Summary
Protein modification by O-linked β-N-acetylglucosamine (O-GlcNAc) is a critical cell signaling modality, but identifying signal-specific O-GlcNAcylation events remains a significant experimental challenge. We describe a new method for visualizing and analyzing organelle- and stimulus-specific O-GlcNAcylated proteins, and use it to identify the mitochondrial voltage-dependent anion channel 2 (VDAC2) as an O-GlcNAc substrate. VDAC2−/− cells resist the mitochondrial dysfunction and apoptosis caused by global O-GlcNAc perturbation, demonstrating a functional connection between O-GlcNAc signaling and mitochondrial physiology through VDAC2. More broadly, our method will enable the discovery of signal-specific O-GlcNAcylation events in a wide array of experimental contexts.
Introduction
O-GlcNAcylation of intracellular proteins has emerged recently as a ubiquitous cell signaling modality in a broad range of organisms, influencing such core cell biological processes as transcription, nutrient sensing, and cell cycle progression (Hanover et al., 2010; Hart et al., 2011). Analogous to phosphorylation, O-GlcNAcylation is a dynamic, rapidly cycling post-translational modification that modulates substrate proteins’ location, function or stability in response to physiological signals (Hanover et al., 2010; Hart et al., 2011). Accordingly, it is of great interest to identify which substrate proteins are O-GlcNAcylated in response to a given biological cue. However, this goal poses a substantial experimental challenge, in part because rare, signal-dependent changes in O-GlcNAc are difficult to detect amidst the large number of abundant, constitutively and multiply glycosylated “background” proteins (e.g., nucleoporins). Although chemical (Wells et al., 2002), chemoenzymatic (Khidekel et al., 2003), lectin-(Vosseller et al., 2006) and antibody- (Teo et al., 2010) based methods for identifying O-GlcNAcylated proteins have been described, these approaches all rely on affinity purification to enrich substrates and are therefore inherently biased towards abundant, unchanging “background” O-GlcNAc, at the expense of the comparatively rare, sub-stoichiometric, signal-dependent changes in O-GlcNAc. Therefore, new, complementary methods are needed for identifying functionally relevant changes in protein O-GlcNAcylation.
We have previously described a strategy for identifying O-GlcNAcylated proteins in living mammalian cells using the unnatural azide-functionalized sugar N-azidoacetylgalactosamine (GalNAz) as a metabolic label (Boyce et al., 2011). Briefly, we showed that GalNAz is converted by endogenous metabolic enzymes to the nucleotide-sugar UDP-GalNAz and then epimerized to UDP-N-azidoacetylglucosamine (GlcNAz), the azido analog of natural UDP-GlcNAc (Boyce et al., 2011; Vocadlo et al., 2003). O-GlcNAc transferase (OGT) accepts UDP-GlcNAz as a nucleotide-sugar donor, resulting in the installation of “O-GlcNAz” on native OGT substrates (Figure 1A) (Boyce et al., 2011; Vocadlo et al., 2003). The resulting azide-functionalized (“O-GlcNAzylated”) proteins can then be chemically tagged using any of several classes of azide-reactive probes (Figure 1A) (Boyce and Bertozzi, 2011; Boyce et al., 2011). Importantly, this strategy allows for selective labeling of new O-GlcNAc moieties (i.e., those formed after GalNAz is added to cells), affording time-resolution control not provided by other methods of O-GlcNAc detection. We have previously used affinity probes to purify bulk endogenous, O-GlcNAzylated proteins, revealing numerous known O-GlcNAc substrates and demonstrating that GalNAz is a faithful and robust metabolic reporter of cellular O-GlcNAc (Boyce et al., 2011). Here, we build upon our GalNAz metabolic labeling strategy to create a new proteomics platform for identifying compartment- or stimulus-specific changes in O-GlcNAcylated proteins across different samples, circumventing major, inherent disadvantages of affinity-based approaches. Furthermore, we use our new approach to identify a novel mitochondrial glycoprotein and demonstrate its functional link to O-GlcNAc signaling.
