Abstract
The common endosymbiotic Wolbachia bacteria influence arthropod hosts in multiple ways. They are mostly recognized for their manipulations of host reproduction, yet, more recent studies demonstrate that Wolbachia also impact host behavior, metabolic pathways and immunity. Besides their biological and evolutionary roles, Wolbachia are new potential biological control agents for pest and vector management. Importantly, Wolbachia-based control strategies require controlled symbiont transfer between host species and predictable outcomes of novel Wolbachia-host associations. Theoretically, this artificial horizontal transfer could inflict genetic changes within transferred Wolbachia populations. This could be facilitated through de novo mutations in the novel recipient host or changes of haplotype frequencies of polymorphic Wolbachia populations when transferred from donor to recipient hosts. Here we show that Wolbachia resident in the European cherry fruit fly, Rhagoletis cerasi, exhibit ancestral and cryptic sequence polymorphism in three symbiont genes, which are exposed upon microinjection into the new hosts Drosophila simulans and Ceratitis capitata. Our analyses of Wolbachia in microinjected D. simulans over 150 generations after microinjection uncovered infections with multiple Wolbachia strains in trans-infected lines that had previously been typed as single infections. This confirms the persistence of low-titer Wolbachia strains in microinjection experiments that had previously escaped standard detection techniques. Our study demonstrates that infections by multiple Wolbachia strains can shift in prevalence after artificial host transfer driven by either stochastic or selective processes. Trans-infection of Wolbachia can claim fitness costs in new hosts and we speculate that these costs may have driven the shifts of Wolbachia strains that we saw in our model system.
Introduction
The ubiquitous intracellular α-proteobacterium Wolbachia pipientis manipulates reproductive biology of many arthropod species in order to warrant its own transmission in host populations (reviewed by [1,2]). Wolbachia are maternally inherited and favor infected females by inducing reproductive phenotypes with cytoplasmic incompatibility (CI) as the most common ([3–5]). Besides reproductive host manipulations, Wolbachia can also affect nutritional and metabolic pathways of hosts ([6,7]), host development and lifespan (reviewed in [8–11]), provide protection of hosts from pathogens and parasites ([12–15]), as well as affect host mating behavior and facilitate host speciation ([16,17]).
Wolbachia have attracted major attention as potential novel biological control agent against the increasing threat that insect populations pose to human health and agriculture either by vectoring pathogens, or by directly causing damage to crops and life stock ([18]). Wolbachia could suppress insect populations through the induction of CI in a way analogous to the Sterile Insect Technique (SIT; reviewed in [19]), manipulate vectorial capacity through host lifespan-shortening effects ([11]) or vector refractoriness to pathogens in mosquitoes ([10,15,20–23]). Wolbachia could also be used in combination with the widely used SIT ([24]) that has encountered some problems with male fitness, mating performance, sperm size and number due to gamma irradiation of individuals ([25,26]). However, all of these Wolbachia applications require that (i) Wolbachia strains are transferable between diverse host systems and insect genera, (ii) the transferred strains are capable of inducing the expected phenotype such as CI, life shortening or resistance against pathogens, and (iii) transferred infection and their expected phenotypes persist stably in the novel host. So far, several authors have reported successful transfer of Wolbachia by microinjection from donor to recipient hosts within the same insect order, followed by confirmation of their phenotype in the novel host ([27–29,11]). The third requirement of phenotypic, and thus genomic stability has not yet been tested extensively, although artificially transferred symbiont strains can potentially experience phenotypic changes ([30]). Genotypic changes might include point mutations and genomic rearrangements triggered and facilitated by symbiont infection dynamics upon arrival and successful establishment in the novel host. In addition, pre-existing variability of Wolbachia in the donor host such as the presence of spurious genomic polymorphism in neotropical Drosophila species ([31]) and tsetse flies ([32]), as well as the existence of low-titer multi-infections ([33]) might affect the outcome of the artificial transfer and the stability of the expected phenotype in the new host. Recent studies have uncovered phenotypic plasticity of Wolbachia over evolutionary short time periods within the same host, and also upon transfer between different host backgrounds. Weeks et al. ([34]) revealed a rapid switch of wRi from a parasitic ([35]) towards a more benign mutualistic state in Californian populations of Drosophila simulans. Adaptation of the symbiont to a novel host cell environment resulted in significant phenotypic changes of wMelPop when transferred between hosts ([36]). In Drosophila melanogaster wMelPop causes early death via over-replication in mainly nervous tissue ([37]). Trans-infection experiments caused the same lifespan reduction in D. simulans and Aedes aegypti ([38,11]). For the latter host, wMelPop was initially pre-adapted to Aedes cell lines, before its successful establishment in mosquitoes via embryonic trans-infection. Re-introduction of the Aedes-cell line adapted Wolbachia from cell lines into their native host D. melanogaster, however, resulted in fluctuations of maternal transmission efficiency, lower titers, and a reduced life shortening effect ([36]). These findings implicate that both host and symbiont interact dynamically and co-evolve rapidly within relatively short time periods.
It is so far unknown how genotypic integrity of Wolbachia is affected by recombination, genetic drift or selection after artificial host transfer. Here, we have monitored Wolbachia genome dynamics and population structure experimentally by utilizing two different Wolbachia-insect host species, D. simulans ([29]) and the Mediterranean fruit fly Ceratitis capitata ([28]). Both hosts were microinjected with Wolbachia of the European cherry fruit fly Rhagoletis cerasi ([29,28]). We tested if the transfer of Wolbachia, in particular of the strain wCer2, (i) induced de novo structural rearrangements, and/or (ii) enhanced sequence polymorphism within the symbiont genome post microinjection. The potential appearance of additional Wolbachia subvariants, hereby designated haplotypes, that are distinctive from the reference sequence of the symbiont in its native host ([39]) can either be assigned as de novo mutations or ancestral haplotypes that had already persisted in the donor at low frequencies and thus had escaped earlier detection. We hypothesized that the structure of the original Wolbachia strain used for trans-infection plays a crucial role in the performance within the new host. It is frequently assumed that Wolbachia strains are monoclonal populations, and thus, only de novo mutations will contribute to potential diversification of the symbiont in the recipient host. In contrast, an alternative situation of Wolbachia strains that represent diverse populations of haplotypes such as in native tsetse flies ([32]) at varying frequencies would allow for detection of sequence variation after host transfer that was not triggered by new mutations. The trans-infection event may solely shift haplotype frequencies and thus enable detection of rare ancestral haplotypes.
To assess the potential for new structural rearrangements of wCer2 in its two new host species, we took advantage of the unusual high numbers of mobile genetic elements in Wolbachia genomes ([40–45]) with their capacity to trigger insertions, inversions as well as ectopic recombination ([46–50]). Our analysis, however, did not reveal rearrangements. To test for sequence polymorphism, we analyzed SNP (single nucleotide polymorphism) frequency of three Multi Locus Sequence Typing (MLST) Wolbachia genes and traced wCer2 sequence heterogeneity in original and new hosts. In the course of this in-depth analysis we found in microinjected D. simulans clear signs of an unexpected infection with wCer1 of the original host that had remained undetected for over 150 generations. We then discuss whether heterogeneity in new hosts is caused by ancestral Wolbachia sequence polymorphism or arises through new mutations.
Materials And Methods
2.1 Insect Lines
Fly stocks of D. simulans and the Mediterranean fruit fly C. capitata known to be infected by wCer2 were used in this study. They had been microinjected with cytoplasm from the Wolbachia-infected cherry fruit fly R. cerasi thirteen ([29]) and eleven years ago ([28]). Rhagoletis cerasi is naturally multi-infected with up to five strains, wCer1 - wCer5 ([51,39]). Based on sequence analysis of MLST genes, wCer1, wCer2 and wCer4 are A supergroup strains, wCer5 a B supergroup strain, and wCer3 a recombinant strain ([39,52]; http://pubmlst.org/wolbachia/). For the first host-transfer experiment, embryos of Wolbachia-free D. simulans (Nouméa TC, generated by tetracycline treatment over three generations; [53]) were injected with egg cytoplasm from Austrian R. cerasi donors in 200 ([29]). From this experiment, six G0 isofemales resulted in wCer2 infected lines RC20, RC21, RC33, RC45, RC50 and RC78 that had to be further selected for Wolbachia in consecutive generations. For selection, DNA was extracted from multiple females and infection status was determined via Wolbachia-specific PCR. Offspring of females that tested positive for Wolbachia was used to proceed into the next generation via sibling mating ([29]). This selection regime was paused between G20 (2001) and G140. In 2007, selection for Wolbachia was continued after only five out of the six initial isofemale lines, RC20, RC21, RC33, RC45, and RC50 tested positive for wCer2. The Wolbachia strain wCer1, however, was not detected since it was considered lost between G1 and G2 ([29]). For the second host-transfer experiment, embryos of the C. capitata Benakeion line were injected with Wolbachia from R. cerasi from Austria and Italy (Sicily) in 2002 ([28]). This resulted in two infected C. capitata lines ([28]) and one of these, WolMed88.6 harboring wCer2, was included in our study. DNA extracts of approximately G50 post-infection were kindly provided by K. Bourtzis' laboratory (University of Ioannina, Greece). Wolbachia-free D. simulans Nouméa TC and D. melanogaster w1118 (Wol neg) were used as negative controls, Wolbachia-infected DSR (D. simulans Riverside, California; [54]) and the D. melanogaster Harwich strain (Wol pos) were used as positive control. All Drosophila lines were kept on standard medium at 24°C.
