Abstract
The dehydration responsive element binding (DREB) transcription factors play an important role in regulating stress-related genes. OsDREB2A, a member of the DREBP subfamily of AP2/ERF transcription factors in rice (Oryza sativa), is involved in the abiotic stress response. OsDREB2A expression is induced by drought, low-temperature and salt stresses. Here, we report the ability of OsDREB2A to regulate high-salt response in transgenic soybean. Overexpressing OsDREB2A in soybeans enhanced salt tolerance by accumulating osmolytes, such as soluble sugars and free proline, and improving the expression levels of some stress-responsive transcription factors and key genes. The phenotypic characterization of transgenic soybean were significantly better than those of wild-type (WT). Electrophoresis mobility shift assay (EMSA) revealed that the OsDREB2A can bind to the DRE core element in vitro. These results indicate that OsDREB2A may participate in abiotic stress by directly binding with DRE element to regulate the expression of downstream genes. Overexpression of OsDREB2A in soybean might be used to improve tolerance to salt stress.
Introduction
Adverse environmental conditions, such as high-salt and drought, affect plant growth and productivity. To survive and adapt in adverse conditions, plants have evolved a variety of stress response mechanisms at the molecular, cellular, physiological, and biochemical levels. Transcription factors play an important role in upstream gene regulation of plant stress response pathways [1]. Among these are the AP2/EREBP transcription factors identified in a variety of higher plants, such as Arabidopsis, Nicotiana tabacum, Solanum lycopersicum L., Oryza sativa, Zea mays L., Ricinus communis L., and Brassica [2]–[4]. AP2/EREBP transcription factor genes are involved in many plant functions including growth, hormone signal transduction, pathogen responses, and responses to stresses such as drought and salt. They are characterized by the presence of the highly conserved AP2/EREBP DNA-binding domain of about 58 or 59 amino acid residues, additionally the AP2/ERF domains bind to the dehydration responsive element (DRE) or GCC-box [3], [5]. Both dehydration responsive element binding (DREB) and ethylene responsive factors (ERF) subfamilies are of particular interest due to their involvement in stress response. The genes in the DREB subfamily play a crucial role in the resistance of plants to abiotic stresses by recognizing DRE with a core motif of A/GCCGAC [6], [7]. The ERF subfamily includes a large number of ERFs [8], [9], which are mainly involved in the plant response to biotic stresses like pathogenesis by recognizing the cis-acting element AGCCGCC, known as the GCC box [8], [10]. Of all the subfamilies, DREBs can regulate expression of downstream genes by binding to cis-acting DRE/CRT elements located in the promoter region of downstream stress-responsive genes. An increasing number of DREB genes have been cloned from various plants [11]. DREB genes cloned from Oryza sativa include OsDREB1A to OsDREB1G, OsDREB2A, and OsDREB2B, although only OsDREB1A, OsDREB1E, OsDREB1G, OsDREB2A, and OsDREB2B can specifically bind to DRE elements [12]–[14]. DREB2A/CBF exist in a wide array of plants, in addition to Atriplex hortensis [15], including plants that are acclimatized to salt, such as perennial Lolium perenne [16], [17]. Overexpressions of DREB orthologs in Arabidopsis and Oryza sativa result in improved resistance to abiotic stresses, such as salt and drought [6], [12], [13].
The cDNAs encoding DRE-binding protein, DREB2A, have been isolated by using yeast one-hybrid screening in Arabidopsis [7]. A OsDREB homologous gene, OsDREB2A, which was isolated from rice classified into A-2 subgroup in DREB subfamily [12]. Overexpression of rice OsDREB2A in transgenic Arabidopsis also enhanced tolerance to dehydration and high-salt stress [12], [18]. To further analyze whether OsDREB2A has a function in abiotic stress in Glycine max, we overexpressed OsDREB2A in soybeans and investigated salt-stress tolerance in seeds and seedlings in present study. Taken together, our results will be helpful in determining the functions of OsDREB2A in different species.
Materials and Methods
Rice Seeds and Salt Treatment of Rice Seedlings
Rice seeds of 9311 were germinated on vermiculite in a light chamber at 25°C for 3 weeks. To determine expression pattern of OsDREB2A gene under high-salt stress, 3-week-old seedlings were treated with 200 mM NaCl, and samples were collected at 0 h, 6 h, 12 h, 24 h, 48 h and 72 h.