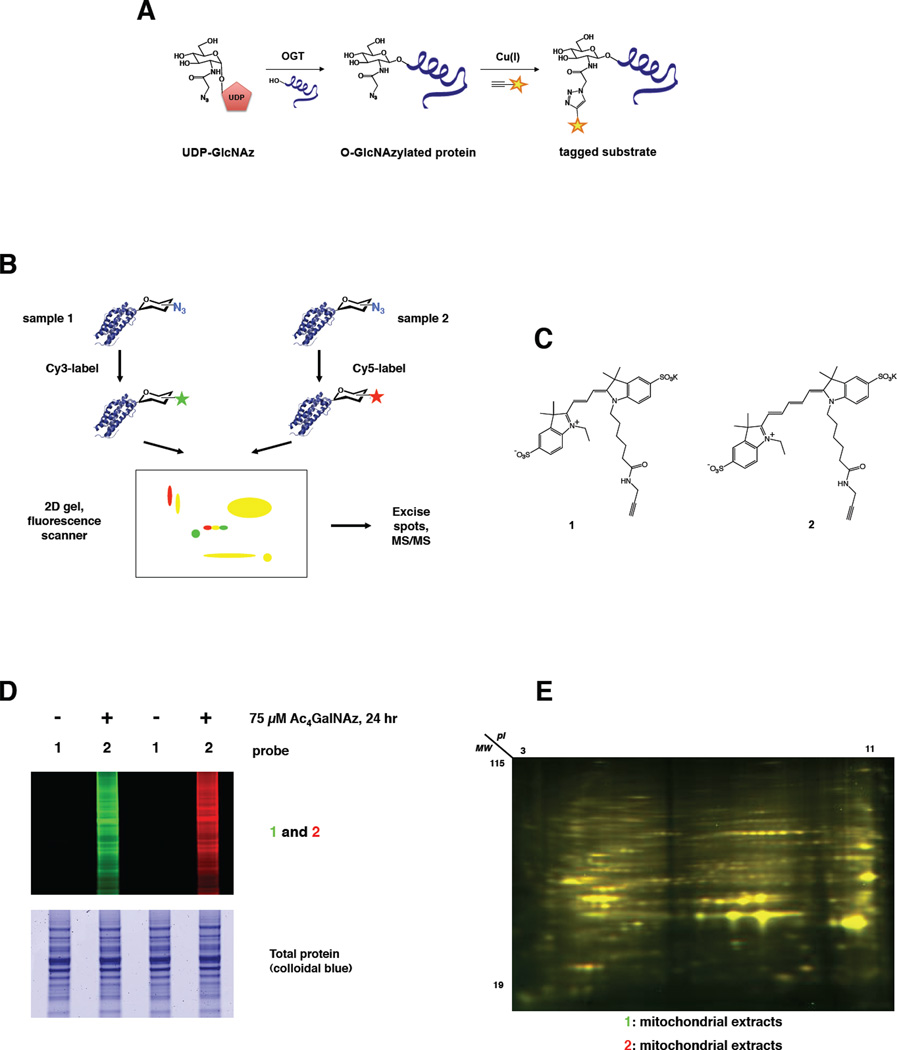
Figure 1. Glyco-DIGE system.
(A) Endogenous O-GlcNAcylated proteins can be metabolically labeled by the conversion of GalNAz to UDP-GlcNAz, as described (Boyce et al., 2011). Such “O-GlcNAzylated” substrates can then be detected by chemical tagging with one of several azide-reactive probes. CuAAC is depicted here. The yellow star represents a useful chemical moiety on the probe molecule, such as a fluorophore or affinity handle. (B) Glyco-DIGE workflow. First, protein extracts are made from two populations of cells (e.g., control and drug-treated) that have been treated with GalNAz to metabolically label O-GlcNAc substrates with an azidoglycan (“O-GlcNAz”), as described (Boyce et al., 2011). The two protein samples are then labeled separately with azide-reactive Cy3- and Cy5-based dyes, mixed and analyzed by 2D gel (horizontal dimension: isoelectric focusing; vertical dimension: SDS-PAGE). When the gel is scanned, protein spots unchanged between samples appear as dual-color overlap (yellow), whereas proteins unique to one sample appear as single color spots (green: Cy3; red: Cy5), thereby permitting ready detection of sample-specific changes in O-GlcNAcylated proteins. (C) Glyco-DIGE alkyne-functionalized fluorescent probes. (D) Glyco-DIGE probes specifically label azidosugar-tagged mammalian proteins. Jurkat cells were metabolically labeled with 100 µM peracetylated GalNAz (Ac4GalNAz) or vehicle only for 24 hours. Whole-cell lysates were reacted with 1 or 2 via CuAAC and analyzed by standard SDS-PAGE and fluorescence scanning. (E) 1 and 2 do not differentially affect protein migration in 2D gels. Jurkat cells were metabolically labeled with 100 µM Ac4GalNAz for 24 hours. Then, mitochondria were purified and extracted into lysis buffer. Half the sample was labeled with 1 and half with 2, and labeled samples were analyzed by glyco-DIGE. 1: Green; 2: Red; 1+2 overlap: Yellow.