Antibiotic Treatment Of Donor And Recipient Hosts
For Wolbachia depletion, embryos of R. cerasi were transferred to larval medium containing a final concentration of 0.02, 0.025 and 0.05% (w/v) tetracycline and incubated at 24°C until reaching the third instar (L3). Antibiotic larval media and R. cerasi individuals were kindly provided by K. Köppler from the Center for Agricultural Technology Augustenberg (LTZ), Stuttgart, Germany. D. simulans RC20 and RC50 were placed on standard Drosophila diet containing 0.03% tetracycline for two consecutive generations before the presence of Wolbachia was tested.
2.2 Molecular Isolation And Characterization Of Wolbachia From R. Cerasi And Their Novel Hosts
Dna Extraction
For PCR, cloning and sequencing, high quality genomic DNA was extracted from individual pupae or adults using the Puregene DNA Purification Kit. For Southern blots, genomic DNA was extracted from individual adult flies and processed following the protocol from [55]. For Wolbachia depletion assays, DNA was extracted from tetracycline-treated adults of trans-infected D. simulans RC lines using the Puregene DNA Purification Kit (Qiagen). DNA was stored at −20°C until use.
Mlst-Pcr, Cloning, Sequencing And Sequence Analysis
We analyzed the frequencies of SNPs in the three Wolbachia MLST genes gatB (WD_0146, wMel), coxA (WD_0301, wMel) and ftsZ (WD_0723, wMel) in donor and recipient hosts ([56],[57]). General coxA and ftsZ primer sets were used as in [56]. For gatB, an additional primer set amplifying a 404-bp fragment, was designed (gatBF 5′-gatttaaatcgtgcaggggtt-3′ and gatB_450R 5′-ttgaattaaatcaattttatcctgg-3′). To selectively target wCer1, we used a strain-specific primer set described in [39] plus the VNTR-141 primer set from [48]. For all PCR reactions a Biometra T300 Thermocycler was used. PCR products were purified using the peqGOLD Gel Extraction Kit, inserted into the pTZ57R/T vector and then transformed into competent DH5α Escherichia coli cells. Clones containing the insert were Sanger sequenced at the University of Chicago Cancer Research Center (UCCRC-DSF). Sequences were analyzed using ApE plasmid editor v1.10.4 (M. Wayne Davis), CodonCode Aligner Version 2.0.3 (CodonCode Corporation) and the BLAST algorithm (www.ncbi.nlm.nih.gov).
For D. simulans recipients, we analyzed SNP frequencies for each line separately (RC20, RC21, RC33, RC45, and RC50) plus for the pool of all trans-infected lines (RC) in order to maximize sample size. The automated base-calling in CodonCode Aligner software detected SNPs in many clones from single individuals. In order to verify their authenticity, we visually inspected all SNPs in the corresponding chromatograms from both forward and reverse reads. All ambiguous SNPs were excluded from the final data set. Confirmed SNPs were then divided into two groups: recurrent SNPs and true singletons. Recurrent SNPs refer to nucleotide positions that were detected in independent clones in either different lines of new hosts, in R. cerasi clones only, or in both systems. Unique SNPs that were found in single clones only, but appeared reliable in the sequence chromatogram, were classified as singletons.
Anticipating that PCR accuracy was strongly impacted by the performance of the enzyme polymerase, we first determined the error base line of the Taq DNA polymerase used for all assays. We did not use a proof-reading enzyme as the Promega Go Taq® Flexi DNA Polymerase used in all experiments is one of the best non-proof-reading high quality and high performance polymerases on the market (see Promega notes available at www.promega.com). Based on re-PCR and re-sequencing of known coxA, ftsZ, and gatB fragments inserted into the pTZ57R/T cloning vector from independent batches of this polymerase (data not shown), we calculated the following error base lines: 0 SNPs in 4.44 kb of coxA (10×444 bp), 0 SNPs in 2.90 kb of ftsZ (10×290 bp), and 2 SNPs in 6.00 kb of gatB (14×429 bp). Compared to the published mean estimate for standard non-proof-reading Taq DNA polymerases of 0.21 SNPs/kb ([58]), our Promega Go Taq® Flexi DNA Polymerase control assay thus resulted in lower DNA polymerase error rates (0.15 SNPs/kb). We deposited 33 coxA, ftsZ, and gatB sequences, which represent rare allelic variants and nonsense mutations at GenBank (accession numbers KF17541-17573).
Restriction Fragment Length Polymorphism (rflp) Analysis Via Single Fly Southern Hybridization
We determined the structural integrity of the bacterial chromosome in the novel hosts via RFLP-analysis with highly dynamic Wolbachia marker sets: Insertion Sequence elements (IS) and Variable Number of Tandem Repeats (VNTRs). 0.5–1 µg genomic DNA from single flies was digested with 6U HindIII (New England Biolabs, USA) for 4 hrs at 37°C. After high-resolution vertical gel electrophoresis ([55]), gels were vacuum-blotted onto a positively charged nylon membrane (Hybond™-N+, GE Healthcare, UK). Membranes were hybridized with [α-32P]dCTP-labeled specific probes of IS and VNTR loci. Probes were prepared with the RediprimeTM II DNA labelling kit (GE Healthcare, UK) and exposed to high sensitivity X-ray films (Kodak, Germany). Probe primers were designed with respect to the annotated genome of wMel of D. melanogaster (NC_002978; [40]). For transposon probing, three repeats greater than 200 bp, belonging to different IS families (IS3, IS5, and ISNew) in the wMel genome of D. melanogaster were chosen ([40]). VNTR probes were VNTR-141, consisting of tandemly repeated 141-bp units, located at coordinates 89,003–90,332 in wMel ([48]) and VNTR-144, consisting of 11.8 copies of a 144 bp repeat unit located at 34,727–37,210 in wMel (MR, unpublished). HindIII-digested DNA of the lambda phage (New England Biolabs, USA) was utilized as size marker. Size of fragments was determined with respect to this standard, allowing the comparison of number and size of fragments between donor R. cerasi and new hosts D. simulans and C. capitata.
2.3 Ovary Screening Assay
We estimated the general fecundity cost of the artificially generated infection by comparing the ovaries of recipient and uninfected control lines of D. simulans. Analysis was performed between G168 and G182 post microinjection and followed the fecundity assay by [59]. Fertilized females were raised on standard food and were dissected in sterile PBS ten days after eclosion. 40 ovaries per recipient Drosophila line were screened. Fecundity status of ovaries was estimated according to number of mature eggs in the ovary: 0 eggs = class I; 1–2 eggs = class II; 3–9 eggs = class III; 10 and more eggs = class IV. Only eggs at stage 14 of oogenesis ([60]; indicated by dorsal appendages) were counted.
2.4 Statistical Analysis
For statistical analysis SPSS 16.0 and GraphPad Software (www.graphpad.com) were used. SNP frequencies were analyzed using χ2 with Yates Correction (2×2 contingency table); two tailed P-values indicated significant differences between values when <0.05. To detect potential traits of positive or negative selection, synonymous to non-synonymous substitutions per site were calculated using SNAP (Synonymous and Non-synonymous Analysis Program) provided at http://www.hiv.lanl.gov. This program uses the Nei-Gojobori corrected path counting method that adjusts for counts via Jukes-Cantor plus the weighting of pathways from one codon to another according to an equi-probable model for each possible codon-to-codon path ([61]).
Results
3.1 Conserved Wolbachia Genome Synteny After Artificial Transfer Into Novel Recipient Hosts
We analyzed the genome synteny of artificially transferred wCer2 infection from R. cerasi into D. simulans and C. capitata with five marker probes for RFLP mapping (Figure S2). Although IS and VNTRs had earlier been reported as hypervariable entities of Wolbachia genomes, we did not detect any structural re-arrangements for the five tested loci in wCer2 of the novel hosts (Figure S2 and Data S1).
