Abstract
Backgrounds
The activation of Toll-like receptors (TLRs) may be an important event in the immune evasion of tumor cell. Recently, numerous studies have investigated the associations between TLR2 −196 to −174 del and two SNPs of TLR4 (rs4986790 and rs4986791) and the susceptibility to different types of cancer; however, the results remain conflicting. The aim of this study was to assess the association between TLR2 and TLR4 polymorphisms and cancer risk in a meta-analysis with eligible published studies.
Methodology/Principle Findings
A dataset composed of 14627 cases and 17438 controls from 34 publications were included in a meta-analysis to evaluate the association between overall cancer risk or cancer-specific risk and three SNPs of TLRs (TLR2 −196 to −174 del, TLR4 rs4986790 and rs4986791). The results showed that all of these three polymorphisms were significantly associated with the increased cancer risk (dominant model: OR = 1.64, 95% CI: 1.04–2.60 for TLR2 −196 to −174 del; OR = 1.19, 95% CI: 1.01–1.41 for TLR4 rs4986790; and OR = 1.47, 95% CI: 1.120–1.80 for TLR4 rs4986791; respectively). In stratified analysis, we found the effect of TLR2 −196 to −174 del on cancer risk remained significant in the subgroup of Caucasians and South Asians, but not in East Asians. However, the association between rs4986791 and cancer risk was significant in both South Asians and East Asians, but not in Caucasians. Furthermore, the association between rs4986790 and cancer risk was statistically significant in digestive cancers (dominant model: OR = 1.76, 95% CI: 1.13–2.73) and female-specific cancers (dominant model: OR = 1.50, 95% CI: 1.16–1.94). However, no significant association with risk of digestive system cancers was observed for TLR2 −196 to −174 del and TLR4 rs4986791.
Conclusions/Significance
This meta-analysis presented additional evidence for the association between TLR2 and TLR4 polymorphisms and cancer risk. Further well-designed investigations with large sample sizes are required to confirm this conclusion.
Introduction
Toll-like receptors (TLRs) are a family of membrane-spanning innate immune receptors that recognize ligands derived from bacteria, fungi, viruses, and parasite [1]. TLRs play a key role in the realization of innate and adaptive immune response, being involved in the regulation of inflammatory reactions and activation of the adaptive immune response to eliminate infectious pathogens and cancer debris [2], [3]. Besides driving inflammatory responses, TLRs also regulate cell proliferation and survival by expanding useful immune cells and integrating inflammatory responses and tissue repair processes [4]. Furthermore, functional TLRs are expressed not only in immune cells, but also in cancer cells, thus implicating a role of TLRs in tumor biology [5], [6]. Increasing bodies of evidence have suggested that TLRs can act as a double-edged sword in cancer cells because uncontrolled TLR signaling provides a microenvironment that is necessary for tumor cells to proliferate and evade the immune response [4], [7]. In addition, activation of TLRs not only leads to the up-regulation of cellular defense mechanisms, but also results in up-regulation of DNA repair genes and increased functional DNA repair [8], [9].
The TLR family includes 2 subgroups, extracellular and intracellular, depending on their cellular localization. TLR1, 2, 5, 6 and 10 are extracellular TLRs, which are largely localized on the cell surface. Conversely, TLR3, 7, 8 and 9 (intracellular TLRs) are localized in intracellular organelles. The subcellular localization of TLR4 is unique because it is localized to both the plasma membrane and endosomal vesicles [10]. TLR2 and TLR4 are major TLRs and have been actively investigated in inflammation and cancer. There is evidence that TLRs, particularly TLR2 and TLR4, directly regulate major proinflammatory and host defense functions of human neutrophils [11]. Additionally, TLR2 recognizes microbial pathogen-associated molecular patterns, such as cell wall peptidoglycan and lipoteichoic acid [12]. Positive TLR2 expression in the tumor microenvironment suggests that immune surveillance is activated against the altered epithelial cells, whereas TLR2 expression by malignant keratinocytes may be indicative of resistance to apoptosis as a prosurvival mechanism [13]. TLR4 ligation on tumor cells can enhance the secretion of immunosuppressive cytokines and induce resistance to TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis [14], [15]. Studies have shown that lipopolysaccharide (LPS) ligation to TLR4 promotes tumor cell adhesion and invasion in a murine model by acting NF-kappa B [16], and the silencing of TLR4 increases tumor progression and metastasis in a murine model of breast cancer [17].
Genetic studies have identified a polymorphism of TLR2 that causes a 22-bp nucleotide deletion (−196 to −174 del) in the promoter region, which may influence the promoter activity of TLR2 and lead to the decreased transcription of TLR2 gene. Additionally, two SNPs in TLR4 have also been identified; one is an A→G substitution at 896 base pair (bp) which results in an aspartic acid to glycine replacement at the codon 299 (D299G, rs4986790) and the other is a C→T substitution at 1196 bp which results in a threonine to isoleucine exchange at codon 399 (T399I, rs4986791). It has been shown that these two polymorphisms (rs4986790 and rs4986791) affect the extracellular domain of the receptor and may cause decreased ligand recognition [18]. The associations of these three polymorphisms with cancer risk have been widely studied, including bladder cancer [19], [20], breast cancer [21], [22], gastric cancer [23]–[31], prostate cancer [32]–[37], hepatocellular cancer [38], [39], gallbladder cancer [40], cervical cancer [41], nasopharyngeal cancer [42], leukemia [43], melanoma [44], endometrial cancer [45], lymphoma [46]–[50], esophageal cancer [31] and colorectal cancer [51], [52]. However, the results remained inconsistent rather than conclusive.
Considering the relatively small sample size in each single study might have low power to detect the effect of the polymorphisms on cancer risk and the underlying heterogeneity among different studies need be explored, we conducted a meta-analysis on all eligible published case-control studies to establish a relatively comprehensive picture of the relationship between these genetic variants (−196 to −174 del in TLR2, rs4986790 and rs4986791 in TLR4) and cancer risk.
Materials and Methods
Selection Criteria and Identification of Eligible Studies
Candidate studies were identified through computer-aided literature searching in PubMed for relevant articles in English and Chinese (last search was in January, 2013). The following keywords were used for this search: ‘TLR2 or Toll like receptor 2′ or ‘TLR4 or Toll like receptor 4′ and ‘cancer’ and ‘polymorphism’. We also included additional studies by a hands-on search of references of original studies. Abstracts, case-only articles, editorials, review articles and repeated literatures were excluded. The inclusion criteria of studies in the current meta-analysis were defined as follows: (1) original papers containing independent data; (2) case-control design on the association of TLR2 (−196 to −174 del) or TLR4 (rs4986790 and rs4986791) polymorphisms and cancer risk; (3) providing sufficient information to calculate the odds ratio (OR) or P-value; (4) written in English or Chinese.