Results
Glyco-DIGE permits simultaneous detection and analysis of hundreds of O-GlcNAcylated proteins
To analyze sample-specific changes in O-GlcNAcylated proteins, we turned to difference gel electrophoresis (DIGE) (Minden et al., 2009). In traditional DIGE experiments, each of two protein samples (e.g., from control versus stimulated cells) is covalently labeled with a protein-nonspecific, cyanine-3 (Cy3)- or cyanine-5 (Cy5)-based fluorophore (Minden et al., 2009). Then, the samples are mixed and analyzed by conventional 2D electrophoresis (isoelectric focusing followed by SDS-PAGE). When these 2D gels are imaged for Cy3 and Cy5, proteins that are unchanged between the two samples show perfect overlap of Cy3 and Cy5 fluorescence, whereas the rare proteins that are different between the two samples are visualized as Cy3-only or Cy5-only signal. These sample-specific proteins of interest can be excised from the gel and identified by mass spectrometry.
We envisioned that DIGE methods could be adapted to analyze glycoproteins, including O-GlcNAcylated substrates tagged via our GalNAz label and appropriate azide-reactive Cy3 and Cy5 derivatives, permitting us to visualize and identify sample-specific changes in O-GlcNAcylated proteins (Figures 1A,B). Furthermore, we hypothesized that this “glyco-DIGE” platform would permit the visualization of rare, sample-dependent changes in O-GlcNAcylated proteins, even in experiments where abundant, unchanging background O-GlcNAc is present elsewhere on the same 2D gel. Importantly, such a glyco-DIGE approach could detect any alteration in an O-GlcNAcylated protein that affected its mobility on a 2D gel, including de novo (de)glycosylation and other changes (e.g., cleavage, ubiquitination), providing broad information on the role of O-GlcNAcylated proteins in dynamic cell signaling.
To test the utility of a glyco-DIGE approach, we synthesized alkyne-functionalized Cy3 (1) and Cy5 (2) (Figure 1C). As expected, 1 and 2 reacted specifically with O-GlcNAcylated proteins from Jurkat cells metabolically labeled with GalNAz via the well-characterized, bioorthogonal copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction (Nwe and Brechbiel, 2009) (Figure 1D). Importantly, while hundreds of 1- and 2-labeled protein spots could be separated and visualized efficiently on the same glyco-DIGE gel, labeling by 1 or 2 did not differentially affect the migration of proteins in a glyco-DIGE experiment (Figure 1E). We concluded that glyco-DIGE can label and compare large numbers of O-GlcNAcylated proteins across different mammalian samples.
Glyco-DIGE identifies VDAC2 as a mitochondrial O-GlcNAc substrate
We next asked whether glyco-DIGE could detect sample-specific differences in O-GlcNAcylated proteins. As a proof of principle experiment, we examined differences in protein glycosylation between mitochondria and cytosol. Although O-GlcNAc is a well-known sentinel of cellular glucose levels (Hanover et al., 2010; Hart et al., 2011) and regulates both metabolic (Dentin et al., 2008; Yang et al., 2008) and mitochondrial (Hu et al., 2009; Wang and Schwarz, 2009) pathways, the mitochondrial glycoproteome has not been analyzed systematically. We used glyco-DIGE to compare O-GlcNAcylated proteins from mitochondrial and cytosolic extracts (Figure S1) (Frezza et al., 2007) of the same human cell line. As expected, we detected numerous differences in the respective glycoproteomes of these two compartments (Figure 2A). Across many experiments and multiple cell types, we noticed one set of especially prominent mitochondrial O-GlcNAcylated protein spots (Figure 2A, arrows). Using fluorescence as a guide, we excised the corresponding spots from preparative gels and identified the voltage-dependent anion channel 2 (VDAC2) protein as the major component (Figure S2).