3.2 Snp Frequency In Gatb, Coxa, And Ftsz Of Recipient Host Populations
Prior to all sequencing experiments, we determined the base line error rate during polymerase chain reaction of the Taq polymerase used in our lab. Consequently, we analyzed SNP frequencies in three Wolbachia loci (gatB, coxA, and ftsZ) in trans-infected D. simulans and C. capitata. For gatB, we sequenced and analyzed 29.1 kb: 12×404 bp from each of RC20, RC21, RC33, RC45, RC50 of D. simulans, plus 12×404 bp from WolMed88.6 of C. capitata (Table S1). In total, 38 SNPs randomly dispersed along the 404 bp gatB gene fragment ( Table 1 ) were detected. Out of these 38 SNPs, 6 (16%) were found recurrently, and 32 (84%) were singletons. Recurrent SNPs were SNP-11 (2×); SNP-42 (4×); SNP-93 (2×); SNP-186 (2×); SNP-250 (2×), and SNP 253 (2× ochre mutation, i.e., CAA to TAA, occurring in RC of D. simulans and in WolMed88.6; Table 1 ). Overall SNP frequency for gatB of wCer2 was 1.01 SNPs/kb ( Table 2A ). For coxA of wCer2 we analyzed 16.4 kb (37×444 bp amplicon) with an average SNP-frequency of 1.64 SNPs/kb ( Table 2B ). For ftsZ, we analyzed 7.6 kb of wCer2 (16×478 bp amplicon) with an average SNP-frequency of 0.78 SNPs/kb ( Table 2C ). Overall SNP-frequencies for all three genes were significantly higher, i.e., 5–11fold, than the error base line we had determined before for the Taq polymerase (compare frequencies of 0.78, 1.01, and 1.64 SNPs/kb to 0.15 SNPs/kb from Taq polymerase).
Table 1. Variable nucleotide positions in gatB (A) and amino acid positions in GATB (B) of wCer2.
| variable nucleotide positions in gatB of w Cer2 | 3 | 11 | 16 | 31 | 32 | 36 | 42 | 61 | 72 | 88 | 89 | 93 | 102 | 112 | 121 | 149 | 186 | 195 | 206 |
| consensus | T | A | G | C | T | C | T | A | A | T | A | T | T | A | A | C | T | A | A |
| w Cer2 R. cerasi | . | . | . | . | . | . | C | G | G | . | . | . | . | . | . | T | . | G | . |
| w Cer2 RC21 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . |
| w Cer2 RC20 | . | . | . | . | . | . | C | . | . | . | . | C | C | . | T | . | C | . | . |
| w Cer2 RC33 | . | . | . | . | A | . | . | . | . | . | G | . | . | . | . | . | . | . | . |
| w Cer2 RC45 | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| w Cer2 RC50 | . | G | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| w Cer2 Wol Med88.6 | . | . | A | T | . | A | . | . | . | . | . | . | . | . | . | . | . | . | G |
| SNP-Frequency | singleton | 2× in RC | singleton | singleton | singleton | singleton | 3× in RC, 1× in R. cerasi | singleton | singleton | singleton | singleton | 2× in RC | singleton | singleton | singleton | singleton | 2× in RC | singleton | singleton |
| variable nucleotide positions in gatB of w Cer2 | 221 | 226 | 238 | 250 | 253 | 257 | 288 | 305 | 321 | 323 | 324 | 343 | 350 | 354 | 355 | 367 | 371 | 390 | 398 |
| consensus | T | C | T | A | C | T | T | T | A | A | A | A | A | A | A | T | T | A | A |
| w Cer2 R. cerasi | C | . | . | . | . | . | . | . | G | . | G | . | . | . | . | . | . | . | . |
| w Cer2 RC21 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| w Cer2 RC20 | . | T | . | . | . | . | . | C | . | G | . | G | . | . | . | . | . | . | . |
| w Cer2 RC33 | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . |
| w Cer2 RC45 | . | . | C | . | . | C | C | . | . | . | . | . | . | G | . | . | . | C | . |
| w Cer2 RC50 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | G | . | C | . | . |
| w Cer2 Wol Med88.6 | . | . | . | G | T | . | . | . | . | . | . | . | G | . | . | . | . | . | G |
| SNP-Frequency | singleton | singleton | singleton | 1× RC, 1× in WolMed88.6 | 1× RC, 1× in WolMed88.6 | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton | singleton |
| variable amino acid positions in GATB of w Cer2 | 1 | 4 | 6 | 11 | 11 | 12 | 14 | 21 | 24 | 30 | 30 | 31 | 34 | 38 | 41 | 50 | 62 | 65 | 69 |
| consensus | A | E | V | L | L | L | S | M | R | Y | Y | I | C | M | G | F | R | I | N |
| aa changes in all RC lines and Wol Med88.6 | . | G | I | F | R | . | . | V | G | H | C | . | . | V | . | S | . | M | S |
| variable amino acid positions in GATB of w Cer2 | 74 | 76 | 80 | 84 | 85 | 86 | 96 | 102 | 107 | 108 | 108 | 115 | 117 | 118 | 119 | 121 | 124 | 130 | 133 |
| consensus | I | Q | D | R | E | I | S | F | G | K | K | L | D | A | S | Y | F | L | E |
| aa changes in all RC lines and Wol Med88.6 | T | * | . | G | * | T | . | S | . | R | . | E | E | . | G | H | S | F | G |
A) Position 1 in the presented 404 bp fragment corresponds to position 981 of the full gatB locus of wRi infecting Drosophila simulans Riverside (GenBank accession number CP001391). Aa position 1 in (B) corresponds to aa position 148 of the full GATB protein of wRi (protein ID:ACN94961.1). Frequency of SNP indicates which SNPs are singletons or occur recurrently in what host system. Nonsense mutations leading to a stop codon are indicated by asterisks. Abbreviations: aa amino acid.(
Table 2. SNP frequencies in wCer of R. cerasi and de novo hosts.
| A | |||||||
| no | gatB of wCer | bases | SNP- | SNP- | assay | P valuec | P valued |
| frequencya | frequencyb | ||||||
| 1 | wCer1 of R. cerasi | 9696 | 0.52 | nd | nd | ||
| 2 | wCer2 of R. cerasi | 8888 | 1.01 | 0.73 | 1 vs. 2 | 0.3346 | 0.8854 |
| 3 | wCer1 of RC | 1212 | 2.48 | nd | 1 vs. 3 | 0.0703* | nd |
| 4 | wCer2 of RC | 29088 | 1.01 | 0.73 | 2 vs. 4 | 0.6057 | 0.6152 |
| 5 | RC20 | 4848 | 2.68 | 2.40 | 4 vs. 5 | 0.0188* | 0.0109* |
| 6 | RC33 | 4848 | 0.83 | 0.55 | 4 vs. 6 | 0.2103 | 0.6386 |
| 7 | RC45 | 4848 | 1.24 | 0.96 | 4 vs. 7 | 0.9097 | 0.8714 |
| 8 | RC50 | 4848 | 1.03 | 0.75 | 4 vs. 8 | 0.9718 | 0.9391 |
| 9 | RC21 | 4848 | 0.00 | −0.28 | 4 vs. 9 | 0.0622* | nd |
| 10 | WolMed88.6 | 4848 | 1.65 | 1.37 | 4 vs. 10 | 0.4453 | 0.3824 |
Frequencies were calculated for (A) coxA, (B) ftsZ, and (C) gatB. Column ‘bases’ gives the total number of sequenced bases.
a SNP-frequency per kilobase;
b SNP-frequency per kilobase minus calculated error base line (0.28/kb) of Taq polymerase;
c two-tailed P values from χ2 calculations with Yates Correction (2×2 contingency table) for a;
d two-tailed P values from χ2 calculations with Yates Correction (2×2 contingency table) for b.
Abbreviations: nd not determined.