Isolation of OsDREB2A
The extremely high sequence homology (99% at the nucleotide level) suggests that the OsDREB1 (accession no. AY064403) cloned belongs to the same cluster with the OsDREB2A (accession no. JQ341059) at nucleotide level. The full-length cDNA of OsDREB1 was amplified by RT-PCR from leaves of rice using gene-specific primers (F: 5′-CTGATAGCCTCCTTGATTTT-3′, R: 5′-AAGACGAAAACCGTAAATG-3′) (accession no. AY064403). The opening reading frame (ORF) was 846 bp, ligated into pMD-18T vector by T4 DNA ligase and sequencing confirmation.
Construction of OsDREB2A Expression Vector
An 846 bp coding region of OsDREB2A was amplified from the OsDREB2A-T vector by PCR. The primers were designed as F: 5′-GGATCCATGCTGTTTCGATTTGTG-3′ and R: 5′-GGTACCCTAATAGGAGAAAAGGCT-3′ (accession no. JQ341059) with the BamH I and Kpn I sites, respectively. After sequencing confirmation, the coding region of OsDREB2A was digested with BamH I/Kpn I and was cloned into the GUS position of the intermediate vector of pUC18-pZY102 (The plasmids pUC18 (TaKaRa, Dalian, China) deleted the sites between BamH I and Kpn I and the segment of pZY102 with 35S-GUS-NOS sequence was digested with restriction endonuclease Hind III, and then the two linearized parts were linked together, named as pUC18-pZY102). After sequencing confirmation, the segment of 35S-OsDREB2A-NOS from pUC18-pZY102 was inserted into pZY101 vector at Hind III site, which was named pZY101-OsDREB2A. The resulting binary vector was introduced into Agrobacterium tumefaciens strain EHA101 by the freeze-thaw method [19], which was then used for further genetic soybean transformation.
Soybean Transformation
Mature soybean seeds of cultivar Huachun 3 bred in Guangdong Subcenter of National Center for Soybean Improvement were surface sterilized for 13.5 h using chlorine gas produced by mixing 4.2 ml of 12 N HCl with 100 ml sodium hypochlorite in tightly sealed desiccators [20]. The cotyledonary node method described previously was used [21]. T3 generation homozygous lines of soybeans were used for the phenotype analysis.
High-salt Tolerance Characterization
For the germination assay, 18 seeds for each transgenic soybean or WT were surface sterilized and placed on half-strength Murashige and Skoog (MS) [22] agar plates containing different concentrations of NaCl (0, 200, 250, and 300 mM) or ABA (0, 1.0, 1.5, and 2.0 µM). Plates were placed in the growth chamber at constant 25°C and 16 h daily exposure to 1000 lux Grolux light for germination for 3 d. For salt germination assay, obvious differences in the phenotypes of the transgenic and WT seedlings were observed. For ABA germination assay, germination rates were then measured daily for one week. The WT seedlings were used as control. Each treatment had 3 replicates.
For hydroponic solution and soil culture experiments according to the previously described method by Angkinand et al. [23], with some slight modification. Briefly, the seeds were cultivated in silica sands in artificial climate incubator, for 25°C /28°C at intervals of 12 hours for 7 days. The seedlings were transplanted in Hoagland’s solution. The phenotypic characterizations were assayed in the seedling with ternately compound leaves, within 48 h after treatment with 300 mM NaCl.
In the soil culture test, transgenic and WT seedlings were grown in sterilizing soil (Jiangmei Horticultural Company, Shanghai, China) supplemented for 4 weeks under normal condition. We irrigated the transgenic and WT plants with 200 mM and 300 mM NaCl three times a week for two consecutive weeks, respectively.
Free Proline and Soluble Sugars Concentration Determinations
For measuring of free proline and soluble sugars contents, the WT and transgenic plants were grown in pots under 16 h light at 28°C and 8 h dark at 22°C. Four-week-old plants were irrigated with 300 mM NaCl three times in one week. At the 12th day after the treatment, free proline and soluble sugar contents of leaves were measured as described previously by Saltzmann et al. [24].
Gene Expression Analysis
The total RNAs were extracted using Trizol® reagent (Invitrogen, USA) from the six-week-old soybean seedlings of transgenic and WT, separately. After RNase-free DNase (TaKaRa, Dalian, China) treatment, approximately 1 µg total RNA was used for reverse transcription using the oligo (dT) primer and MMLV (Invitrogen, USA). qRT-PCR was performed using CFX96 (Bio-Rad, USA) and SYBR Green I (Bio-Rad, USA). Each of the cDNA samples was subjected to a qRT-PCR analysis in triplicate. The data were normalized using the reference gene β-tubulin. The relative expressions of specific genes were quantified using the 2−△△Ct calculation. The primer pairs used for qRT-PCR are listed in Table 1.