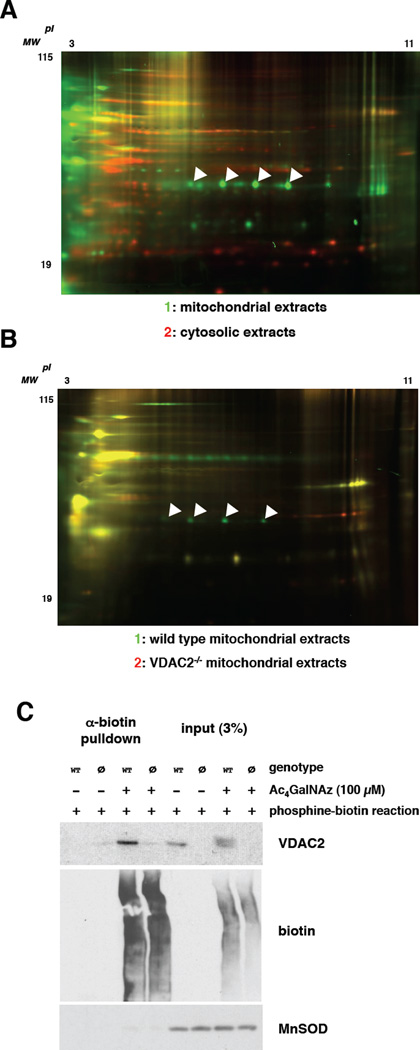
Figure 2. Glyco-DIGE identifies VDAC2 as a mitochondrial glycoprotein.
(A) Jurkat cells were metabolically labeled with 100 µM Ac4GalNAz for 24 hours, and mitochondrial and cytosolic extracts were prepared. Mitochondrial extracts were reacted with 1 (green), and cytosolic extracts with 2 (red). Then, the samples were mixed and analyzed by glyco-DIGE. 1+2 overlap: yellow. Arrows indicate VDAC2 spots. The characteristic “charge train” pattern of VDAC2 spots likely reflects the presence of multiple phosphorylated forms of the protein. (B) Wild type and VDAC2−/− MEFs were metabolically labeled with 100 µM Ac4GalNAz for 24 hours. Mitochondrial extracts were prepared and labeled with 1 (wild type, green) or 2 (VDAC2−/−, red) and then analyzed by glyco-DIGE. Arrows indicate VDAC2 spots. (C) Wild type (“WT”) and VDAC2−/− (“ø”) MEFs were metabolically labeled with 100 µM Ac4GalNAz or vehicle only for 24 hours and mitochondrial extracts were prepared and reacted with phosphine-biotin. Then, biotin-tagged proteins were affinity-purified essentially as described (Boyce et al., 2011) and analyzed by immunoblot. Left: Affinity-purified material. Right: 3% total input of material (loading control). MnSOD serves as a loading control for total mitochondrial protein and demonstrates the removal of unglycosylated proteins during affinity purification.
VDAC2 is a member of a family of multipass channel proteins residing in the mitochondrial outer membrane, with important roles in organelle metabolite flux, nutrient metabolism and apoptotic signaling (Cheng et al., 2003; Ren et al., 2009; Shoshan-Barmatz et al., 2010). While some work has suggested that VDAC family proteins might be glycosylated (Jones et al., 2008), VDAC2 had not been described or validated as a specific O-GlcNAc substrate. We performed two experiments to confirm our glyco-DIGE results with VDAC2. First, we compared mitochondrial extracts from wild type and VDAC2−/− mouse embryonic fibroblasts (MEFs) (Cheng et al., 2003) in a glyco-DIGE experiment (Figure 2B). As expected, we found that spots corresponding to the ones identified in human cells were present in wild type MEF mitochondrial samples but absent from the VDAC2−/− samples, indicating that these spots are VDAC2 (Figure 2B). Furthermore, these fluorescent spots correlated with anti-VDAC2 immunoreactivity on a 2D immunoblot of wild type mitochondrial extracts (Figure S3). Second, we used an affinity approach (Boyce et al., 2011) to confirm our glyco-DIGE results with VDAC2. We labeled wild type or VDAC2−/− MEFs with GalNAz, made mitochondrial extracts and reacted them with phosphine-biotin to tag azide-bearing proteins. Then, we enriched for GalNAz-labeled proteins via anti-biotin affinity chromatography. As expected, anti-VDAC2 immunoblotting showed that VDAC2 was affinity-purified only from wild type mitochondrial samples from cells labeled with GalNAz (Figure 2C), demonstrating the specificity of GalNAz labeling of VDAC2. As further confirmation, we analyzed similar biotin affinity-purified samples by mass spectrometry and detected enrichment of VDAC2 in mitochondrial extracts from GalNAz-treated, but not vehicle-treated, wild type MEFs (Figure S4). Taken together, these results indicate that VDAC2 is an O-GlcNAcylated mitochondrial protein in human and mouse cells.