3.3 Uncovering Cryptic Co-Infection With The Wcer1 Strain
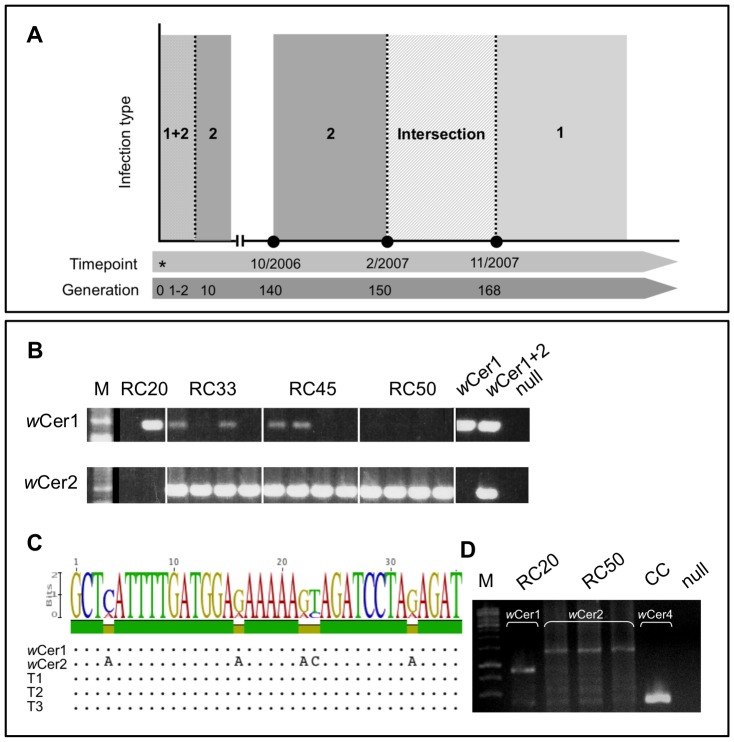
Unexpectedly, we detected sequence traces of wCer1 in three lines of microinjected D. simulans, although this strain had previously been considered as lost from the microinjected lines between G1 and G2 ([29]; and Figure 1A ). To confirm the presence of wCer1 independently, we performed PCR analyses utilizing primer sets specifically targeting wsp of either wCer1 or wCer2, respectively ([39]; Figure 1B ). According to the signal intensity of the wsp amplicon during electrophoresis, RC20 harbored wCer1 at high densities whereas wCer2 could no longer be tracked ( Figure 1B ). Wsp sequence reads from RC20 revealed exclusively the wCer1 haplotype ( Figure 1C ). In RC33 and RC45, we traced wCer1 co-infecting wCer2 ( Figure 1B ). RC50 showed no signals of wCer1 but of a single infection with wCer2 ( Figure 1B ). In addition to wsp, the presence of wCer1 in RC20 and its absence in RC50 was confirmed by the diagnostic VNTR-141 locus via PCR ( Figure 1D ).
Figure 1. Cryptic co-infection with wCer1 in wCer2 carrying trans-infected lines.
(A) Switch of strain prevalence from wCer2 to wCer1 in RC20. Asterisk represents time point of line establishment via microinjection in 200. Generations are indicated on x-axis. (B) Presence of wCer1 in random samples of RC lines determined via strain-specific wsp PCR. First RC20 sample seems to carry Wolbachia at extremely low density below detection limit of wsp PCR whereas the other one gives a bright band with wCer1-specific wsp primer set. DNA extracted from adult Rhagoletis cerasi served as positive controls (wCer1 and wCer1+2). (C) Random 36-bp fragment of the general wsp amplicon showing diagnostic wCer1/wCer2 sites. (D) Differentiation between wCer1, wCer2 and wCer4 Wolbachia using VNTR-141 PCR. Abbreviations: CC C. capitata, M DNA size marker, T1-3 trans-infected RC line sample.
After verification of the wCer1 sequences we analyzed the SNP frequency of this strain using gatB and coxA in a small sample of new hosts. We tested 1,212 bp for gatB with an average SNP-frequency of 2.48 SNPs/kb ( Table 2A ). For coxA we sequenced 4.9 kb (3×305 bp amplicons from RC20 and 9×444 bp from WolMed88.6 and RC20 and RC33; Table 2B ) with an average SNP frequency of 1.63 SNPs/kb. SNP frequencies for ftsZ of wCer1 in novel hosts were not determined due to the low coverage ( Table 2C ).
Regarding the heterogeneity detected in wCer, we hypothesized that the SNPs can (i) either represent ancestral, hidden sequence polymorphism, i.e., multiple Wolbachia haplotypes already present in the donor host, or (ii) have arisen de novo following microinjection. To test this hypothesis, we compared SNP frequencies in the novel hosts with the frequencies in the donor of the Wolbachia strains. Since original donor specimens for the microinjection experiments into D. simulans and C. capitata were not kept as voucher material, we sequenced gatB, coxA, and ftsZ from a broader representative range of other R. cerasi populations across Europe instead.
3.4 Snp Frequency Baseline In Gatb, Coxa, And Ftsz Of Donor And Comparison With Recipient Host Populations
We analyzed Wolbachia SNP frequencies from R. cerasi samples collected all over Europe ([39]). In total, we sequenced 8.9 kb (22×404 bp amplicons) of wCer2 gatB from R. cerasi derived from individuals from more than ten collection sites across Europe (Table S2 and Table S4). As shown in Table 2A the average SNP frequency for gatB of wCer2 from the donor R. cerasi was 1.01/kb (9 variable positions in 8.9 kb). Comparing these data with those of the novel hosts, we did not observe an increase in SNP-frequencies per kb of wCer2 gatB in the recipients ( Table 2 ). Only one (SNP-42) out of the 38 SNPs, however, determined in wCer2 gatB of the novel hosts, occurred also in the original donor R. cerasi.
Since we unexpectedly detected wCer1 in three trans-infected host lines we included this Wolbachia strain in our SNP analyses (however at a lower coverage). For gatB of wCer1, we sequenced 9.7 kb consisting of a 24×404 bp amplicons data set and calculated an average SNP-frequency for gatB of wCer1 in R. cerasi that was not statistically significant from wCer2 in R. cerasi (0.52 vs. 1.01; P = 0.3346; Table 2A ). The average SNP-frequency in gatB of wCer1 in novel hosts lines was also not higher than in the native host R. cerasi (2.48 vs. 0.52; P = 0.0703), but the small sample set (1,212 bp) impeded statistical testing and thus did not allow a reliable comparison between both wCer 1 and wCer2.
For the coxA locus, we sequenced 5.7 kb of wCer1 (13×444 bp amplicons), and 888 bp of wCer2. Average SNP-frequencies for wCer1 and wCer2 infections were rather low (0.69 SNPs/kb and 0 SNPs/kb, respectively; Table 2B ). It must be taken into account that the data set for wCer2 consisted only of two clone reads and can thus not be regarded as highly representative.
For the ftsZ locus, we sequenced 10.5 kb of wCer1 (23×478 bp amplicons) and 10 kb of wCer2 (21×478 bp amplicons). At this locus SNP frequencies of the two Wolbachia strains were quite similar with 1.14 SNPs/kb and 1.10 SNPs/kb for wCer1 and wCer2, respectively ( Table 2C ). One SNP in ftsZ of wCer1 resulted in a transversion from guanine to thymine in the first position of the consensus triplet GGA (Gly), thus introducing the stop codon TGA to the sequence. We detected this stop codon twice and independently in ftsZ of wCer1.
3.5 Presence Of Stop Codons In Wcer1 And Wcer2 Of Essential Wolbachia Genes In Both Donor And Recipient Host Populations
The recurrent finding of SNPs causing nonsense mutations in essential Wolbachia housekeeping genes was highly unexpected and hence considered with extreme caution. However, similar to the ochre mutation in gatB of wCer2 in D. simulans and C. capitata (see 3.2), we found additional nonsense mutations in coxA and ftsZ of wCer1 Wolbachia ( Table 3 ). A recurrent SNP (4×) in ftsZ of wCer1 of its native host R. cerasi caused a transition of guanine to thymine in the first position of a GGA triplet in wCer1 (opal mutation). Finally we also uncovered an ochre mutation in coxA of wCer1 in recipient line RC20 but as a singleton only ( Table 3 ). In order to test whether such Wolbachia pseudogenes might stem from translocations onto the host chromosome, tetracycline-treated individuals of RC20, RC50, and R. cerasi, plus their corresponding untreated controls were tested via gatB PCR for the presence of potential nuclear Wolbachia copies. As shown in Figure S1, both recipient lines lost the gatB PCR signal after two generations of antibiotic treatment, which makes a lateral gene transfer event unlikely. Hence alternative scenarios will be necessary for explaining these counter intuitive findings, i.e., the persistence of nonsense mutations in essential Wolbachia genes (see discussion).
Table 3. Stop codons in gatB, coxA, and ftsZ of wCer1 and wCer2 Wolbachia.
| Line | Gene | Position | Mutation | Wol-infection |
| CAA = consensus | wCer2 | |||
| RC20 | gatB | 226/404 | TAA (ochre) | wCer2 |
| RC50 | gatB | 253/404 | TAA (ochre) | wCer2 |
| WolMed88.6 | gatB | 253/404 | TAA (ochre) | wCer2 |
| AAA = consensus | wCer1 | |||
| RC20 | coxA | 22/444 | TAA (ochre) | wCer1 |
| GGA = consensus | wCer1 | |||
| F37 eastern Sicily | ftsZ | 25/478 | TGA (opal) | wCer1 |
| F38 eastern Sicily | ftsZ | 25/478 | TGA (opal) | wCer1 |
| F40 western Sicily | ftsZ | 25/478 | TGA (opal) | wCer1 |
| F42 western Sicily | ftsZ | 25/478 | TGA (opal) | wCer1 |
Lane three lists the position of the mutation corresponding to the size of the amplified MLST-gene fragment. Lines F37 to F42 represent R. cerasi individuals from different populations sampled in Sicily, Italy.