Table 1. Primers used in this article.
| Gene | Accession number | Primer sequence |
| β-tubulin | NM _178014 | F:5′-CCTCGTTCGAATTCGCTTTTTG-3′R:5′-CAACTGTCTTGTCGCTTGGCAT-3′ |
| GmNHX1 | NM_001250237 | F:5′- GTGCCTTGCTTACGACT -3′R:5′- GGTGAGCCAGGTTCTAC-3′ |
| GmDREB1 | NM_001250325 | F:5′- CGGGTTTAGGAGATTGT-3′R:5′- TATTCCTCTGTATGGCTTC-3′ |
| GmDREB3 | NM_001251571 | F:5′- GCAAATGGGTATCCGAAAT -3′R:5′- GGCCACGTAAGCAGAACA-3′ |
| GmDREB5 | EF583447 | F:5′-GCCATTGTTTAGGCTTCC -3′R:5′- AAGGCTTCTCGGTCGTAG-3′ |
| GmDREB6 | EF551166 | F:5′-ACTCCCTCTACCTCCTCTTC -3′R:5′- TTTCGGATACCCATTTGC-3′ |
| GmERF3 | EU681278 | F:5′- GTCTACGGCAGCGAAAT -3′R:5′- CCAAGCCAGACACGAAC -3′ |
| GmERF7 | JN416602 | F:5′- ATCTCCGACTTCATTCC-3′R:5′- CTCTGAACCCTGCCTC-3′ |
| Gmcor47-like | NM_001253177 | F:5′-GGCAGACGAGACCCAGAACA-3′R:5′-CTTTTTGAAACTCGGTGGCG-3′ |
| GmKin | NM_001250504 | F:5′-AATCTTGTGCTCGCTTAC-3′R:5′-TCCGCATTTGGGCTTTAT-3′ |
| GmP5CS | NM_001251224 | F:5′-TTGGGACTGCTGTGGT-3′R:5′-CAACTTGCGGCTTCTG-3′ |
Electrophoretic Mobility Shift Assay (EMSA)
Glutathione-S-transferase (GST) fusion proteins preparation and gel mobility shift assay were conducted. An 846 bp fragment of OsDREB2A containing DNA-binding domain was amplified using the primer pair: OsDREB2A-F: 5′- GGATCCATGCTGTTTCGATTTGTGTCTTGC-3′; OsDREB2A-R: 5′- CTCGAGCCCAAATCGGAAAAGAGGATAATC-3′ (accession no. JQ341059). BamH I and Xho I recognition sites were introduced and underlined. This fragment was cloned into the BamH I/Xho I sites of the pGEX-4T-2 vector (Amersham Biosciences, Piscataway, USA) and transformed into Escherichia coli BL21 cells (Amersham Biosciences, Piscataway, USA) to produce the GST-fusion protein. The GST-fusion protein was purified using a glutathione Sepharose 4B column according to the manufacturer’s instruction (Amersham Biosciences, Piscataway, USA). The following double-stranded oligonucleotides were synthesized as WT or mutated (m) DRE element in EMSA: DRE forward, 5′-ATTTCATGGCCGACCTGCTTTCATGGCCGACCTGCTT-3′; and mGCC forward, 5′-ATTTCATGGAAGACCTGCTTTCATGGAAGACCTGCTT-3′. The gel mobility shift assay was conducted as described previously [7].
Promoter Analysis
The promoter fragments of 10 abiotic stress-response genes were isolated from the soybean genome by searching the GmGDB database (http://www.plantgdb.org/GmGDB/), and cis-elements in promoters were searched in the PLACE database (http://www.dna.affrc.go.jp/PLACE/) [12], [18].
Statistical Analysis
All data were processed by analysis of SPSS13.0 Data Editor (SPSS Inc., Chicago, Illinosis, USA). Values of P<0.05 were considered to be statistically significant.