Data Extraction
Two investigators (Zhu LB and Jiang T) independently extracted data and reached a consensus on all items. For each study, the following information was extracted: first author, publication date, country, ethnicity, total number of cases and controls, the numbers of cases and controls grouped by different genotypes and Hardy-Weinberg equilibrium test in control subjects.
Statistical Analysis
The crude odds ratios (ORs) and 95% confidence intervals (95% CIs) of TLR2 (−196 to −174 del) and TLR4 (rs4986790 and rs4986791) polymorphisms and cancer risk were estimated for each study. In addition, we also performed stratification analyses by cancer types and races. Digestive system included gastric, esophageal, colorectal, gallbladder and hepatocellular cancer; blood system included leukemia and lymphoma; female-specific included endometrial, breast and cervical cancer; male-specific included prostate cancer. If one cancer type contained less than three individual studies, they were combined into the ‘other’ group. All subjects were categorized as Caucasian, East Asian (China and Japan), South Asian (India) and mixed. The pooled ORs were performed by allele comparisons and genetic models comparisons. The HWE was assessed via χ2 test. A Chi-square based Q test and I2-statistic test were performed to assess the potential heterogeneity among the studies [53]. If the result of heterogeneity test was p>0.05, ORs were pooled according to the fixed-effect model [54]. Otherwise, the random-effect model was used [55]. The significance of the pooled ORs was determined by the Z-test. The sensitivity analysis was carried out to test the stability of the pooled effect by excluding each study individually and recalculating the ORs and 95% CI. To further explore the potential sources of heterogeneity among studies, meta regression was performed with some study characteristics, including ethnicity, genotyping methods, tumor types, sample size(≥500 or <500), minor allele frequency (MAF) in control subjects, and source of controls (population-based or hospital-based). Additionally, the inverted funnel plots and Begg’s funnel plot were used to evaluate publication bias [56]. The statistical analyses were performed by STATA 12.0 software. All P values were two-sided.
Results
Characteristics of Studies
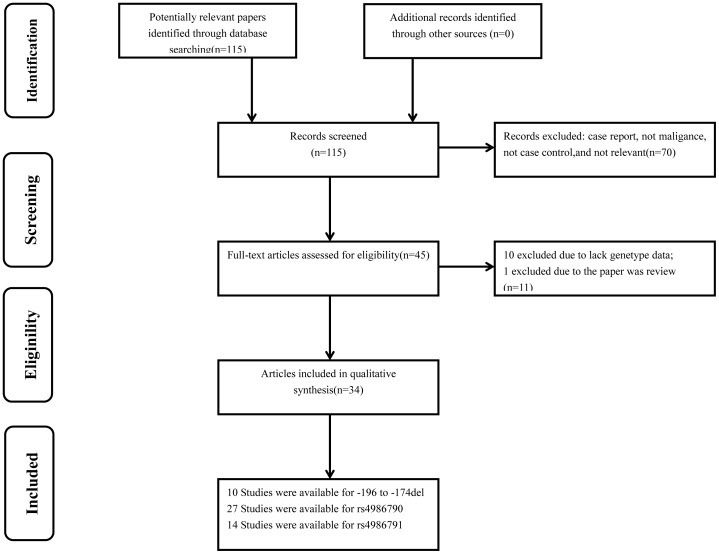
115 articles were initially identified. Among them, 70 papers did not meet our criteria and were excluded. After reading the full texts of the remaining 45 papers, we found 10 papers had not enough genotype data and 1 paper was a review. Therefore, a total of 34 publications including 51 studies were remained (Figure 1). All studies were of case-control design, including fourteen kinds of cancers. Among them, 10 case-control studies focused on TLR2 −196 to −174 del (2521 cases and 3247 controls), 27 on TLR4 rs4986790 (9743 cases and 10839 controls), and 14 on TLR4 rs4986791 (2363 cases and 3352 controls). Moreover, three publications focused on all three SNPs, ten publications focused on two SNPs, and twenty-one publications focused on only one SNP of all. The detailed characteristics of these studies, including first author, year of publication, country, ethnicity, cancer type, numbers of cases and controls, minor allele frequency (MAF) and HWE for all studies were summarized in Table 1. The distribution of genotypes in the controls of the studies was all in agreement with HWE except for four studies [20], [35], [40], [42].
Figure 1. Study selection process.
Table 1. Characteristics of literatures included in the meta-analysis.
| Reference | Year | Country | Ethnicity | Cancer type | Genotype-case | Genotype-control | MAF | HWE | ||||
| TLR2 −196 to −174 del | ins/ins | ins/del | del/del | ins/ins | ins/del | del/del | ||||||
| Singh V17 | 2012 | India | South Asian | Bladder | 110 | 79 | 11 | 119 | 73 | 8 | 0.223 | 0.437 |
| Theodoropoulos GE19 | 2012 | Greece | Caucasian | Breast | 120 | 113 | 28 | 432 | 46 | 2 | 0.052 | 0.518 |
| de Oliveira JG21 | 2012 | Brazil | Caucasian | Gastric | 116 | 50 | 8 | 189 | 34 | 2 | 0.084 | 0.733 |
| Mandal RK30 | 2012 | India | South Asian | Prostate | 135 | 54 | 6 | 193 | 52 | 5 | 0.124 | 0.500 |
| Zeng HM22 | 2011 | China | East Asian | Gastric | 119 | 110 | 19 | 187 | 246 | 63 | 0.375 | 0.195 |
| Nischalke HD36 | 2011 | Germany | Caucasian | Hepatocellular | 115 | 63 | 11 | 248 | 92 | 7 | 0.153 | 0.649 |
| Hishida A23 | 2010 | Japan | East Asian | Gastric | 243 | 267 | 73 | 304 | 316 | 79 | 0.339 | 0.819 |
| Srivastava K38 | 2010 | India | South Asian | Gallbladder | 132 | 94 | 6 | 163 | 87 | 4 | 0.187 | 0.044 |
| Pandey S39 | 2009 | India | South Asian | Cervical | 102 | 43 | 5 | 114 | 35 | 1 | 0.123 | 0.333 |
| Tahara T28 | 2007 | Japan | East Asian | Gastric | 126 | 112 | 51 | 73 | 65 | 8 | 0.277 | 0.182 |
| TLR4 rs4986790 | AA | AG | GG | AA | AG | GG | ||||||
| Theodoropoulos GE19 | 2012 | Greece | Caucasian | Breast | 201 | 57 | 3 | 412 | 63 | 5 | 0.076 | 0.148 |