3.6 Trans-Infection With Wcer Is Costly For The Recipient
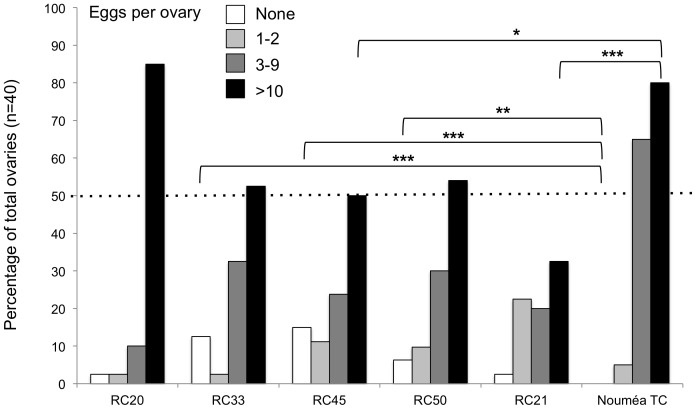
Between G168 and G182 post-microinjection we performed an ovary screening assay ([59]) to estimate the costs of establishing an artificial infection with Wolbachia. Based on the number of mature eggs per ovary, individuals were sorted into the four fecundity classes. In total, we screened 40 individuals per D. simulans line including 40 individuals from Wolbachia-uninfected D. simulans Nouméa TC, which had been used as recipient for the microinjection. As shown in Figure 2 , percentage of individuals in both extreme classes, I and IV, varies highly among new hosts. Compared to 80% in the control (Nouméa TC), RC21 shows only 32.5% of individuals in class IV, suggesting poor fecundity (P = 0.004***). Except for RC20, also the other new hosts exhibited reduced number of mature eggs (ranging from 52.5 to 60% in class IV) pointing towards a fitness cost effect related to the new symbiont infection. Interestingly, in RC20 percentage of class IV ovaries is higher than in all other new hosts and even slightly higher then in the control (85% vs. 80%). This might represent a positive correlation of fecundity with Wolbachia infection in this special case, but would require further testing for verification (see discussion).
Figure 2. Ovary screen in trans-infected RC lines.
Size of ovaries was compared on basis of number of mature eggs in one ovary. Bars represent ovary size per line determined for each ovary class: white is class I with no mature eggs; light grey class II (1–2 eggs); dark grey class III (3–9 eggs); and black is class IV with 10 or more eggs. Y-axis shows percent of ovaries per class; x-axis shows RC lines plus the Wolbachia-unifected D. simulans Nouméa TC control. Significances based on two-tailed P values from Fisher's exact test are indicated by asterisks.
Discussion
4.1 No Traces Of Structural Re-Arrangements Detectable By Means Of Wolbachia Mobile Genetic Elements Upon Artificial Transfer Into Novel Recipient Hosts
We looked at two highly informative VNTR markers and three Wolbachia transposons (IS3, IS5, and ISNew) via Southern hybridization, and thus covered at least 30 loci dispersed in the wCer2 genome. We did, however, not detect any transpositions in these loci. Recent studies demonstrated that Wolbachia carry a high percentage of functional transposable elements that can display transpositional activity during short term Wolbachia evolution ([47–49]). A study of polymorphism in IS element insertions sites and VNTRs of wMel revealed that the previously assumed homogeneous Wolbachia infection of D. melanogaster is a set of different variants, such as wMel and wMelCS ([48,62]). However, given that so far 20 major families of insertion sequences have been classified from 171 bacterial and archeal species ([63,64]) we cannot rule out that any transposition effect occurred in our study system with any other mobile elements. Thus, as a next step, whole genome sequencing of wCer will be the most adequate strategy to obtain sufficient information on potential structural rearrangements in the symbiont genome upon transfer into recipients.
4.2 Wolbachia Strains Are Most Likely Bacterial Populations Of Diverse Haplotypes With Shifting Frequencies
We determined SNP frequencies for gatB, ftsZ, and coxA, between native and recipient hosts and did not detect any SNP differences that could have arisen after microinjection. The SNP-frequencies in ftsZ, and gatB of wCer2 in the native host R. cerasi were with approximately only 1 SNP/kb low. For coxA, the frequency was even lower (0 SNPs/kb) but this might not be highly representative since we tested a very small sample size. In the recipient hosts D. simulans and C. capitata, we determined SNP-frequencies in wCer2 that were not accelerated compared to R. cerasi (see Table 2 ). Although not statistically significant, we observed a trend towards an increase in SNP-frequency, at least in gatB of one of the recipients (RC20), at the time when wCer2 was unambiguously present). We calculated the ratio of the number of non-synonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS) for gatB of wCer2, a general indicator for selective pressure acting on protein-coding genes. This ratio for gatB, in R. cerasi and RC lines (Table S3) corroborated our finding that SNP-frequencies did not change upon arrival of Wolbachia in the new host systems.
Similar to wCer2, we found overall low SNP-frequencies in gatB, coxA, and ftsZ of wCer1 in the native host R. cerasi, ranging from 0.5 to 1.1 SNP/kb. For the trans-infected hosts, we only analyzed the frequency of gatB and coxA, not for ftsZ. Both of these data sets are rather small and thus still inconclusive. Unexpectedly, we revealed cryptic double infections with wCer1, but this Wolbachia strain persisted in lower densities than the predominant wCer2. Since the focus of this study was from its onset primarily on wCer2 and not wCer1, a more extensive analysis of wCer1 sequences deriving from the trans-infected host systems was not carried out. Our coxA sequence data, however, did not indicate an increased wCer1 SNP-frequency in the de novo hosts. In particular, in wCer1-gatB, we did not find a statistically significant increase in the SNP frequency within RC compared to R. cerasi.
As shown in Table 1 , the variable sites we detected in gatB of wCer2 were either present in the native and/or in the trans-infected hosts. This finding raises the question whether the observed polymorphism represented de novo mutation events or ancestral cryptic infection polymorphism. [65] recently showed that new Wolbachia haplotypes might be generated by point mutations in outer membrane proteins. In our study 68% of the SNPs revealed in wCer2 of gatB (26/38) were not detected in the donor of the infection but exclusively in the trans-infected hosts, suggesting de novo mutation events. Our PCR, cloning and Sanger sequencing based approach resulted in relatively low sample size, so that this study is not sensitive enough to rule out the existence of rare haplotypes in the donor host. Deep-sequencing strategies of donor and recipient with much higher coverage will be of pivotal interest to finally uncover potential de novo mutation rates of the endosymbiont upon artificial host switch. An alternative reason for our failure of detecting shared SNPs in both donor and recipients is genetic drift. Through drift effects, allele frequencies change and this may result in the loss of certain haplotypes followed by consequent reduction of genetic variation within a population. As a third explanation, selection for certain, beneficial haplotypes can be envisaged. This would lead to subsequent fixation of these haplotypes and loss of others.
We found, however, that SNP-42 in gatB of wCer2 ( Table 1 ) occurred recurrently in both donor and recipient systems (RC20), clearly suggesting an ancestral origin. It is possible that the mutation in position 42 in the 404-bp gatB fragment represents a rare haplotype of wCer2 that coexists with the canonical haplotype in the R. cerasi donor ([39]; this study). Moreover, SNP-250 of gatB of wCer2 that results in the replacement of Arg with Gly ( Table 1 ) was found in both heterologous hosts D. simulans and C. capitata, but not in the donor. This situation is similar to SNP-253 of gatB of wCer2 that results in a nonsense mutation (see below). SNP-11, SNP-93 and SNP-186 provide three additional cases for the existence of distinctive gatB haplotypes of the wCer2 infection since all three were repeatedly isolated from hosts that were independently microinjected ( Table 1 ). Hence the wCer2 infection cannot be considered monoclonal but a bacterial population of diverse haplotypes at varying frequencies. This idea is supported by the recent finding from Symula and colleagues who proposed the existence of high Wolbachia sequence variation between and within individuals of the tsetse fly Glossina fuscipes fuscipes ([32]).