Results
Overexpression of OsDREB2A Increase Tolerances to High-salt Stress in Transgenic Soybeans
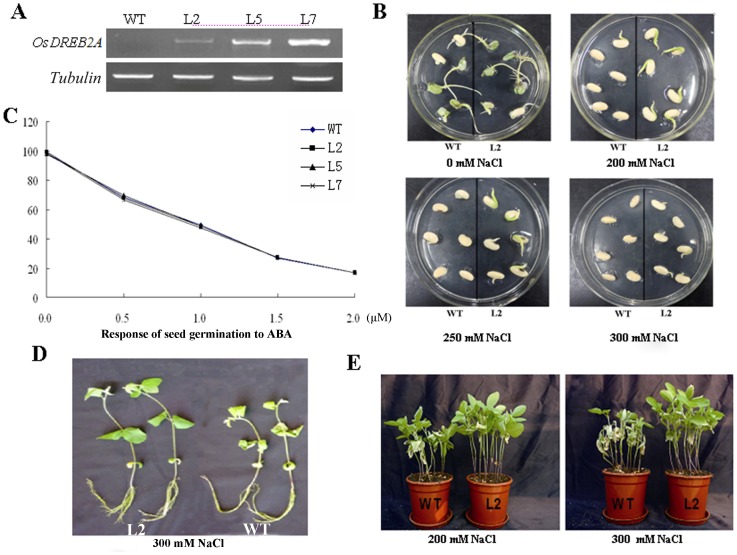
Overexpressing of OsDREB2A was done to examine the biological function in transgenic soybeans. RT-PCR analysis showed that OsDREB2A mRNA accumulated in all transgenic lines, but not in WT plants (Figure 1A). To examine the effects of the overexpression of OsDREB2A on salt tolerance, transgenic and WT seedlings were grown in normal and high-salt conditions. In the germination assays under normal conditions, no obvious differences in the phenotypes of the transgenic and WT seedlings were observed. After incubation in 200 mM NaCl, the germination rate of transgenic seeds was significant higher than that of WT seedlings (100% against 33.3%), but discernible phenotypic differences were observed. After incubation in 250 mM NaCl, the growth of the WT seedlings were completely inhibited, while, the bulk transgenic seedlings remained green and continued to grow, and the germination rate was 100%. After incubation in 300 mM NaCl, the transgenic seedlings began to show a lack of greening (Figure 1B). The germination of both WT and transgenic seedlings showed a similar sensitivity to exogenous ABA (Figure 1C).
Figure 1. Response to salt stress of OsDREB2A overexpressing in plants.
(A) OsDREB2A overexpression in soybean. Transcripts of OsDREB2A in transgenic lines (L2, L5, L7) were detected using RT-PCR, WT were used as control. Tubulin was used as an internal control and added 1 µg in 20 ul RT-PCR system. (B) Salt tolerances of germination assay. Eighteen seeds from the WT and transgenic lines were germinated on MS agar medium containing different concentrations of NaCl. The representative pictures were taken on the 7th day. (C) Response of seed germination to ABA. Seeds on 1/2 MS solid medium with different concentrations of ABA were incubated at 25°C for 3 d and scored. (D) For hydroponic culture experiment, the taproot, lateral root and fibrous root were assayed between transgenic soybean (L2) and WT at 48 h after treatment with 300 mM NaCl, respectively. (E) Representative images of transgenic lines grown in soil under salt conditions. Four-week-old plants growing in soil irrigated with 200 mM NaCl and 300 mM NaCl three times a week for two consecutive weeks, respectively. The photographs were taken 14 d after salt treatment.
Post-development assays were used as a growth indicator using high concentration of NaCl in both hydroponic solution and soil culture experiments. In the hydroponic solution assay, we found that the taproot had no obvious difference in circumference along the plant’s longitudinal axis between transgenic soybean and WT, but the transgenic lines had more branching lateral and fibrous root system than WT (Figure 1D), and the leaves of WT gradually became withered and turned yellow after incubation with 300 mM NaCl. In the soil culture experiment, the results showed that the leaves of WT plants became brown, wilted and curled up, in contrast, the transgenic leaves exhibited healthy growth under the treatment of 200 mM NaCl. However, under the treatment of 300 mM NaCl, most of WT leaves began to albino, and the transgenic leaves began to show a lack of greening (Figure 1E).
Physiological Changes in OsDREB2A Transgenic Soybean
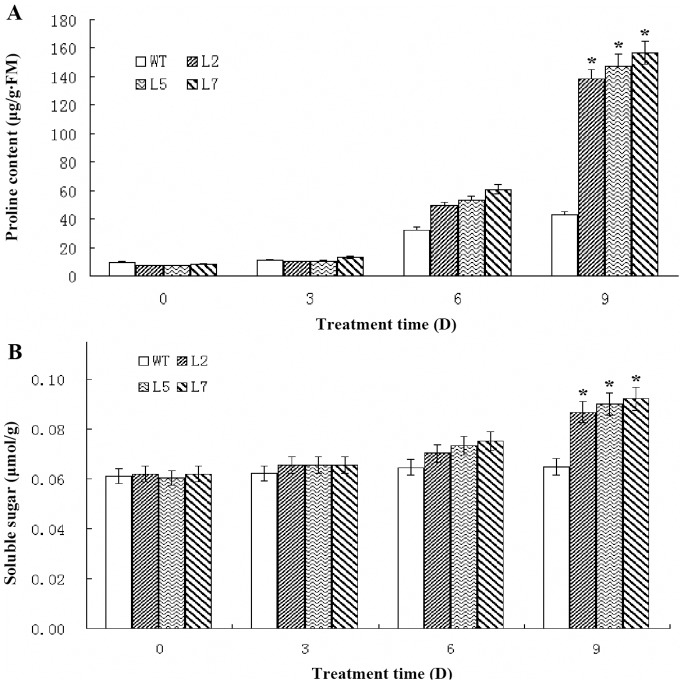
To evaluate physiological changes of transgenic soybean, contents of free proline and soluble sugars were measured following high-salt treatment. Under normal conditions, there were no evident morphological differences between the transgenic lines and WT in terms of their germination rates, seedling sizes, flowering times, primary root lengths and lateral root numbers (data not shown). At 0 d, 3 d, 6 d and 9 d, the concentrations of free proline and soluble sugars were investigated in the transgenic lines. Three transgenic lines had significantly higher proline concentrations and soluble sugars as compared to the WT (Figure 2).