| de Oliveira JG21 | 2012 | Brazil | Caucasian | Gastric | 154 | 20 | 0 | 215 | 10 | 0 | 0.022 | 0.773 |
| Yang ZH40 | 2012 | China | East Asian | Nasopharyngeal | 205 | 29 | 2 | 250 | 33 | 4 | 0.071 | 0.024 |
| Shen Y18 | 2012 | China | East Asian | Bladder | 431 | 2 | 3 | 519 | 1 | 2 | 0.005 | 0.000 |
| Miedema KG41 | 2011 | Netherlands | Caucasian | Leukemia | 168 | 20 | 0 | 151 | 28 | 0 | 0.078 | 0.256 |
| Gast A42 | 2011 | Germany | Caucasian | Malignant Melanoma | 665 | 91 | 0 | 659 | 73 | 3 | 0.054 | 0.525 |
| Ashton KA43 | 2010 | Australia | Caucasian | Endometrial | 163 | 25 | 3 | 258 | 31 | 2 | 0.060 | 0.326 |
| Balistreri CR31 | 2010 | Italy | Caucasian | Prostate | 49 | 1 | 0 | 111 | 13 | 1 | 0.060 | 0.383 |
| Rigoli L27 | 2010 | Italy | Caucasian | Gastric | 42 | 18 | 0 | 80 | 7 | 0 | 0.023 | 0.696 |
| Etokebe GE20 | 2009 | Croatia | Caucasian | Breast | 110 | 20 | 0 | 84 | 15 | 0 | 0.076 | 0.449 |
| Pandey S39 | 2009 | India | South Asian | Cervical | 114 | 35 | 1 | 123 | 26 | 1 | 0.093 | 0.767 |
| Purdue MP44 | 2009 | US | Mixed | Non-Hodgkin lymphoma | 1195 | 133 | 6 | 1126 | 131 | 8 | 0.058 | 0.055 |
| Wang MH32 | 2009 | US | Caucasian | Prostate | 230 | 24 | 0 | 216 | 35 | 0 | 0.070 | 0.235 |
| Trejo-de la OA24 | 2008 | Mexico | Mixed | Gastric | 34 | 4 | 0 | 138 | 6 | 0 | 0.021 | 0.798 |
| Ture-Ozdemir F46 | 2008 | Greece | Caucasian | Gastric MALT lymphoma | 38 | 18 | 0 | 39 | 12 | 0 | 0.118 | 0.341 |
| Santini D25 | 2008 | Italy | Caucasian | Gastric | 159 | 11 | 1 | 140 | 11 | 0 | 0.036 | 0.642 |
| Garza-Gonzalez E26 | 2007 | Mexico | Mixed | Gastric | 72 | 6 | 0 | 175 | 14 | 0 | 0.037 | 0.518 |
| Hold GL29 | 2007 | Poland, US | Caucasian | Gastric | 414 | 79 | 3 | 451 | 47 | 2 | 0.041 | 0.518 |
| Hold GL29 | 2007 | US | Mixed | Oesophageal | 97 | 10 | 0 | 194 | 16 | 1 | 0.043 | 0.299 |
| Cheng I35 | 2007 | US | Mixed | Prostate | 439 | 66 | 1 | 456 | 48 | 2 | 0.051 | 0.544 |
| Nieters A45 | 2006 | Germany | Caucasian | Lymphoma | 590 | 84 | 1 | 596 | 71 | 1 | 0.055 | 0.456 |
| Boraska Jelavic T49 | 2006 | Croatia | Caucasian | Colorectal | 77 | 10 | 2 | 84 | 4 | 0 | 0.023 | 0.827 |
| Landi S50 | 2006 | Spain | Caucasian | Colorectal | 251 | 31 | 0 | 232 | 37 | 0 | 0.069 | 0.226 |
| Forrest MS47 | 2006 | US/UK | Caucasian | Non-hodgkin lymphoma | 794 | 106 | 3 | 1254 | 172 | 6 | 0.064 | 0.969 |
| Hellmig S48 | 2005 | Germany/Austria | Caucasian | Gastric MALT lymphoma | 83 | 4 | 0 | 313 | 45 | 0 | 0.063 | 0.204 |
| Chen YC33 | 2005 | USA | Caucasian | Prostate | 588 | 66 | 3 | 605 | 59 | 5 | 0.052 | 0.011 |
| Zheng SL34 | 2004 | Sweden | Caucasian | Prostate | 1241 | 136 | 1 | 693 | 79 | 5 | 0.057 | 0.103 |
| TLR4 rs4986791 | CC | CT | TT | CC | CT | TT | ||||||
| Singh V17 | 2012 | India | South Asian | Bladder | 163 | 35 | 2 | 173 | 26 | 1 | 0.070 | 0.983 |
| Theodoropoulos GE19 | 2012 | Greece | Caucasian | Breast | 253 | 8 | 0 | 466 | 14 | 0 | 0.015 | 0.746 |
| de Oliveira JG21 | 2012 | Brazil | Caucasian | Gastric | 165 | 9 | 0 | 219 | 6 | 0 | 0.013 | 0.839 |
| Yang ZH40 | 2012 | China | East Asian | Nasopharyngeal | 188 | 45 | 3 | 254 | 32 | 1 | 0.059 | 0.994 |
| Agundez JA37 | 2012 | Spain | Caucasian | Hepatocellular | 143 | 12 | 0 | 341 | 47 | 2 | 0.065 | 0.783 |
| Shen Y18 | 2012 | China | East Asian | Bladder | 433 | 1 | 2 | 517 | 3 | 2 | 0.007 | 0.000 |
| Srivastava K38 | 2010 | India | South Asian | Gallbladder | 195 | 32 | 5 | 232 | 24 | 1 | 0.051 | 0.656 |
| Balistreri CR31 | 2010 | Italy | Caucasian | Prostate | 48 | 2 | 0 | 118 | 7 | 0 | 0.028 | 0.747 |
| Rigoli L27 | 2010 | Italy | Caucasian | Gastric | 57 | 13 | 0 | 81 | 6 | 0 | 0.034 | 0.739 |
| Pandey S39 | 2009 | India | South Asian | Cervical | 127 | 21 | 2 | 133 | 16 | 1 | 0.060 | 0.505 |
| Trejo-de la OA24 | 2008 | Mexico | Mixed | Gastric | 57 | 4 | 0 | 193 | 9 | 0 | 0.022 | 0.746 |
| Santini D25 | 2008 | Italy | Caucasian | Gastric | 155 | 15 | 1 | 147 | 4 | 0 | 0.013 | 0.869 |
| Garza-Gonzalez E26 | 2007 | Mexico | Mixed | Gastric | 77 | 1 | 0 | 179 | 10 | 0 | 0.026 | 0.709 |
| Boraska Jelavic T49 | 2006 | Croatia | Caucasian | Colorectal | 77 | 12 | 0 | 82 | 5 | 0 | 0.029 | 0.783 |
Meta-analysis Results
The main results of this meta-analysis were listed in Table 2 and Figure S1. For TLR2 polymorphism (−196 to −174 del), the meta-analysis showed a significantly increased risk for all cancers (allele comparison: OR = 1.62, 95% CI: 1.09–2.43, P<0.001 for heterogeneity test; dominant model: OR = 1.64, 95% CI: 1.04–2.60, P<0.001 for heterogeneity test; recessive model: OR = 2.28, 95% CI: 1.23–4.20, P<0.001 for heterogeneity test). Similarly, both of TLR4 rs4986790 (allele comparison: OR = 1.17, 95% CI: 1.00–1.37, P<0.001 for heterogeneity test; dominant model: OR = 1.19, 95% CI: 1.01–1.41, P<0.001 for heterogeneity test) and rs4986791 (allele comparison: OR = 1.47, 95% CI: 1.21–1.78, P = 0.070 for heterogeneity test; dominant model: OR = 1.47, 95% CI: 1.20–1.80, P = 0.078 for heterogeneity test) also significantly increased the overall cancer risk.