If wCer2 infection is a population of haplotypes, rare haplotypes within this population might be difficult to detect. Any change in the structure of this bacterial population, however, massively impacts the frequency of haplotypes. Events that impact the population structure as well as population size in such a crucial way are referred to as bottleneck events. The artificially transfer of wCer2 from its native host R. cerasi into two new hosts, was such a bottleneck event and thus manipulated the structure of the original wCer2 population. We argue that this resulted in the shift of haplotype frequencies in the trans-infected lines. It is likely that the polymorphism that we observed in gatB upon arrival in D. simulans and C. capitata represented rare haplotypes that already persisted in the ancestral and native wCer2 population of R. cerasi and are only detectable after the artificially induced bottleneck scenarios.
4.3 Wolbachia Strains Accumulate Nonsense Mutations Upon Arrival In New Hosts
We tested if SNP frequency in Wolbachia genes is increased after microinjection, thus suggesting relaxation of purifying constraints on these genes. Our results did not explicitly support such an effect although a slight trend towards diversifying selection was still observed (see 4.2). We revealed, however, that three out of 38 SNPs (8%) detected in gatB of wCer2 introduced novel pre-mature stop codons caused by in-frame ochre mutations, i.e. a transition of cytosine to thymine in the first position of a CAA triplet. SNP-226 was found uniquely in transinfected RC20, whereas SNP-253 was found in both recipient hosts independently, i.e. D. simulans (line RC50) and C. capitata (line WolMed88.6). Novel stop codons were not restricted to wCer2 since we also traced them in coxA and ftsZ of wCer1. SNP-22 in coxA of wCer1 was found uniquely in line RC20. In contrast, SNP-45 in ftsZ of wCer1 seemed to be of ancestral origin, occurring in Sicilian R. cerasi populations only.
A growing body of empirical evidence has demonstrated that Wolbachia genes and even complete genomes are being transferred onto insect host chromosomes ([66–69]). Such lateral gene transfer events can explain the accumulation of nonsense mutations when fully functional copies of these genes are still present in the symbiont genome. In order to test for lateral gene transfer, we cleared the recipient lines with antibiotics. Symbiont genes transferred into the host genome would be not be affected and thus still detectable. Our screen of treated RC lines did not indicate any gene transfer event, suggesting that the detected nonsense mutations are present in cytoplasmic Wolbachia.
In total, we found two types of nonsense mutations in recipient and donor hosts, ochre and opal. In the recipient systems, we revealed two in-frame ochre mutations in wCer2 of gatB, and two in-frame opal mutations in wCer1 of coxA. In the original donor system we determined one in-frame mutation in ftsZ of wCer1. Mutant tRNA is able of suppressing some stop codons in E. coli ([70,71]), and allele-specific super-suppressor mutants have been reported for the yeast Saccharomyces cerevisiae ([72]). We performed a PCR-based pilot screen for Wolbachia candidate tRNA suppressor mutants that would be able to rescue both ochre mutations in wCer1 but did not find any (data not shown). It can hence not yet be explained why unexpectedly high frequencies of nonsense mutations occurred in two Wolbachia housekeeping genes of both Wolbachia strains. We propose, however, several ideas that might explain the compensation of these mutations. First, the ‘codon capture model’ allows a bacterial codon that has fallen to low frequencies to be reassigned without triggering fitness implications ([73,74]). In the case of the ochre mutation in wCer2, TGA would be re-coded into a synonym and hence not affect protein length. Alternatively, in concert with our theory of co-existing wCer2 haplotypes in the population of one single host, it is possible that one haplotype carries the nonsense mutation whereas another one does not. A fully functional wCer2 haplotype could then potentially compensate the mutation in the non-functional haplotype. Generally, bacteria are assumed to be monoploid i.e., they carry only one copy of a circular chromosome. Recent publications have demonstrated that this is not necessarily case. [75] have shown that Neisseira gonorrhoeae are polyploidy and carry three genome copies in average. [76] have added striking new findings by stating that monoploidy is not typical for bacteria. In contrast, polyploidy is very common in proteobacteria with up to even 20 genome copies. Following these interesting findings, it might be possible that Wolbachia also contain more than one genome copy per cell. If those copies are different, i.e., one carries the mutation and the other does not, the Wolbachia sequence polymorphism we detected in this study can be explained. Alternatively, but highly unlikely, the formation of paralogues via intrachromosomal duplications of the three Wolbachia genes coxA, ftsZ and gatB can be employed. Finally, it could also be speculated that alternative genetic codes might support compensating the nonsense mutations. Contrasting to the bacterial code, the TAA triplet does for example not lead to a termination signal in the ciliate, dasycladacean and Hexamita nuclear genomes as well as in the alternative flatworm mitochondrial code. The TGA codon can be compensated by even nine alternative codes (source: www.ncbi.nlm.nih.go). So far, we do not know how stably these mutant haplotypes are maintained within the wCer population but an ongoing deep sequencing project of the wCer2 genome will allow us to screen, in detail, for the presence and maintenance of these mutations.
4.4 Maintenance And Frequency Shifts Of Diversity After Trans-Infection
Multiple Wolbachia strains can coexist within single host individuals e.g. ants ([77]) and tephritid fruitflies ([78,39]). However, simultaneous persistence of more than one Wolbachia strain within a single host raises the question as to whether inter-strain competition for survival and stable persistence does occur. In the case of the observed strain replacement in RC20, inter-strain competition between wCer1 and wCer2 is expected. We collected evidence that the initially common wCer2 was no longer traceable in G168 after the trans-infection event, supporting the idea of strain replacement following inter-strain competition. Recent studies demonstrated that Wolbachia infections can occur at extreme low titer levels. The persistence of such natural low titer Wolbachia infections have been reported in bark beetles ([33]), neotropical Drosophila species ([17]), aphids ([79]), D. simulans ([80]) and tsetse flies ([81]). We hence assume that wCer2 density is extremely variable, making detection of the symbiont impossible even by using highly sensitive techniques. However, the artificial double infection in the new host background might have triggered ongoing inter-strain competition. An explanation for wCer1 displacing prevalent wCer2 in RC20 might be a negative symbiont-host productivity correlation (see 4.5). Beaumont and colleagues recently reported on the experimental evolution of bet hedging strategies in bacterial populations ([82]). Bet hedging is defined as stochastic switching between phenotypic stages ([83,84]) in order to facilitate persistence in fluctuating environmental conditions. The new host background, representing changed environmental conditions for the symbionts, could have led to the evolution of a bet hedging-like strategy in RC20, switching between two Wolbachia variants. Switch from wCer2 to wCer1 is correlated with enhanced fecundity in RC20, and it cannot be ruled out that such adaptive bet hedging results in switching back to wCer2 as main infection variant.
In RC33 we found clear co-existence of wCer1 and wCer2, obviously not subjected to inter-strain competition. Densities of both strains seem to be equal in this system, suggesting competition-free co-existence. This was already shown for the parasitic wasp Leptopilina heterotoma where density of different Wolbachia strains was not affected by the presence of other strains ([85]). Stable multiple infections with Wolbachia were reported from D. simulans ([86]), R. cerasi ([39]), and Ae. albopictus hosts ([87]). The idea of stable co-existence without inter-strain competition of the bacteria is supported by significantly increased rates of maternal transmission determined in this line in comparison to the rate evaluated shortly after microinjection. This is in agreement with our study where wCer2 in D. simulans exhibited an initial prevalence of 65% ([29]), and has now reached almost complete transmission (95%; data not shown).
4.5 Trans-Infection Events Claim Reproduction Costs In Novel Hosts
[88] demonstrated that wMelPop triggers severe phenotypic changes such as decrease of fecundity in the mosquito Ae. albopictus. The authors report a clear correlation between host phenotype and the endosymbiont Wolbachia. Similar to the situation in mosquitoes, we observed that fecundity of trans-infected D. simulans lines is affected by the artificially introduced Wolbachia infection when measured ten generations after microinjection ([29]). Data obtained from an ovary screening assay more than 150 generations post microinjection suggested that female flies were still not adapted to the infection. Compared to the uninfected D. simulans Nouméa TC strain, 80% of the trans-infected RC lines displayed decreased ovary sizes. Most interestingly, ovaries of RC20 females were significantly larger, as they contained more mature eggs than the other RC lines and slightly larger than the uninfected control. The fecundity of RC20 seemed to change from very poor at the beginning to enhanced in later host generations in the course of this study (DS, personal observation). Hence we might observe a correlation between female fecundity and Wolbachia infection in RC20. We have tracked a switch in wCer2/wCer1 prevalence in RC20 that most likely occurred between generations 150 and 167. It might be possible that this change of female fecundity is correlated with a Wolbachia strain switch in this line. Although we have no direct evidence yet, we speculate that wCer2 might be negatively correlated with female fecundity in this special case of RC20. This line was reported as mono-infected with wCer2 in earlier passages ([29]), later diagnosed as wCer1&2 double-infected (this study), and eventually the wCer1 infection has outcompeted wCer2, since the latter strain was no longer traceable by PCR in later generations ( Figure 1B ). In order to determine when this shift from wCer2 to wCer1 took place, we analyzed RC20 DNA extracts from seven, randomly picked, non-consecutive generations between G140 (beginning of this study) and G168 (see time course in Figure 1A ). We found a switch from wCer2 to wCer1 during a transition period between generations F150 and F167, followed by replacement of wCer2 by wCer1 in F168. However, to directly prove the correlation between wCer2 and host fecundity, and to rule out that the microinjection-caused bottleneck did not just lead to accumulation of negative effects, further experiments are needed. Re-evaluation of our data in a homogenized host nuclear background obtained through outcrossing the RC20 line will allow for better analysis of wCer-triggered fitness costs in the host.
