo nucifera Gaertn. Chp 2; pp. 9–12. [Google Scholar]
- Kasting JF, Howard MT. Atmospheric composition and climate on the early Earth. Philos Trans R Soc Lond Bio Sci. 2006;361(1474):1733–1742. doi: 10.1098/rstb.2006.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Lai L, Lee HH, et al. On the mechanism of chaperone activity of the small heat-shock protein of Methanococcus jannaschii. PNAS-USA. 2003;100(14):8151–8155. doi: 10.1073/pnas.1032940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Rose R, Comai L. Developmental expression of tomato heat-shock cognate Protein 80. Pl Physiol. 1992;100:801–811. doi: 10.1104/pp.100.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JM, Kelly RM, Konisky J, et al. Purification and functional characterization of a charperone from Methanococcus jannaschii System. Appl. Microbiol. 1998;21:173–178. doi: 10.1016/S0723-2020(98)80021-0. [DOI] [PubMed] [Google Scholar]
- Kwon SY, Jeong YJ, Lee HS, et al. Enhanced tolerances of transgenic tobacco plants expressing both superoxide dismutase and ascorbic peroxidase in chloroplasts against methyl viologen-mediated oxidative stress. Plant Cell & Environ. 2002;25:873–882. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of the bacteriophage. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lal SK, Lee C, Sachs MM. Differential regulation of enolase during anaerobiosis in maize. Pl Physiol. 1998;118:1285–1293. doi: 10.1104/pp.118.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling ZQ, Xie BJ, Yang EL. Isolation, characterization, and determination of antioxidative activities of oligomeric procyanindins from the seedpot of Nelumbo nucifera Gaertn. J Agri Food Chem. 2005;53:2442–2445. doi: 10.1021/jf040325p. [DOI] [PubMed] [Google Scholar]
- Mahanty S, Kaul T, Pandey P, et al. Biochenical and molecular analyses of copper-zinc superoxide dismutase from a C4 plant Pennisetum glaucum reveals an adaptive role in response to oxidative stress. Gene. 2012;505:309–317. doi: 10.1016/j.gene.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Marcus JP, Green JL, Goulter KC, Manners JM. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999;19(6):699–710. doi: 10.1046/j.1365-313x.1999.00569.x. [DOI] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Biology. 2013;14(5):R41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PW, Drew MC. Ethylene and plant responses to stress. Physiol Plantarum. 1997;100:620–630. [Google Scholar]
- Nguyen BA, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear signal prediction. BMC bioinformatics. 2009 Jun 29;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohga I. On structure of some ancient, still viable fruits of Indian lotus, with special reference to their prolonged dormancy. Japanese J Bot. 1926;3:1–20. [Google Scholar]
- Ohga I. Supramaximal temperature and life duration of the ancient fruits of Indian lotus. Bot Magazine. 1927;41:161–172. [Google Scholar]
- Pennington SR, Dunn MJ. Proteomics, from protein sequence to function. UK: BIOS Sci Publ Ltd; 2001. p. 313. [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Priestley DA. Seed Aging, implication of seed storage and persistence in the soil. Ithaca, NY: Comstock Publ Assoc; 1986. 304 pp. [Google Scholar]
- Priestley DA, Posthumus MA. Extreme longevity of lotus seeds from Pulantien. Nature. 1982;299(9):148–149. [Google Scholar]
- Qu CP, Xu ZR, Liu GJ, et al. Differential expression of copper-zinc superoxide dismutase gene of Polygonum sibiricum leaves, stems and underground stems, subjected to high-salt stress. Int J Mol Sci. 2010;11:5234–5245. doi: 10.3390/ijms11125234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R. The specific features of Methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Chaussidon M. A paleotemperature curve for the Precambrian oceans based on silicon isotopes in chert. Nature. 2006;443:969–972. doi: 10.1038/nature05239. [DOI] [PubMed] [Google Scholar]