Table 2. Associations between TLRs polymorphisms and overall cancer risk by races.
| Polymorphism | Ethnicities | Studies | Allele comparison | Dominant model | Recessive model | |||
| OR(95% CI) | p * | OR(95% CI) | p * | OR(95% CI) | p * | |||
| −196 to −174 del | Total | 10 | 1.62(1.09–2.43) | <0.001 | 1.64(1.04–2.60) | <0.001 | 2.28(1.23–4.20) | <0.001 |
| Caucasian | 3 | 3.29(1.14–9.51) | <0.001 | 3.56(1.10–11.51) | <0.001 | 7.29(1.75–30.37) | 0.029 | |
| East Asian | 3 | 1.04(0.71–1.52) | <0.001 | 0.96(0.66–1.40) | <0.001 | 1.27(0.55–2.95) | <0.001 | |
| South Asian | 4 | 1.32(1.11–1.58) | 0.785 | 1.37(1.11–1.68) | 0.870 | 1.72(0.94–3.14) | 0.751 | |
| rs4986790 | Total | 27 | 1.17(1.00–1.37) | <0.001 | 1.19(1.01–1.41) | <0.001 | – | – |
| Caucasian | 19 | 1.17(0.95–1.45) | <0.001 | 1.18(0.95–1.47) | <0.001 | – | – | |
| East Asian | 2 | 1.04(0.79–1.36) | 0.770 | 1.08(0.81–1.45) | 0.797 | – | – | |
| South Asian | 1 | 1.37(0.82–2.30) | <0.001 | 1.44(0.82–2.52) | <0.001 | – | – | |
| Mixed | 5 | 1.05(0.87–1.27) | 0.348 | 1.08(0.89–1.32) | 0.320 | – | – | |
| rs4986791 | Total | 14 | 1.47(1.21–1.78) | 0.070 | 1.47(1.20–1.80) | 0.078 | – | – |
| Caucasian | 7 | 1.51(0.84–2.71) | 0.023 | 1.55(0.85–2.83) | 0.023 | – | – | |
| East Asian | 2 | 1.72(1.14–2.62) | 0.198 | 1.77(1.12–2.77) | 0.192 | – | – | |
| South Asian | 3 | 1.58(1.16–2.16) | 0.718 | 1.55(1.11–2.17) | 0.846 | – | – | |
| Mixed | 2 | 0.75 (0.28–2.01) | 0.117 | 0.75(0.28–2.02) | 0.114 | – | – | |
The results were in bold, if the P<0.05.
P values for heterogeneity test. If the result of the heterogeneity test was p>0.05, ORs were pooled according to the fixed-effect model. Otherwise, the random-effect model was used.
We further performed stratification analysis by ethnicity and cancer types. The results indicated that variant genotypes of TLR2 −196 to −174 del tended to be associated with overall cancer risk in Caucasians (allele comparison: OR = 3.29, 95% CI: 1.14–9.51, P<0.001 for heterogeneity test; dominant model: OR = 3.56, 95% CI: 1.10–11.51, P<0.001 for heterogeneity test) and South Asians (allele comparison: OR = 1.32, 95% CI: 1.11–1.58, P = 0.785 for heterogeneity test; dominant model: OR = 1.37, 95% CI: 1.11–1.68, P = 0.870 for heterogeneity test), but not in East Asians (Table 2). However, the association between rs4986791 and cancer risk was significant in both South Asians (allele comparison: OR = 1.58, 95% CI: 1.16–2.16, P = 0.718 for heterogeneity test; dominant model: OR = 1.55, 95% CI: 1.11–2.17, P = 0.846 for heterogeneity test) and East Asians (allele comparison: OR = 1.72, 95% CI: 1.14–2.62, P = 0.198 for heterogeneity test; dominant model: OR = 1.77, 95% CI: 1.12–2.77, P = 0.192 for heterogeneity test), but not in Caucasians (Table 2). When stratified by cancer types, significantly increased risks of TLR4 rs4986790 were found in digestive cancers (allele comparison: OR = 1.79, 95% CI: 1.14–2.81, P = 0.001 for heterogeneity test; dominant model: OR = 1.76, 95% CI: 1.13–2.73, P = 0.003 for heterogeneity test) and female-specific cancers (allele comparison: OR = 1.44, 95% CI: 1.14–1.83, P = 0.641 for heterogeneity test; dominant model: OR = 1.50, 95% CI: 1.16–1.94, P = 0.537 for heterogeneity test), but not in blood cancers or male-specific cancers (Table 3). However, no significant association with risk of digestive cancers was observed for TLR2 −196 to −174 del and TLR4 rs4986791 (Table 3). We also further investigated the associations between three SNPs and gastric cancer or prostate cancer (involved in more than three studies) and found that both TLR4 rs4986790 (allele comparison: OR = 2.18, 95% CI: 1.67–2.84, P = 0.068 for heterogeneity test; dominant model: OR = 2.20, 95% CI: 1.67–2.89, P = 0.104 for heterogeneity test) and rs4986791 (allele comparison: OR = 1.90, 95% CI: 1.20–3.12, P = 0.193 for heterogeneity test; dominant model: OR = 1.98, 95% CI: 1.22–3.21, P = 0.104 for heterogeneity test) were associated with a significantly increased risk of gastric cancer, but not TLR2 −196 to −174 del (Table 4). Furthermore, we did not observe significant association between rs4986790 and prostate cancer risk.