Conclusion
In this study we aimed at testing if artificial symbiont transfer triggers structural rearrangements, and acceleration of SNPs in the symbiont genome. Analysis of mobile genetic elements within Wolbachia did not reveal rearrangements after arrival of the symbiont in the recipient hosts. By assessing SNP frequency in three essential Wolbachia genes before and after microinjection, we determined that the purifying constraint operating on these loci is hardly relaxed after more than 150 host generations. Instead of tracing new mutations upon transfer in the recipients, we discovered ancestral polymorphic sites in symbiont genes deriving from the donor, pinpointing that both wCer1 and wCer2 exhibit ancestral and cryptic sequence polymorphism in its original host R. cerasi. We further uncovered multiple strains in D. simulans lines that were previously typed as singly infected. This may have been due the co-existence of Wolbachia strains, where one of these persisted at low titer and thus had escaped standard detection techniques. We demonstrated that infections by multiple strains are prone to shifts in strain prevalence upon artificial host transfer. This reflects the population-like structure of Wolbachia within and between different hosts and thus will have consequences for symbiont population dynamics. Persistence of cryptic multiple infections after transfer from a multiply infected donor, captures the importance of studying, in detail, the integrity of Wolbachia infections prior to application as tools in modern pest and disease control management.
Supporting Information
Extended methodology for RFLP-mapping via genomic Southern blot analysis. Detailed information about RFLP mapping can be found in Data S1.
(DOCX)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOC)
(DOCX)
Acknowledgments
We are thankful to Lisa Klasson (Sweden) for critically reading the manuscript. We thank Kostas Bourtzis (Greece) for kindly providing DNA from C. capitata strain WolMed88.6, and Kirsten Köppler (Germany) for technical assistance with antibiotic treatment of R. cerasi. We are obliged to Traude Kehrer (Austria) for excellent fly care.
Funding Statement
WJM and DS were partly supported by the research grant FWF P19206-B17 and P22634-B17 from the Austrian Science Fund and the EU-COST Action FA0701 “Arthropod Symbiosis: From Fundamental Studies to Pest and Disease Management”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stouthamer R, Breeuwer JA, Hurst GD (1999) Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53: 71–102. [DOI] [PubMed] [Google Scholar]
- 2. Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulator of invertebrate biology. Nat Rev Microbiol 6: 741–51. [DOI] [PubMed] [Google Scholar]
- 3. Yen JH, Barr AR (1971) New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L . Nature 232: 657–8. [DOI] [PubMed] [Google Scholar]
- 4. Hoffmann AA, Turelli M, Simmons GM (1986) Unidirectional incompatibility between populations of Drosophila simulans . Evolution 40: 692–701. [DOI] [PubMed] [Google Scholar]
- 5. Bordenstein SR, Werren JH (2007) Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia . Heredity 99: 278–87. [DOI] [PubMed] [Google Scholar]
- 6. Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, et al. (2009) Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog 5: e100368 10.1371/journal.ppat.100368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kremer N, Voronin D, Charif D, Mavingui P, Mollereau B, et al. (2009a) Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog 5: e100630 10.1371/journal.ppat.100630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perlman SJ, Hunter MS, Zchori-Fein E (2006) The emerging diversity of Rickettsia . Proc Biol Sci 273: 2097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suh E, Mercer DR, Fu Y, Dobson SL (2009) Pathogenicity of life-shortening Wolbachia in Aedes albopictus after transfer from Drosophila melanogaster . Appl Environ Microbiol 75 (24) 7783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kambris Z, Cook PE, Phuc HK, Sinkins SP (2009) Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. 2: 134–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McMeniman CJ, Lane RV, Cass BN, Fong AWC, Sidhu M, et al. (2009) Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti . Science 323: 141–4. [DOI] [PubMed] [Google Scholar]
- 12. Hedges LM, Brownlie JC, O'Neill SL, Johnson KN (2008) Wolbachia and virus protection in insects. Science 322: 702 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- 13. Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster . PLoS Biol 6: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bian G, Xu Y, Lu P, Xie Y, Xi Z (2010) The endosymbiotic bacterium Wolbachia pipientis induces resistance to dengue virus in Aedes aegypti . PLoS Pathog 6: e100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bian G, Joshi D, Dong Y, Lu P, Zhou G, et al. (2013) Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 340: 748–51. [DOI] [PubMed] [Google Scholar]
- 16. Koukou K, Pavlikaki H, Kilias G, Werren JH, Bourtzis K, et al. (2006) Influence of antibiotic treatment and Wolbachia curing on sexual isolation among Drosophila melanogaster cage populations. Evolution 60: 87–96. [PubMed] [Google Scholar]
- 17. Miller WJ, Ehrman L, Schneider D (2010) Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum . PLoS Pathog 6: e1001214 10.1371/journal.ppat.1001214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brelsfoard CL, St Clair W, Dobson SL (2009) Integration of irradiation with cytoplasmic incompatibility to facilitate a lymphatic filariasis vector elimination approach. Parasit Vectors 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bourtzis K (2008) Wolbachia-based technologies for insect pest population control. Adv Exp Med Biol 627: 104–13. [DOI] [PubMed] [Google Scholar]
- 20. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139: 1268–78. [DOI] [PubMed] [Google Scholar]
- 21. Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, et al. (2011) Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476: 454–7. [DOI] [PubMed] [Google Scholar]
- 22. Iturbe-Ormaetxe I, Walker T, O' Neill SL (2011) Wolbachia and the biological control of mosquito-borne disease. EMBO Rep 12: 508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, et al. (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476: 450–3. [DOI] [PubMed] [Google Scholar]
- 24. Knipling EF (1955) Possibilities of insect control or eradication through the use of sexually sterile males. J Econ Entomol 48: 459–462. [Google Scholar]
- 25. Helinski ME, Parker AG, Knols BG (2009) Radiation biology of mosquitoes. Malar J 8 Suppl 2: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ben Ami E, Yuval B, Jurkevitch E (2010) Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J 4: 28–37. [DOI] [PubMed] [Google Scholar]
- 27. Poinsot H, Bourtzis K, Markakis G, Savakis C, Merçot H (1998) Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics 150: 227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, et al. (2004) Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci U S A 101: 15042–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riegler M, Charlat S, Stauffer C, Merçot H (2004) Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: investigating the outcomes of host-symbiont coevolution. Appl Environ Microbiol 70: 273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaenike J (2007) Spontaneous emergence of a new Wolbachia phenotype. Evolution 61: 2244–52. [DOI] [PubMed] [Google Scholar]
- 31. Miller WJ, Riegler M (2006) Evolutionary dynamics of wAu-like Wolbachia variants in neotropical Drosophila spp. Appl Environ Microbiol 72: 826–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Symula RE, Alam U, Brelsfoard C, Wu Y, Echodu R, et al. (2013) Wolbachia association with the tsetse fly, Glossina fuscipes fuscipes reveals high levels of genetic diversity and complex evolutionary dynamics. BMC Evol Biol 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arthofer W, Riegler M, Avtzis D, Stauffer C (2009a) Evidence for low-titre infections in insect symbiosis: Wolbachia in the bark beetle Pityogenes chalcographus (Coleoptera, Scolytinae). Environ Microbiol 11: 1923–33. [DOI] [PubMed] [Google Scholar]
- 34. Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA (2007) From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol 5: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoffmann AA, Turelli M, Harshman LG (1990) Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans . Genetics 126: 933–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McMeniman CJ, Lane AM, Fong AW, Voronin DA, Iturbe-Ormaetxe I, et al. (2008) Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl Environ Microbiol 74: 6963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Min KT, Benzer S (1997) Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci U S A 94: 10792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGraw EA, Merritt DJ, Droller JN, O'Neill SL (2002) Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A 99: 2918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arthofer W, Riegler M, Schneider D, Krammer M, Miller WJ, et al. (2009b) Hidden Wolbachia diversity in field populations of the European cherry fruit fly, Rhagoletis cerasi (Diptera, Tephritidae). Mol Ecol 18: 3816–30. [DOI] [PubMed] [Google Scholar]