- Schopf JW. The oldest known records of life: stromatolites, microfossils, and organic matter from the early Archaean of South Africa and Western Australia. In: Bengtsen S, editor. Early Life on Earth. NY: Columbia Univ Press; 1994. pp. 193–206. [Google Scholar]
- Schopf JW. The paleobiological record of photosynthesis. Photosynth Res. 2011;107:87–101. doi: 10.1007/s11120-010-9577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MF. A microchemical analysis study of the fruit coat of Nelumbo lutea. Am J Bot. 1929;16:259–276. [Google Scholar]
- Shen-Miller J, Aung LH, Turek J, et al. Centuries-old viable fruit of Nelumbo nucifera Gaertn, var. China Antique. Tropical Plant Biology. doi: 10.1007/s12042-013-9124-2. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, et al. Exceptional seed longevity and robust growth: Ancient sacred lotus from China. Am J Bot. 1995;82(11):1367–1380. [Google Scholar]
- Shen-Miller J, Schopf JW, Harbottle G, et al. Long-living lotus: germination and soil γ-irradiation of centuries-old fruits, and cultivation, growth, and phenotypic abnormality of offspring. Am J Bot. 2002;89(2):236–247. doi: 10.3732/ajb.89.2.236. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gells. Annal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Small T, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4(6):1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Stetter KO. Hyperthermophilic prokaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- Tatsuka M, Mitsui H, Wada M, et al. Elongation factor-1 alpha gene determines susceptibility to transformation. Nature. 1992;359(6393):333–336. doi: 10.1038/359333a0. [DOI] [PubMed] [Google Scholar]
- Thapar N, Kim AK, Clarke S. Distinct patterns of expression but similar biochemical properties of protein L-isoaspartyl methyltransferase in higher plants. Plant Physiol. 2001;125(2):1023–1035. doi: 10.1104/pp.125.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tele Images-Nature. Power Plants (Series #4) Paris, France: Dir L. Frapat, Assist Dir I. Han, Camera P. Moreau, Sound P. Fleurant; 2003. Eternal Seeds, a documentary of Nelumbo nucifera: Xipaozi, China, USA, Japan & Africa. 50 min. [Google Scholar]
- Thirumalia D, Lorimer G. Chaperonine-mediated protein folding. Ann Rev Biophys Biochem Struc. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- Tiedmann J, Neubohn B, Muntz K. Different functions of vicilin and legumin are reflected in the histopattern of globulin mobilization during germination of vetch (Vicia sativa L.) Planta. 2000;211:1–12. doi: 10.1007/s004250000259. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures ans some applications. Proc Nat Acad Sci (USA) 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth LD, Van Der Ost J, Koutsopoulos S. Hyperthermopholic enzymes – stability, activity and implementation strategies for high temperature applications. FESB J. 2007;274:4044–4056. doi: 10.1111/j.1742-4658.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- Van Der Straeten D, Rodrigues-Pousada A, Goodman HM, Van Montagu M. Plant enolase: gene structure, experession, and evolution. The Plant Cell. 1991;3:719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. Hyperthermophlic enzymes sources, uses, and molecular mechanisms for thermostability. Microbiol Mole Biol Rev. 2002;65:1, 1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa ST, Xu Q, Downie AB, Clarke SG. Arabidopsis protein repair L-isoaspartyl methyltrransferase: predominant activities at lethal temperatures. Physiol Plantarum. 2006;128:581–592. doi: 10.1111/j.1399-3054.2006.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen PV, Schmidt M, Bucher J, et al. Functional characterization of the higher plant chloroplast chaperonins. J Biol Chem. 1995;270:18158–18164. doi: 10.1074/jbc.270.30.18158. [DOI] [PubMed] [Google Scholar]
- Xu Q, Belcastrp MP, Villa ST, et al. A second protein L-isoaspartyl transferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Pl Physiol. 2004;136:2652–2664. doi: 10.1104/pp.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.