Figure 2. Measurement of soluble sugars and proline content of transgenic and WT plants after treatment with 300.
The WT and transgenic lines (L2, L5 and L7) were grown in pots under normal condition for 4 weeks and then leaves of plants were harvested as controls. Then, the WT and transgenic soybeans were treated with 300 mM NaCl three times with 48 hours interval, while leaves were harvested at 0, 3rd, 6th and 9th days for free proline and soluble sugars content analysis, Data represented the average of three replicates ± SE. *indicated significant difference in comparison to the WT at P<0.05, respectively.
The Expression of Salt-responsive Genes in Transgenic Soybean
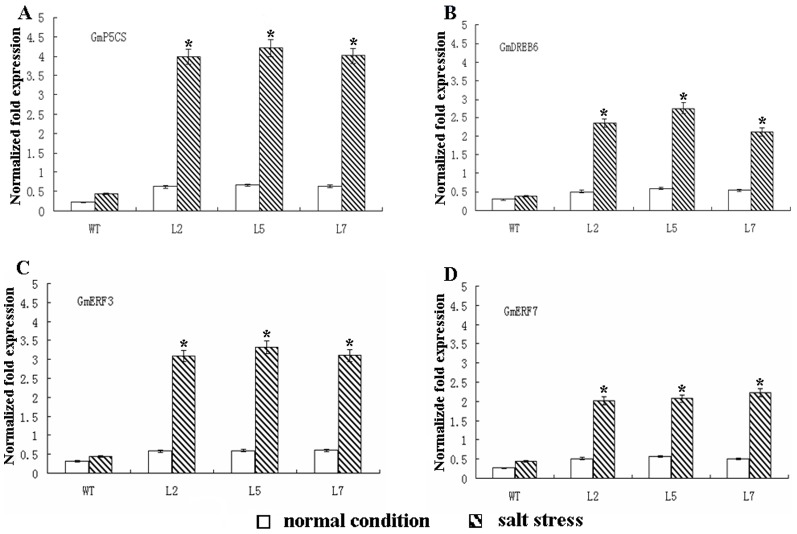
To elucidate the putative molecular mechanisms of OsDREB2A in the high-salt response, we investigated the expression of 10 abiotic stress-response genes, including GmNHX1, GmDREB1, GmDREB3, GmDREB5, GmDREB6, GmERF3, GmERF7, Gmcor47-like, GmKin and GmP5CS in both transgenic and WT plants under normal and 300 mM salt stress conditions, respectively. According to the qRT-PCR analyses, four genes, GmDREB6, GmP5CS, GmERF3, and GmERF7 were shown to have significantly (P<0.05) increased expression in the transgenic soybean under normal and salt stress condition, especially, the increasing tendency of GmP5CS was the most obvious effect in the four genes (P<0.05) (Figure 3). The other six genes were not differentially expressed in the WT and transgenic soybeans (Data not shown).
Figure 3. Expression patterns of salt stress-response genes.
Tubulin was used as an internal control. Four genes, GmP5CS, GmDREB6, GmERF3, and GmERF7 showed increased expression levels in the transgenic soybeans compared to the WT. Values are the mean of four biological replicates ± SE.