Loss of VDAC2 protects cells from mitochondrial dysfunction and apoptosis following global perturbation of O-GlcNAcylation
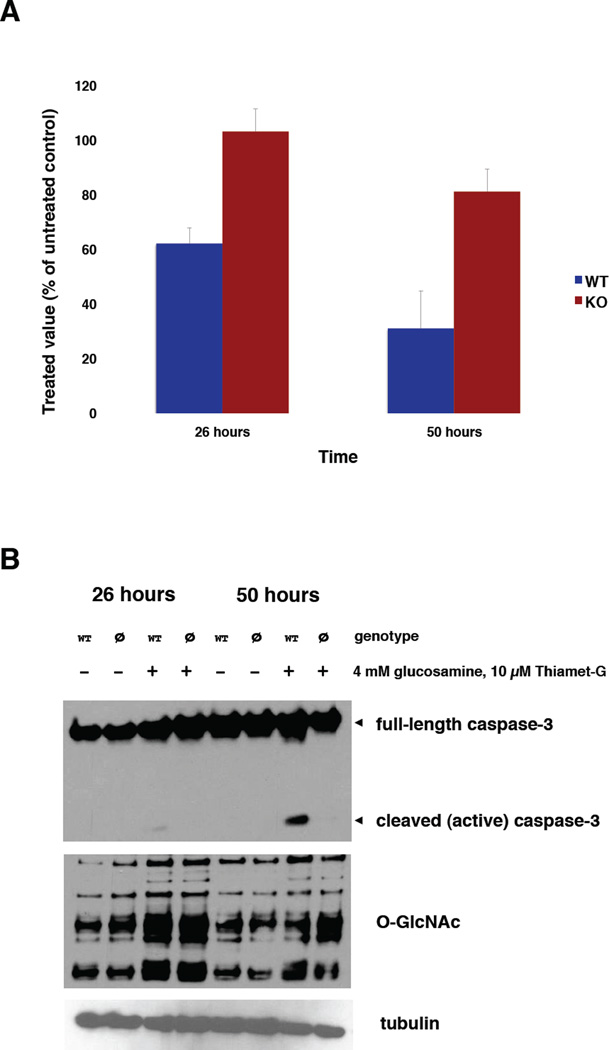
Intriguingly, VDAC2 (Cheng et al., 2003; Ren et al., 2009; Shoshan-Barmatz et al., 2010) and O-GlcNAc (Hu et al., 2009; Wang and Schwarz, 2009) are both critical regulators of mitochondrial metabolism and cell death, but no experimental evidence had established a functional connection between them. Given our finding that VDAC2 is a glycoprotein, we asked whether a phenotypic relationship existed between O-GlcNAc signaling and VDAC2. We found that potentiating global O-GlcNAc levels using a combination of glucosamine, to increase levels of UDP-GlcNAc (Vosseller et al., 2002), and Thiamet-G, a specific small molecule inhibitor of the glycoside hydrolase O-GlcNAcase (OGA) (Yuzwa et al., 2008) resulted in mitochondrial dysfunction and apoptosis in MEFs (Figure 3). Consistent with previous reports (Gloster et al., 2011; Slawson et al., 2005), both wild type and VDAC2−/− MEFs altered OGA and OGT protein levels to compensate for the pharmacological perturbation of O-GlcNAc, especially at later time points (Figures 3B and S5). Nevertheless, we found that VDAC2−/− MEFs were resistant to the mitochondrial dysfunction (Figure 3A) and apoptosis (Figure 3B) caused by Thiamet-G and glucosamine. Furthermore, this difference was caused by loss of VDAC2 per se and not an adventitious or irreversible downstream effect of gene deletion, because stable re-expression of VDAC2 in VDAC2−/− MEFs restored sensitivity to Thiamet-G/glucosamine treatment (Figure S6). We concluded that global O-GlcNAc perturbation induces mitochondrial dysfunction and apoptosis, and that VDAC2 is required for these effects, demonstrating a critical functional connection between VDAC2 and O-GlcNAc signaling in organelle physiology and cell death.
Figure 3. Loss of VDAC2 protects cells from mitochondrial dysfunction and cell death induced by global O-GlcNAc perturbation.