Table 3. Associations between TLRs polymorphisms and overall cancer risk by cancer types.
| Polymorphism | Cancer type | Studies | Allele comparison | Dominant model | Recessive model | |||
| OR(95% CI) | p * | OR(95% CI) | p * | OR(95% CI) | p * | |||
| −196 to −174 del | Digestive | 6 | 1.32(0.97–1.79) | <0.001 | 1.29(0.92–1.80) | <0.001 | 1.74(0.91–3.34) | <0.001 |
| Others | 4 | 2.19(0.82–5.82) | <0.001 | 1.32(0.80–6.77) | <0.001 | 3.93(0.89–17.47) | 0.002 | |
| rs4986790 | Digestive | 9 | 1.79(1.14–2.81) | 0.001 | 1.76(1.13–2.73) | 0.003 | – | – |
| Blood | 6 | 0.95(0.83–1.10) | 0.170 | 0.95(0.83–1.11) | 0.140 | – | – | |
| Female-specific | 4 | 1.44(1.14–1.83) | 0.641 | 1.50(1.16–1.94) | 0.537 | – | – | |
| Male-specific | 5 | 0.95(0.80–1.13) | 0.068 | 0.99(0.82–1.18) | 0.062 | – | – | |
| other | 3 | 1.11(0.87–1.43) | 0.535 | 1.16(0.89–1.52) | 0.666 | – | – | |
| rs4986791 | Digestive | 8 | 1.58(0.93–2.69) | 0.014 | 1.60(0.94–2.74) | 0.017 | – | – |
| Others | 6 | 1.47(1.13–1.92) | 0.607 | 1.47(1.11–1.96) | 0.589 | – | – | |
The results were in bold, if the P<0.05.
P values for heterogeneity test. If the result of the heterogeneity test was p>0.05, ORs were pooled according to the fixed-effect model. Otherwise, the random-effect model was used.
Table 4. Main result of pooled odds ratios (ORs) in gastric and prostate cancer.
| Polymorphism | Cancer type | Studies | Allele comparison | Dominant model | Recessive model | |||
| OR(95% CI) | p * | OR(95% CI) | p * | OR(95% CI) | p * | |||
| −196 to −174 del | Gastric cancer | 4 | 1.27(0.83–1.95) | <0.001 | 1.21(0.75–1.94) | <0.001 | 1.58(0.70–3.59) <0.001 | |
| rs4986790 | Gastric cancer | 6 | 2.18(1.67–2.84) | 0.068 | 2.20(1.67–2.89) | 0.104 | – | – |
| Prostate cancer | 5 | 0.95(0.80–1.13) | 0.068 | 0.99(0.82–1.18) | 0.062 | – | – | |
| rs4986791 | Gastric cancer | 5 | 1.93(1.20–3.12) | 0.193 | 1.98(1.22–3.21) | 0.177 | – | – |
The results were in bold, if the P<0.05.
P values for heterogeneity test. If the result of the heterogeneity test was p>0.05, ORs were pooled according to the fixed-effect model. Otherwise, the random-effect model was used.
Test of Heterogeneity
A meta-regression was conducted to explore the possible source of heterogeneity for −196 to −174 del and rs4986790 because both of P values for heterogeneity test were less than 0.05 in the comparisons. We identified that MAFs of −196 to −174 del and rs4986790 were significant sources of heterogeneity (P = 0.008 for −196 to −174 del, P = 0.039 for rs4986790, respectively). We also found that ethnicity was a significant source of heterogeneity for −196 to −174 (P = 0.036). However, genotyping methods, tumor types, sample size, and source of controls could not substantially influence the initial heterogeneity.
Sensitivity Analyses and Publication Bias
The leave-one-out sensitivity analysis indicated that no single study changed the pooled ORs qualitatively (data not shown). Furthermore, we also conducted a sensitivity analysis on the TLR2 and TLR4 polymorphism and risk of cancer by excluding all four studies departure from HWE among controls [20], [35], [40], [42] and their exclusion did not substantially affect the results of the meta-analysis (dominant model: OR = 1.68, 95% CI: 1.00–2.81 for −196 to −174del; dominant model: OR = 1.20, 95% CI: 1.00–1.44 for rs4986790; dominant model: OR = 1.49, 95% CI: 1.21–1.83 for rs4986791).
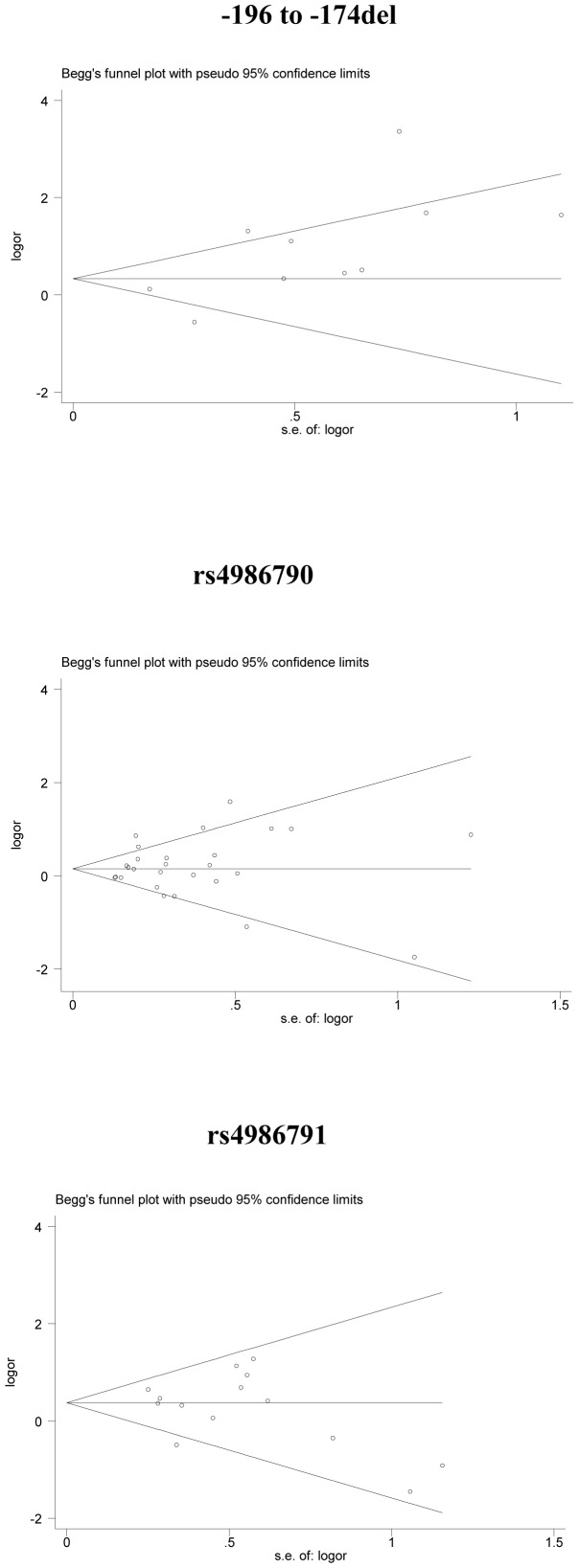
The inverted funnel plots (Figure 2) and Begg’s test were performed to assess the publication bias, and the results did not suggest any obvious evidence of asymmetry for TLR2 and TLR4 polymorphisms (P = 0.152 for −196 to −174 del; P = 0.505 for rs4986790; P = 0.324 for rs4986791, respectively).