DNA-binding Activity Analysis of OsDREB2A using Gel Mobility Shift Assay
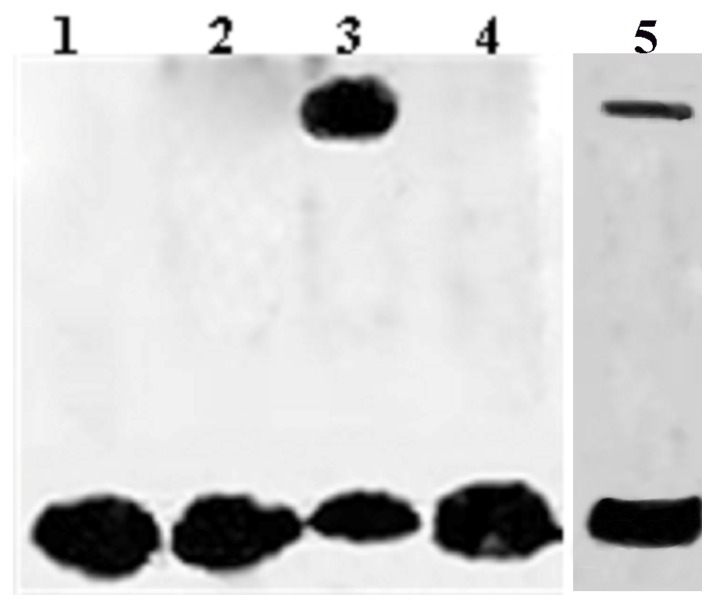
Gel mobility shift assay revealed that the DRE element could interact with OsDREB2A-GST fusion protein and was retarded on SDS-PAGE (Figure 4), the DNA-protein complex migrated more slowly than free DNA, which indicated that the OsDREB2A protein was able to specifically bind to the DRE element. In contrast, the OsDREB2A protein had no interaction with the mutated DRE element. These results suggested that OsDREB2A could specifically bind to DRE element containing -CCGAC-core sequence in vitro.
Figure 4. EMSA showing OsDREB2A specific binding to the DRE element.
Lane 1, free labeled DRE probes; lane 2, GST proteins control; lane 3, GST-OsDREB2A fusion proteins plus labeled the WT DRE element; lane 4, GST-OsDREB2A fusion proteins plus labeled the mutant DRE element; and lane 5, GST-OsDREB2A fusion proteins plus unlabeled and labeled DRE element.
Promoters of OsDREB2A-activated Genes Contain GCC and DRE cis-elements
It was previously shown that DREB is a trans-acting factor that can bind to the DRE/CRT(C-repeat) sequence which contains an A/GCCGAC motif to activate the gene expression in the stress-signaling pathway in plants. To determine whether promoters of OsDREB2A-activated genes contain putative cis-elements possibly binding to OsDREB2A, we searched for cis-elements in the 2-kb promoter region upstream of ATG in the PLACE database [25]. The promoter regions of GmDREB6, GmERF3 and GmERF7 all contain the cis-elements of GT-1 and DRE (Table 2). The GmP5CS promoter has GT-1, but no DRE. Although the promoter region of GmDREB1 has one DRE cis-element (Table 2), it seems that OsDREB2A did not activate GmDREB1 expression. Consistent with the GmNHX1, GmDREB3, GmDREB5, Gmcor47-like and GmKin genes expression, we did not find DRE in the promoter regions.
Table 2. cis-elements in promoters of OsDREB2A-activated genes.
| Gene | ID | cis-elements |
| GmNHX1 | Glyma10g30020 | GT-1(−592, −817, −875, −1392) |
| GmDREB1 | Glyma14g09320 | DRE(−1085), GT-1(−213) |
| GmDREB3 | Glyma17g05240 | GT-1(−325, −633, −1012, −1120), GCC(−1610) |
| GmDREB5 | Glyma12g33020 | GT-1(−402, −705, −740, −1124) |
| GmDREB6 | Glyma5g04920 | DRE(−1113), GT-1(−133, −1398, −1488, −1560, −1993) |
| GmERF3 | Glyma03g42440 | DRE(−487), GT-1(−25, −1112, −1293, −1507, −1530, −1988) |
| GmERF7 | Glyma07g04950 | DRE(−1873), GT-1(−1782), G-box(−36) |
| Gmcor47-like | Glyma04g01130 | GT-1(−779, −1700), GCC(−119) |
| GmKin | Glyma14g07880 | GT-1(−80) |
| GmP5CS | Glyma18g03820 | GT-1(−56, −1243, 1641), GCC(−1899) |
Discussion
DRE cis-acting element is mainly involved in regulation of genes by drought, salt and cold stress under ABA-independent pathways [26]. Previous studies have shown that DREB genes are resistant to different abiotic stresses, including salt, drought, oxidation, and freezing [12], [27], [28]. DREB2 are involved in drought and salt stresses but not in cold stress while DREB1/CBF-type transcription factors function in response to cold stress in another ABA-independent pathway [7], [11], [17], [29]. In the present study, our results showed that OsDREB2A overexpression improved high-salt tolerance in transgenic soybeans. And interestingly, OsDREB2A does not affect the developmental growth of soybean under normal growth conditions (Data not shown). Effect of salt stress on both seed germination and seedling growth showed that the transgenic lines increased tolerance to salt stress than the WT (Figure 1B, 1D and 1E). The results demonstrated that OsDREB2A acted as a positive regulator in response to salt stress at the germination and seedling stages in transgenic soybeans.