(A) Wild type or VDAC2−/− cells were treated with 10 µM Thiamet-G and 4 mM glucosamine or vehicle control for 26 or 50 hours and analyzed by MTS assay to measure mitochondrially produced reducing equivalents (i.e., NADH and NADPH). Treated samples were normalized to their corresponding vehicle-only controls. Error bars represent standard deviations. (B) Wild type (“WT”) or VDAC2−/− (“ø”) cells were treated as in (A), and whole-cell lysates were prepared and analyzed by immunoblot as indicated.
Discussion
We have developed a new proteomics platform, termed glyco-DIGE, which can detect differences in O-GlcNAcylated proteins across samples. Importantly, the combination of GalNAz labeling and glyco-DIGE permits time-resolved examination of new cellular O-GlcNAc and allows the detection of rare, sub-stoichiometric, sample-dependent changes in O-GlcNAcylated substrates, without incurring the bias towards abundant, unchanging, multiply glycosylated “background” substrates that is endemic to affinity-based analysis of O-GlcNAc. Therefore, while affinity-based approaches will remain valuable for the bulk identification of O-GlcNAc substrates, we anticipate that GalNAz labeling and glyco-DIGE will provide an important complementary method to examine subtle, functionally relevant changes in O-GlcNAc. In a proof of principle experiment, we used glyco-DIGE to perform the first unbiased search for mitochondria-specific O-GlcNAcylated proteins (Figure 2). We expect that our method will also lend itself to numerous applications for examining signal-dependent changes in O-GlcNAc. For example, in preliminary experiments, we used glyco-DIGE to examine differences in nuclear O-GlcNAcylated proteins in control versus DNA-damaged cells, and found genotoxic stress-specific changes conserved across disparate human cell lines (Figure S7). The identification of these and other signal-specific glyco-DIGE protein spots is ongoing. More generally, we note that conventional DIGE methods can resolve ~2500 cyanine-labeled protein spots from samples of 1,000–10,000 mammalian cells (Meyer and Stuhler, 2007; Minden et al., 2009), and we anticipate that glyco-DIGE capabilities will be comparable. In addition, glyco-DIGE experiments can be easily tailored to specific pI and molecular weight (MW) ranges (narrow or broad, as appropriate) using commercial reagents, and glyco-DIGE is compatible with a wide variety of biochemical procedures for sample pre-fractionation. Therefore, glyco-DIGE will be a useful in analyzing even more complex biological samples in the future.
In the current work, we used glyco-DIGE to identify VDAC2 as a novel mitochondrial O-GlcNAc substrate and showed that the mitochondrial dysfunction and cell death caused by global O-GlcNAc perturbation depend on VDAC2. Interestingly, while VDAC2 has a previously described role in apoptosis, its loss was shown to sensitize cells to such stimuli as protein kinase inhibition, genotoxic stress (Cheng et al., 2003) and T cell receptor ligation (Ren et al., 2009), whereas we find that loss of VDAC2 protects cells from global O-GlcNAc perturbation. Given the well-known roles of O-GlcNAc and VDAC2 in apoptotic signaling and cell metabolism, it will be interesting to dissect the molecular explanation for VDAC2’s various pro- and anti-apoptotic effects in response to distinct stimuli, and determine whether these phenotypes are controlled directly by O-GlcNAcylation of VDAC2 itself, and/or by other mechanisms. To this end, we are working to identify the glycosylation site(s) on VDAC2 and will use unglycosylatable mutants to address these questions in future work. Beyond mitochondrial O-GlcNAc, we believe glyco-DIGE will serve as a useful new tool for analyzing rare, signal-dependent changes in O-GlcNAcylated proteins, providing a powerful complement to existing affinity-based approaches for studying O-GlcNAc signaling in a wide range of experimental contexts.
Experimental Procedures
Compound synthesis and small molecule reagents
Glyco-DIGE probes 1 and 2 were synthesized from propargyl amine and N-hydroxysuccinamide esters of Cy3 and Cy5, respectively (GE Healthcare) and purified by reversed-phase high-performance liquid chromatography. See Supplemental Information for details. Syntheses and purification of peracetylated GalNAz (Laughlin and Bertozzi, 2007), phosphine-biotin (Saxon and Bertozzi, 2000) and Thiamet-G (Yuzwa et al., 2008) were performed essentially as described. Other chemical reagents were obtained from Sigma-Aldrich unless otherwise indicated.