Figure 2. Begg’s funnel plot for publication bias test.
Each point represents a separate study for the indicated association. s.e., standardized effect.
Discussion
In this meta-analysis of 34 independent publications, we found that three genetic variants of TLRs (TLR2 −196 to −174 del, TLR4 rs4986790 and rs4986791) were significantly associated with an increased risk of overall cancers. Furthermore, the stratification analysis showed that the risk effect of polymorphisms was more prominent in subjects with some special races or cancer types. All these findings suggested that polymorphisms of TLR2 and TLR4 might contribute to risk of human cancer.
The −196 to −174 del polymorphism in TLR2 located on chromosome 4 causes a 22-bp nucleotide deletion and it has been recently proposed to reflect differential trans-activation of TLR2 promoter constructs and expression levels of TLR2 [38]. However, population studies showed that TLR2 −196 to −174 del polymorphism might play conflicting roles for the risk of different types of cancer. For example, it was reported that the TLR2 −196 to −174 del polymorphism was associated with risk of several cancers, such as cervical cancer, gastric cancer, breast cancer and hepatocellular cancer [21], [23], [24], [38], [41], but not associated with other cancers including bladder, prostate cancer and gallbladder cancer [19], [32], [40]. And even the same kind of cancer, the results were inconsistent [23], [25]. To comprehensively investigate the effect of this polymorphism on the risk of overall cancers, we conducted this meta-analysis and found that TLR2 −196 to −174 del polymorphism significantly increased risk of cancers, supporting the hypothesis that this SNP plays a role in changed expression of TLR2 and cancer development.
The TLR4 gene is mapped on chromosome 9 and consists of three exons. In exon 3, two non-synonymous SNPs (+896A/G rs4986790 and +1196C/T rs4986791) induces the substitution of amino acids Asp299Gly and Thr399Ile, respectively. The substitution of Asp299Gly disrupts the normal structure of the extracellular region of the TLR4, which may cause decreased ligand recognition or protein interaction and decreased responsiveness to lipopolysaccharide [57]. Consequently, such change can affect the transport of TLR4 to the cell membrane and lead to an exaggerated inflammatory response with severe tissue destruction. The results of previous studies regarding the association between these two SNPs and cancer risk were inconsistent. These pooled analysis did not find any significant association between the two SNPs and risk of prostate cancer [58] or gastric cancer [59]. However, a recent meta-analysis of 22 publications on six selected SNPs (rs1927914, rs4986790, rs4986791, rs11536889, rs1927911 and rs2149356) in TLR4 and cancer risk reported that TLR4 rs4986790 and rs4986791 were significantly associated with increased risk of overall cancer and significantly elevated risk of gastric cancer was observed for rs4986790 in a stratification study [60]. Our meta-analysis including more studies (27 studies for rs4986790 and 14 studies for rs4986791) and more cancer types provided additional evidence that these two SNPs may play a role in the development of cancer. In the stratification analysis by cancer types, we found that the effect of rs4986790 on cancer risk was more evident in female-specific cancers and digestive cancers, especially for gastric cancer. Similarly, the risk effect of rs4986791 was also prominent in gastric cancer. Studies have shown that H.pylori activates TLR4 expression in gastric epithelial cells and TLR4 can serve as a receptor for H.pylori binding [61], [62]. Thus, potentially functional polymorphisms of TLR4 may affect the function of TLR4 and contribute to H. pylori-associated carcinogenesis. An important reason for the different findings by previously performed studies may be the insufficient study power to detect modest effects of polymorphisms.
In term of stratified analyses by races, our findings indicated that TLR2 −196 to −174 del had an significant association with cancer risk in Caucasians and South Asians, but not in East Asians. However, the association between TLR4 rs4986791 and cancer risk was significant in both South Asians and East Asians, but not in Caucasians. These differences may be induced by different genetic backgrounds and environmental exposures, as indicated by the difference of minor allele frequency in controls among the two populations (Table 1). For example, the MAF of TLR2 −196 to −174 del in Caucasian controls varied from 0.05 to 0.15, but that in Asians was from 0.12 to 0.38. Allele frequency might reflect the natural selection pressures or a balance by other related functional genetic variants and/or environmental exposures. We also searched some public databases, such as Hapmap (http://hapmap.ncbi.nlm.nih.gov/) and SNPinfo (http://snpinfo.niehs.nih.gov/), and found that rs4986790 was in high linkage disequilibrium (LD) with rs4986791 in Caucasians (r2 = 1), but not data was available in Asians because of low allele frequency of these two SNPs. In our analysis, the associations of rs4986790 and rs4986791 with cancer risk were consistent in Caucasians, but inconsistent in Asians. These findings further indicate that the effect of genetic variants on cancer risk may be different between multiple ethnic groups. Some limitations and potential bias should be addressed. First, the subgroups may have a relatively lower power based on a small number of studies. Second, a more precise analysis should be conducted, if individual data were available, allowing for the adjustment by some co-variants such as age, gender and other environmental factors. However, these information were unavailable from most of studies. Third, the controls in the included studies were recruited from different ways and not uniformly defined, which may have induced some bias for the meta-analysis. Last, the gene-gene interaction is important for the development of complex diseases including cancer because single genetic variation may only have a modest effect [63], [64]. However, the original genotyping data of each publication was unavailable and we could not carry out gene-gene interaction analysis in this study.
In conclusion, this meta-analysis provided statistical evidence that the TLR2 and TLR4 polymorphisms were associated with cancer risk, particularly for gastric cancer. However, due to the limitations of original studies included in the meta-analyses, well-designed prospective studies with larger samples are needed to confirm these findings.