While many abiotic-stress-inducible genes are controlled by ABA, but some are not, which indicates that both ABA-dependent and ABA-independent regulatory pathways are involved in stress-responsive gene expression [30]. ABA has been suggested to be involved in the adaptation of plants to salt stress [31]. Cross-talk between salt and ABA signaling pathways has been demonstrated [6], [17], [32]. To investigate whether OsDREB2A is involved in ABA-dependant or independent salt-stress signaling, we determined the effect of ABA on seed germination and found it having a similar effect on the germination of both transgenic and WT seeds (Figure 1C), which suggested that OsDREB2A did not depend on ABA to improve plant salt tolerance in soybean. In this study, overexpression of OsDREB2A in soybean increased tolerance to salt and osmotic stress in an ABA-independent manner.
Previous studies have found that plants may enhance stress tolerance by accumulating osmolytes, such as soluble sugars and free proline to adjust the osmotic potential and protect cell structures [33], [34]. Our results showed that the contents of free proline and soluble sugars in the transgenic lines were higher than that in WT plants under high-salt stress (Figure 2), suggesting that OsDREB2A can regulate free proline and soluble sugars biosynthesis. Consistent with our results, previous studies have shown that overexpression of DREB family genes could elevate proline content [33], [35]–[37]. Indeed, it is well known that proline accumulation can increase the osmotic pressure, thus, improve the salt tolerance of plants [38].
Overexpression of P5CS also increased stress tolerance of transgenic potato, rice and wheat as a result of the increased proline content [39]–[42]. We analyzed the expression of GmP5CS, the key enzyme in proline synthesis [43], [44], and some other genes known to be related to salt resistance. Similarly, qRT-PCR analysis revealed that the expression of OsDREB2A increased GmP5CS transcript level and thus led to higher level of free proline accumulation (Figures 2 and 3). Promoter analysis of the GmP5CS revealed that no DRE region exists. The interaction of OsDREB2A with GmP5CS promoter must not contribute to the increased expression of GmP5CS. With the discovery of miRNAs, a new mechanism to regulate protein expression has been revealed. Considering the inconsistency between P5CS mRNA and protein expression and the importance of miRNAs in cancer, the regulation of miRNAs on P5CS represent a very promising hypothesis [45]. In this study, we propose that OsDREB2A might be involved in many pathways, in addition to combination with DRE region, including regulation of miRNAs and other pathways. A lot of efforts are still required to uncover in detail of each products of gene induced by P5CS and their interacting partners to understand the complexity of the high salinity stress signal transduction pathways.
In order to dissect the enhanced salt tolerance at the molecular level, expression of 10 stress-responsive genes were monitored between the transgenic soybeans and WT. Our results showed that four out of ten, such as GmDREB6, GmP5CS, GmERF3, and GmERF7 were higher expression in transgenic soybeans compared to WT (Figure 3).
We inferred that OsDREB2A might act as an activator to increase the expression of the above stress responsive genes to enhance tolerance of the transgenic soybeans under salt stress. The EMSA indicated that OsDREB2A can bind to DRE. However, it is unknown if OsDREB2A has other binding specificities (Figure 4). Although the promoter of GmDREB1 has one DRE cis-elements (Table 2), it seems that OsDREB2A did not activate GmDREB1 expression (Data not shown). It is not clear why it occurs.
In conclusion, overexpression of OsDREB2A led to the up-regulated expression of several key stress responsive genes, increased accumulation of soluble sugars and free proline, and enhanced tolerance to salt stress in transgenic soybean. Our data suggested that OsDREB2A probably functions as a crucial positive transcription factor in the complex regulatory systems for salt stress response. These results may provide valuable insight into the role of OsDREB2A in abiotic stress tolerance of different species.
Acknowledgments
The authors highly appreciate Prof. Eskild Petersen of Aarhus University and Dr. Nick Florek of University of Wisconsin-Madison for editing and revising on our MS.