Cell culture, treatment and protein sample preparation
Cells were maintained in a 5% CO2, humidified atmosphere at 37 °C in medium (Jurkat: RMPI 1640; MEFs, HT1080, 293T: Dulbecco’s modified Eagle’s Medium) plus 10% fetal bovine serum and penicillin/streptomycin (Invitrogen). 100 mM DMSO stock solution of peracetylated GalNAz was added to achieve final treatment conditions.
For whole-cell lysates, cells were treated as indicated, washed in cold phosphate-buffered saline (PBS) and resuspended in Buffer L (1% Triton, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4) plus protease inhibitor cocktail III (Calbiochem). Lysates were probe-sonicated (Misonix) on ice for one minute and cleared by centrifugation.
For subcellular fractionation, mitochondria were prepared as described (Frezza et al., 2007) and mitochondrial protein extracts were made from purified organelles as outlined above. For cytosolic protein extracts, supernatant from the final spin of the mitochondrial preparation was cleared by high-speed centrifugation and brought to Buffer L reagent concentrations. Highly purified nuclear extracts were prepared as described (Boyce et al., 2011). Where indicated (Figure S1), nuclear/cytoplasmic fractions were prepared by recombining the nuclear and cytoplasmic material after isolation of mitochondria. The quality of subcellular fractionations was verified by immunoblot for known organelle marker proteins (Figures 2C and S1).
For DNA damage experiments, cells were treated with 100 µM GalNAz and either 1 µg/ml doxorubicin or vehicle control (DMSO) for 24 hours. Then, nuclear extracts were made and analyzed pairwise (i.e., with versus without doxorubicin) in glyco-DIGE experiments as described below.
In all cases, protein concentrations were quantified by bicinchoninic acid assay (Thermo-Fisher) and all samples were normalized with buffer to the same total protein concentration. Where indicated, equal protein loading was verified by control immunoblot (e.g., tubulin, MnSOD) or colloidal blue stain (Invitrogen).
Bioorthogonal reactions of azides
For CuAAC reactions, protein extracts were mixed with 25 µM 1 or 2 as indicated, 5 mM sodium ascorbate, 100 µM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) and 1 mM CuSO4 (final concentrations) from the following stock solutions: 10 mM 1 or 2 (in water); 167 mM sodium ascorbate (in water, prepared fresh); 30 mM TBTA (in DMSO); 50 mM CuSO4 (in water). Reactions were incubated in the dark with end-over-end inversion for one hour and then stopped by addition of 10 mM (final) EDTA.
For Staudinger ligation reactions, protein extracts were mixed with 500 µM phosphine-biotin (final concentration, from 5 mM stock in 30% dimethylformamide in PBS) and incubated overnight with end-over-end inversion. NeutrAvidin anti-biotin affinity chromatography was performed as described (Boyce et al., 2011).
Glyco-DIGE sample analysis and spot picking
To perform a glyco-DIGE experiment, equal protein amounts of 1- and 2-labeled samples were combined, and excess, unreacted alkyne probe was removed by exchange into Buffer L + 10 mM EDTA using Bio-Spin P-6 columns (Bio-Rad). Labeled protein was precipitated and washed using the 2D Clean-Up Kit (GE Healthcare) and dissolved in DeStreak Rehydration Solution (GE Healthcare) with 20 mM dithiothreitol (DTT) and 0.5% IPG Buffer pH 3–10 NL (GE Healthcare).
For the isoelectric focusing (IEF) dimension, samples (200 µg – 1 mg of protein) were rehydration-loaded onto 11 cm pH 3–11 NL Immobiline DryStrip IEF strips (GE Healthcare) using an Ettan IPGphor 3 IEF unit according to the manufacturer’s instructions. For the second (i.e., SDS-PAGE) dimension, IEF strips were equilibrated by incubation in SDS equilibration buffer (6M urea, 75 mM Tris-HCl pH 8.8, 29.3% glycerol, 2% SDS, bromophenol blue) with 10 mg/ml DTT for 15 minutes, followed by incubation in SDS equilibration buffer with 25 mg/ml iodoacetamide for 15 minutes. Equilibrated strips were laid over precast 10% or 4–20% gradient Criterion IEF SDS-PAGE gels (Bio-Rad) and sealed into place using 1% agarose and bromophenol blue in 1× MES buffer (Bio-Rad). Standard SDS-PAGE was then performed using the Criterion gel system (Bio-Rad).