Supporting Information
(DOC)
(DOC)
Funding Statement
This work was supported in part by National Natural Science Foundation of China (81270044 and 81302361), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Jiangsu Natural Science Foundation (BK2011764) and Jiangsu Province Clinical Science and Technology Projects (BL2012008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 2. Iwasaki A, Medzhitov R (2010) Regulation of adaptive immunity by the innate immune system. Science 327: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang RF, Miyahara Y, Wang HY (2008) Toll-like receptors and immune regulation: implications for cancer therapy. Oncogene 27: 181–189. [DOI] [PubMed] [Google Scholar]
- 4. Basith S, Manavalan B, Yoo TH, Kim SG, Choi S (2012) Roles of toll-like receptors in cancer: a double-edged sword for defense and offense. Arch Pharm Res 35: 1297–1316. [DOI] [PubMed] [Google Scholar]
- 5. Takeda K, Kaisho T, Akira S (2003) Toll-like receptors. Annu Rev Immunol 21: 335–376. [DOI] [PubMed] [Google Scholar]
- 6. Aderem A, Ulevitch RJ (2000) Toll-like receptors in the induction of the innate immune response. Nature 406: 782–787. [DOI] [PubMed] [Google Scholar]
- 7. Drexler SK, Foxwell BM (2010) The role of toll-like receptors in chronic inflammation. Int J Biochem Cell Biol 42: 506–518. [DOI] [PubMed] [Google Scholar]
- 8. Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, et al. (2011) The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet 7: e1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harberts E, Gaspari AA (2013) TLR signaling and DNA repair: are they associated? J Invest Dermatol 133: 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kagan JC, Su T, Horng T, Chow A, Akira S, et al. (2008) TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol 9: 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sabroe I, Dower SK, Whyte MK (2005) The role of Toll-like receptors in the regulation of neutrophil migration, activation, and apoptosis. Clin Infect Dis 41 Suppl 7S421–426. [DOI] [PubMed] [Google Scholar]
- 12. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, et al. (1999) Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11: 443–451. [DOI] [PubMed] [Google Scholar]
- 13. Huang B, Zhao J, Shen S, Li H, He KL, et al. (2007) Listeria monocytogenes promotes tumor growth via tumor cell toll-like receptor 2 signaling. Cancer Res 67: 4346–4352. [DOI] [PubMed] [Google Scholar]
- 14. Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G (2008) Cancers take their Toll–the function and regulation of Toll-like receptors in cancer cells. Oncogene 27: 225–233. [DOI] [PubMed] [Google Scholar]
- 15. He W, Liu Q, Wang L, Chen W, Li N, et al. (2007) TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol Immunol 44: 2850–2859. [DOI] [PubMed] [Google Scholar]
- 16. Wang JH, Manning BJ, Wu QD, Blankson S, Bouchier-Hayes D, et al. (2003) Endotoxin/lipopolysaccharide activates NF-kappa B and enhances tumor cell adhesion and invasion through a beta 1 integrin-dependent mechanism. J Immunol 170: 795–804. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed A, Wang JH, Redmond HP (2012) Silencing of TLR4 Increases Tumor Progression and Lung Metastasis in a Murine Model of Breast Cancer. Ann Surg Oncol. [DOI] [PubMed]
- 18. Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ, et al. (2008) Functional consequences of toll-like receptor 4 polymorphisms. Mol Med 14: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh V, Srivastava N, Kapoor R, Mittal RD (2013) Single-nucleotide polymorphisms in genes encoding toll-like receptor −2, −3, −4, and −9 in a case-control study with bladder cancer susceptibility in a North Indian population. Arch Med Res 44: 54–61. [DOI] [PubMed] [Google Scholar]
- 20. Shen Y, Liu Y, Liu S, Zhang A (2013) Toll-like receptor 4 gene polymorphisms and susceptibility to bladder cancer. Pathol Oncol Res 19: 275–280. [DOI] [PubMed] [Google Scholar]
- 21. Theodoropoulos GE, Saridakis V, Karantanos T, Michalopoulos NV, Zagouri F, et al. (2012) Toll-like receptors gene polymorphisms may confer increased susceptibility to breast cancer development. Breast 21: 534–538. [DOI] [PubMed] [Google Scholar]
- 22. Etokebe GE, Knezevic J, Petricevic B, Pavelic J, Vrbanec D, et al. (2009) Single-nucleotide polymorphisms in genes encoding toll-like receptor −2, −3, −4, and −9 in case-control study with breast cancer. Genet Test Mol Biomarkers 13: 729–734. [DOI] [PubMed] [Google Scholar]
- 23. de Oliveira JG, Silva AE (2012) Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol 18: 1235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zeng HM, Pan KF, Zhang Y, Zhang L, Ma JL, et al. (2011) Genetic variants of toll-like receptor 2 and 5, helicobacter pylori infection, and risk of gastric cancer and its precursors in a chinese population. Cancer Epidemiol Biomarkers Prev 20: 2594–2602. [DOI] [PubMed] [Google Scholar]
- 25. Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, et al. (2010) No associations of Toll-like receptor 2 (TLR2) −196 to −174del polymorphism with the risk of Helicobacter pylori seropositivity, gastric atrophy, and gastric cancer in Japanese. Gastric Cancer 13: 251–257. [DOI] [PubMed] [Google Scholar]
- 26. Trejo-de la OA, Torres J, Perez-Rodriguez M, Camorlinga-Ponce M, Luna LF, et al. (2008) TLR4 single-nucleotide polymorphisms alter mucosal cytokine and chemokine patterns in Mexican patients with Helicobacter pylori-associated gastroduodenal diseases. Clin Immunol 129: 333–340. [DOI] [PubMed] [Google Scholar]
- 27. Santini D, Angeletti S, Ruzzo A, Dicuonzo G, Galluzzo S, et al. (2008) Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in gastric cancer of intestinal and diffuse histotypes. Clin Exp Immunol 154: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garza-Gonzalez E, Bosques-Padilla FJ, Mendoza-Ibarra SI, Flores-Gutierrez JP, Maldonado-Garza HJ, et al. (2007) Assessment of the toll-like receptor 4 Asp299Gly, Thr399Ile and interleukin-8–251 polymorphisms in the risk for the development of distal gastric cancer. BMC Cancer 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rigoli L, Di Bella C, Fedele F, Procopio V, Amorini M, et al. (2010) TLR4 and NOD2/CARD15 genetic polymorphisms and their possible role in gastric carcinogenesis. Anticancer Res 30: 513–517. [PubMed] [Google Scholar]
- 30. Tahara T, Arisawa T, Wang F, Shibata T, Nakamura M, et al. (2007) Toll-like receptor 2–196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci 98: 1790–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, et al. (2007) A functional polymorphism of toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology 132: 905–912. [DOI] [PubMed] [Google Scholar]