- 40. Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, et al. (2004) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klasson L, Walker T, Sebaihia M, Sanders MJ, Quail MA, et al. (2008) Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol 25: 1877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klasson L, Westberg J, Sapountzis P, Näslund K, Lutnaes Y, et al. (2009) The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans . Proc Natl Acad Sci U S A 106: 5725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mavingui P, Valiente Moro C, Tran-Van V, Wisniewski-Dyé F, Raquin V, et al. (2012) Whole-genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus . J Bacteriol 194: 1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duplouy A, Iturbe-Ormaetxe I, Beatson SA, Szubert JM, Brownlie JC, et al. (2013) Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellegaard KM, Klasson L, Näslund K, Bourtzis K, Andersson SGE (2013) Comparative Genomics of Wolbachia and the Bacterial Species Concept. PLoS Genet 9 (4) e1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Masui S, Kamoda S, Sasaki T, Ishikawa H (1999) The first detection of the insertion sequence ISW1 in the intracellular reproductive parasite Wolbachia . Plasmid 42: 13–9. [DOI] [PubMed] [Google Scholar]
- 47. Iturbe-Ormaetxe I, Burke GR, Riegler M, O'Neill SL (2005) Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis . J Bacteriol 187: 5136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Riegler M, Sidhu M, Miller WJ, O'Neill SL (2005) Evidence for a global Wolbachia replacement in Drosophila melanogaster . Curr Biol 15: 1428–33. [DOI] [PubMed] [Google Scholar]
- 49. Cordaux R, Pichon S, Ling A, Pérez P, Delaunay C, et al. (2008) Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol Biol Evol 25: 1889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cerveau N, Leclercq S, Leroy E, Bouchon D, Cordaux R (2011) Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol 3: 1175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Riegler M, Stauffer C (2002) Wolbachia infections and superinfections in cytoplasmically incompatible populations of the European cherry fruit fly Rhagoletis cerasi (Diptera, Tephritidae). Mol Ecol 11: 2425–34. [DOI] [PubMed] [Google Scholar]
- 52. Arthofer W, Riegler M, Schuler H, Schneider D, Moder K, et al. (2011) Allele intersection analysis: a novel tool for multi locus sequence assignment in multiply infected hosts. PLoS One 6: e22198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Merçot H, Llorente B, Jacques M, Atlan A, Montchamp-Moreau C (1995) Variability within the Seychelles cytoplasmic incompatibility system in Drosophila simulans . Genetics 141: 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O'Neill SL, Karr TL (1990) Bidirectional incompatibility between conspecific populations of Drosophila simulans . Nature 348: 178–180. [DOI] [PubMed] [Google Scholar]
- 55. Junakovic N (2004) Southern blot analysis of individual Drosophila flies. Methods Mol Biol 260: 41–57. [DOI] [PubMed] [Google Scholar]
- 56. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, et al. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95: 3140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, et al. (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Appl Environ Microbiol 72: 7098–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keohavong P, Thilly WG (1989) Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci 86: 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Starr DJ, Cline TW (2002) A host parasite interaction rescues Drosophila oogenesis defects. Nature 418: 76–9. [DOI] [PubMed] [Google Scholar]
- 60.King RC (1970) Ovarian Development in Drosophila melanogaster. Academic Press, New York. [Google Scholar]
- 61.Korber B (200) HIV Signature and Sequence Variation Analysis. Computational Analysis of HIV Molecular Sequences, Chapter 4, pages 55–72. Allen G. Rodrigo and Gerald H. Learn, eds. Dordrecht, Netherlands: Kluwer Academic Publishers. [Google Scholar]
- 62. Richardson MF, Weinert LA, Welch JJ, Linheiro RS, Magwire MM, et al. (2012) Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster . PLoS Genet 8 (12) e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mahilllon J, Chandler M (1998) Insertion sequences. Microbiol Mol Biol Rev 62: 725–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34: D32–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baldo L, Desjardins CA, Russell JA, Stahlhut JK, Werren JH (2010) Accelerated microevolution in an outer membrane protein (OMP) of the intracellular bacteria Wolbachia . BMC Evol Biol 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T (2002) Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci U S A 99: 1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dunning Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, et al. (2007) Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317: 1753–6. [DOI] [PubMed] [Google Scholar]
- 68. Zabalou S, Apostolaki A, Pattas S, Veneti Z, Paraskevopoulos C, et al. (2008) Multiple resuce factors within a Wolbachia strain. Genetics 178: 2145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Doudoumis V, Tsiamis G, Wamwiri F, Brelsfoard C, Alam U, et al. (2012) Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol 12 Suppl 1: S3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Garen A (1968) Sense and nonsense in the genetic code. Three exceptional triplets can serve as both chain-terminating signals and amino acid codons. Science 160: 149–59. [DOI] [PubMed] [Google Scholar]
- 71. Goodman HM, Abelson J, Landy A, Brenner S, Smith JD (1986) Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature 217: 1019–24. [DOI] [PubMed] [Google Scholar]
- 72. Gilmore RA, Stewart JW, Sherman F (1971) Amino acid replacements resulting from super-suppression of nonsense mutants of iso-1-cytochrome c from yeast. J Mol Biol 61: 157–73. [DOI] [PubMed] [Google Scholar]
- 73. Osawa S, Jukes TH (1989) Codon reassignment (codon capture) in evolution. J Mol Evol 28: 271–8. [DOI] [PubMed] [Google Scholar]
- 74. Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56: 229–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tobiason DM, Seifert HS (2006) The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol 4: e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pecoraro V, Zerulla K, Lange C, Soppa J (2011) Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS One 31;6: e16392 doi: 10.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Van Borm S, Wenseleers T, Billen J, Boomsma JJ (2003) Cloning and sequencing of wsp encoding gene fragments reveals a diversity of co-infecting Wolbachia strains in Acromyrmex leafcutter ants. Mol Phylogenet Evol 26 (1) 102–9. [DOI] [PubMed] [Google Scholar]
- 78. Jamnongluk W, Kittayapong P, Baimai V, O'Neill SL (2002) Wolbachia infections of tephritid fruit flies: molecular evidence for five distinct strains in a single host species. Curr Microbiol 45 (4) 255–60. [DOI] [PubMed] [Google Scholar]
- 79. Augustinos AA, Santos-Garcia D, Dionyssopoulou E, Moreira M, Papapanagiotou A, et al. (2011) Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One 6: e28695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Casper-Lindley C, Kimura S, Saxton DS, Essaw J, Simpson I, et al. (2011) Rapid fluorescent-based method for rapid Wolbachia detection in the Drosophila germline and somatic tissues. Appl Environ Microbiol 77: 4788–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schneider DI, Garschall KI, Parker AG, Abd-Alla AM, Miller WJ (2013) Global Wolbachia prevalence, titer fluctuations and their potential of causing cytoplasmic incompatibilities in tsetse flies and hybrids of Glossina morsitans subgroup species. J Invertebr Pathol 112 Suppl: S104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB (2009) Experimental evolution of bet hedging. Nature 462: 90–3. [DOI] [PubMed] [Google Scholar]
- 83. Cohen D (1966) Optimizing reproduction in a randomly varying environment. J Theor Biol 12: 119–29. [DOI] [PubMed] [Google Scholar]
- 84.Seger J, Brockman HJ (1987) What is bet-hedging? In: Harvey PH, Partridge L eds. Oxford Surveys in Evolutionary Biology. Oxford University Press. Oxford, pp. 182–211. [Google Scholar]
- 85. Mouton L, Henri H, Bouletreau M, Vavre F (2003) Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol Ecol 12 (12) 3459–65. [DOI] [PubMed] [Google Scholar]
- 86. Rousset F, Braig HR, O'Neill SL (1999) A stable triple Wolbachia infection in Drosophila with nearly additive incompatibility effects. Heredity (Edinb) 82: 620–7. [DOI] [PubMed] [Google Scholar]
- 87. Dobson SL, Marsland EJ, Rattanadechakul W (2001) Wolbachia-induced cytoplasmic incompatibility in single- and superinfected Aedes albopictus (Diptera: Culicidae). J Med Entomol 38: 382–7. [DOI] [PubMed] [Google Scholar]
- 88. Suh E, Merver DR, Fu Y, Dobson SL (2009) Pathogenicity of life-shortening Wolbachia in Aedes albopictus after transfer from Drosophila melanogaster . Appl Environ Microbiol 75: 7783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended methodology for RFLP-mapping via genomic Southern blot analysis. Detailed information about RFLP mapping can be found in Data S1.
(DOCX)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOC)
(DOCX)