Finished glyco-DIGE gels were washed briefly in ultrapure water and immobilized by sealing with 1% agarose in water onto a custom-built low-fluorescence glass stage marked with fluorescent stickers (GE Healthcare). Immobilized gels were scanned on a Typhoon flatbed gel scanner (GE Healthcare) using the factory pre-set excitation and emission settings for Cy3 and Cy5 and DIGE data acquisition mode. Data were analyzed using ImageQuant TL software (GE Healthcare). Fluorescent protein spots of interest were identified by inspection and marked via the ImageQuant 2D gel analysis tool to make a pick list, using the fluorescent guide stickers as reference coordinates. Gel coordinates of spots on the pick list were exported to Excel (Microsoft) and converted to a text file using an in-house macro. Spots from the pick list were excised from the gel using an Ettan Spot Picker robot (GE Healthcare) according to the manufacturer’s instructions into 96-well plates. Gel picks from the same spot were pooled by hand for proteomic analysis.
Immunoblotting
Immunoblotting was performed essentially as described (Boyce et al., 2011). See Supplemental Information for details.
Mass spectrometric analysis
Samples were subjected to reversed-phase chromatography with an Agilent 1200 liquid chromatography system connected in-line to either an LTQ XL mass spectrometer or an LTQ Orbitrap XL hybrid mass spectrometer, and processed using customized data-dependent acquisition methods. Protein identities were obtained using the SEQUEST search algorithm within Proteome Discoverer 1.3 (Thermo-Fisher). See Supplemental Information for details.
Assay of mitochondrial function
Wild type and VDAC2−/− MEFs were plated in 96-well plates (5000 cells/well) in phenol red-free medium (Invitrogen). Cells were treated as indicated and analyzed by Cell Titer Aqueous One Solution Cell Proliferation Assay (to measure reducing equivalents, NADH and NADPH) or Caspase-Glo 3/7 Assay (to measure apoptotic protease activity) (Promega) according to the manufacturer’s instructions. Results were read using a Molecular Devices SpectraMax automated plate reader using the appropriate factory pre-set parameters.
Supplementary Material
Highlights.
Glyco-DIGE identifies changes in O-GlcNAc substrates between two samples
Glyco-DIGE demonstrates that VDAC2 is a mitochondrial glycoprotein
Genetic ablation of VDAC2 protects cells from global O-GlcNAc perturbation
Acknowledgements
We thank J. Jewett for reagent synthesis and S. Siegrist and members of the Bertozzi lab for helpful comments. This work was supported by NIH grant GM066047 and the US Department of Defense Grant W81XWH-09-1-0627 to C.R.B. M.B. is a former Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation and a current Rita Allen Foundation Scholar and a Sydney Kimmel Foundation for Cancer Research Kimmel Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyce M, Bertozzi CR. Bringing chemistry to life. Nature Methods. 2011;8:638–642. doi: 10.1038/nmeth.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nature Protocols. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nature Chemical Biology. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochimica et Biophysica Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Suarez J, Fricovsky E, Wang H, Scott BT, Trauger SA, Han W, Hu Y, Oyeleye MO, Dillmann WH. Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. The Journal of Biological Chemistry. 2009;284:547–555. doi: 10.1074/jbc.M808518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marban E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. Journal of the American Chemical Society. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nature Protocols. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- Meyer HE, Stuhler K. High-performance proteomics as a tool in biomarker discovery. Proteomics. 2007;7(Suppl 1):18–26. doi: 10.1002/pmic.200700183. [DOI] [PubMed] [Google Scholar]
- Minden JS, Dowd SR, Meyer HE, Stuhler K. Difference gel electrophoresis. Electrophoresis. 2009;30(Suppl 1):S156–S161. doi: 10.1002/elps.200900098. [DOI] [PubMed] [Google Scholar]
- Nwe K, Brechbiel MW. Growing applications of "click chemistry" for bioconjugation in contemporary biomedical research. Cancer Biother Radiopharm. 2009;24:289–302. doi: 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Kim H, Tu HC, Westergard TD, Fisher JK, Rubens JA, Korsmeyer SJ, Hsieh JJ, Cheng EH. The VDAC2-BAK rheostat controls thymocyte survival. Sci Signal. 2009;2:ra48. doi: 10.1126/scisignal.2000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. The Journal of Biological Chemistry. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nature Chemical Biology. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nature Chemical Biology. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.