- 32. Mandal RK, George GP, Mittal RD (2012) Association of Toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol Biol Rep 39: 7263–7269. [DOI] [PubMed] [Google Scholar]
- 33. Balistreri CR, Caruso C, Carruba G, Miceli V, Campisi I, et al. (2010) A pilot study on prostate cancer risk and pro-inflammatory genotypes: pathophysiology and therapeutic implications. Curr Pharm Des 16: 718–724. [DOI] [PubMed] [Google Scholar]
- 34. Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, et al. (2009) Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate 69: 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, et al. (2005) Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res 65: 11771–11778. [DOI] [PubMed] [Google Scholar]
- 36. Zheng SL, Augustsson-Balter K, Chang B, Hedelin M, Li L, et al. (2004) Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the CAncer Prostate in Sweden Study. Cancer Res 64: 2918–2922. [DOI] [PubMed] [Google Scholar]
- 37. Cheng I, Plummer SJ, Casey G, Witte JS (2007) Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev 16: 352–355. [DOI] [PubMed] [Google Scholar]
- 38. Nischalke HD, Coenen M, Berger C, Aldenhoff K, Muller T, et al. (2012) The toll-like receptor 2 (TLR2) −196 to −174 del/ins polymorphism affects viral loads and susceptibility to hepatocellular carcinoma in chronic hepatitis C. Int J Cancer. 130: 1470–1475. [DOI] [PubMed] [Google Scholar]
- 39. Agundez JA, Garcia-Martin E, Devesa MJ, Carballo M, Martinez C, et al. (2012) Polymorphism of the TLR4 gene reduces the risk of hepatitis C virus-induced hepatocellular carcinoma. Oncology 82: 35–40. [DOI] [PubMed] [Google Scholar]
- 40. Srivastava K, Srivastava A, Kumar A, Mittal B (2010) Significant association between toll-like receptor gene polymorphisms and gallbladder cancer. Liver Int 30: 1067–1072. [DOI] [PubMed] [Google Scholar]
- 41. Pandey S, Mittal RD, Srivastava M, Srivastava K, Singh S, et al. (2009) Impact of Toll-like receptors [TLR] 2 (−196 to −174 del) and TLR 4 (Asp299Gly, Thr399Ile) in cervical cancer susceptibility in North Indian women. Gynecol Oncol 114: 501–505. [DOI] [PubMed] [Google Scholar]
- 42. Yang ZH, Dai Q, Gu YJ, Guo QX, Gong L (2012) Cytokine and chemokine modification by Toll-like receptor polymorphisms is associated with nasopharyngeal carcinoma. Cancer Sci 103: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miedema KG, Tissing WJ, Te Poele EM, Kamps WA, Alizadeh BZ, et al. (2012) Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia 26: 1203–1210. [DOI] [PubMed] [Google Scholar]
- 44. Gast A, Bermejo JL, Claus R, Brandt A, Weires M, et al. (2011) Association of inherited variation in Toll-like receptor genes with malignant melanoma susceptibility and survival. PLoS One 6: e24370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, et al. (2010) Toll-like receptor (TLR) and nucleosome-binding oligomerization domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer 10: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Purdue MP, Lan Q, Wang SS, Kricker A, Menashe I, et al. (2009) A pooled investigation of Toll-like receptor gene variants and risk of non-Hodgkin lymphoma. Carcinogenesis 30: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nieters A, Beckmann L, Deeg E, Becker N (2006) Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun 7: 615–624. [DOI] [PubMed] [Google Scholar]
- 48. Ture-Ozdemir F, Gazouli M, Tzivras M, Panagos C, Bovaretos N, et al. (2008) Association of polymorphisms of NOD2, TLR4 and CD14 genes with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Anticancer Res 28: 3697–3700. [PubMed] [Google Scholar]
- 49. Forrest MS, Skibola CF, Lightfoot TJ, Bracci PM, Willett EV, et al. (2006) Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br J Haematol 134: 180–183. [DOI] [PubMed] [Google Scholar]
- 50. Hellmig S, Fischbach W, Goebeler-Kolve ME, Folsch UR, Hampe J, et al. (2005) Association study of a functional Toll-like receptor 4 polymorphism with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Leuk Lymphoma 46: 869–872. [DOI] [PubMed] [Google Scholar]
- 51. Boraska Jelavic T, Barisic M, Drmic Hofman I, Boraska V, Vrdoljak E, et al. (2006) Microsatelite GT polymorphism in the toll-like receptor 2 is associated with colorectal cancer. Clin Genet 70: 156–160. [DOI] [PubMed] [Google Scholar]
- 52. Landi S, Gemignani F, Bottari F, Gioia-Patricola L, Guino E, et al. (2006) Polymorphisms within inflammatory genes and colorectal cancer. J Negat Results Biomed 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127: 820–826. [DOI] [PubMed] [Google Scholar]
- 54. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 55. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 56. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 57. Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, et al. (2000) TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet 25: 187–191. [DOI] [PubMed] [Google Scholar]
- 58. Lindstrom S, Hunter DJ, Gronberg H, Stattin P, Wiklund F, et al. (2010) Sequence variants in the TLR4 and TLR6-1-10 genes and prostate cancer risk. Results based on pooled analysis from three independent studies. Cancer Epidemiol Biomarkers Prev 19: 873–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castano-Rodriguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM (2013) The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: a case-control study and meta-analysis. PLoS One 8: e60327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang K, Zhou B, Wang Y, Rao L, Zhang L (2013) The TLR4 gene polymorphisms and susceptibility to cancer: a systematic review and meta-analysis. Eur J Cancer 49: 946–954. [DOI] [PubMed] [Google Scholar]
- 61. Su B, Ceponis PJ, Lebel S, Huynh H, Sherman PM (2003) Helicobacter pylori activates Toll-like receptor 4 expression in gastrointestinal epithelial cells. Infect Immun 71: 3496–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wroblewski LE, Peek RM Jr (2013) Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterol Clin North Am 42: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu L, Wu C, Wang Y, Zhong R, Wang F, et al. (2011) Association of candidate genetic variations with gastric cardia adenocarcinoma in Chinese population: a multiple interaction analysis. Carcinogenesis 32: 336–342. [DOI] [PubMed] [Google Scholar]
- 64. Zhong R, Liu L, Zou L, Sheng W, Zhu B, et al. (2013) Genetic variations in the TGFbeta signaling pathway, smoking and risk of colorectal cancer in a Chinese population. Carcinogenesis 34: 936–942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)