Funding Statement
This work was supported, in part, by grants from National Key Project for Research on Transgenic Biology in China (2014ZX0800921B-002), the China Agricultural Research System (CARS-04-PS09), National Key Project for Research on Transgenic Biology in China (2011ZX08004-002-003) and the Scientific and Technological Planning Project of Guandong Province (2009A020102004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang C, Iu B, Singh J (1996) Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus . Plant Mol Biol 30: 679–684. [DOI] [PubMed] [Google Scholar]
- 3. Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA 2 defines a large new family of DNA binding proteins in Arabidopsis . Proc Natl Acad Sci U S A 94: 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 3: 633–646. [DOI] [PubMed] [Google Scholar]
- 5. Tang M, Sun J, Liu Y, Chen F, Shen S (2007) Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas . Plant Mol Biol 63: 419–428. [DOI] [PubMed] [Google Scholar]
- 6. Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, et al. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain, separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis . Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohme-takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators and repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hao DY, Ohme-Takagi M, Sarai A (1998) Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem 273: 26857–26861. [DOI] [PubMed] [Google Scholar]
- 11. Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–4748. [DOI] [PubMed] [Google Scholar]
- 12. Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763. [DOI] [PubMed] [Google Scholar]
- 13. Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30: 2191–2198. [DOI] [PubMed] [Google Scholar]
- 14. Morran S, Eini O, Pyvovarenko T, Parent B, Singh R (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9: 230–249. [DOI] [PubMed] [Google Scholar]
- 15. Shen YG, Zhang WK, Yan DQ, Du BX, Zhang JS (2003) Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis . Theor Appl Genet 107: 155–161. [DOI] [PubMed] [Google Scholar]
- 16. Xiong Y, Fei SZ (2006) Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224: 878–888. [DOI] [PubMed] [Google Scholar]
- 17. Chen M, Wang QY, Cheng XG, Xu ZS, Li LC (2007) GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem Biophys Res Commun 353: 299–305. [DOI] [PubMed] [Google Scholar]
- 18. Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K (2011) Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol J 9: 736–746. [DOI] [PubMed] [Google Scholar]
- 19.An G, Ebert PR, Mitra A, Ha S (1988) Binary Vectors. In: Gelvin SB, Schilperoort RA, Verma DSP (eds): Kluwer, Dordrecht 1–19.
- 20. Di R, Purcell V, Collins G (1996) Production of transgenic soybean lines expressing the bean pod mottle virus coat protein precursor gene. Plant Cell Rep 15: 746–750. [DOI] [PubMed] [Google Scholar]
- 21. Zeng P, Vadnais D, Zhang Z, Polacco JC (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merr.]. Plant Cell Rep 22: 478–482. [DOI] [PubMed] [Google Scholar]
- 22. Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 15: 473–497. [Google Scholar]
- 23.Angkinand N, Chadchawan S (2000) “Plant physiology laboratory”, Department of Botany, Chulalongkorn University: Bangkok.
- 24. Saltzmann KD, Giovanini MP, Zheng C, Williams CE (2008) Virulent Hessian fly larvae manipulate the free amino acid content of host wheat plants. J Chem Ecol 34: 1401–1410. [DOI] [PubMed] [Google Scholar]
- 25. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, et al. (2005) Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 110: 1355–1362. [DOI] [PubMed] [Google Scholar]
- 27. Liu N, Zhong Q, Wang GL, Li LJ, Liu XL, et al. (2007) Cloning and functional char-acterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens . Planta 226: 827–838. [DOI] [PubMed] [Google Scholar]
- 28. Gutha LR, Reddy AR (2008) Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Biol 68: 533–555. [DOI] [PubMed] [Google Scholar]
- 29. Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration and high-salinity-responsive gene expression. Plant Mol Biol 42: 657–665. [DOI] [PubMed] [Google Scholar]
- 30. Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417. [DOI] [PubMed] [Google Scholar]
- 31. Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008. [DOI] [PubMed] [Google Scholar]
- 32. Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A 97: 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97. [DOI] [PubMed] [Google Scholar]
- 34. Cui M, Zhang W, Zhang Q, Xu Z, Zhu Z, et al. (2011) Induced overexpression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol Biochem 49: 1384–1491. [DOI] [PubMed] [Google Scholar]
- 35. Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, et al. (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47: 141–153. [DOI] [PubMed] [Google Scholar]
- 36. Zhao J, Ren W, Zhi D, Wang L, Xia G (2007) Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant Cell Rep 26: 1521–1528. [DOI] [PubMed] [Google Scholar]
- 37. Cong L, Chai TY, Zhang YX (2008) Characterization of the novel gene BjDREB1B encoding a DRE-binding transcription factor from Brassica junceaL . Biochem Biophys Res Commun 371: 702–706. [DOI] [PubMed] [Google Scholar]
- 38. Armengaud P, Thiery L, Buhot N, Grenier-De MG, Savoure A (2004) Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol Plant 120: 442–450. [DOI] [PubMed] [Google Scholar]
- 39. Hmida-Saya ri A, Gargouri-Bouzid R, Bidani A, Jaoua L, Savoure A, et al. (2005) Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci 169: 746–752. [Google Scholar]
- 40. Anoop N, Gupta AK (2003) Transgenic indica rice cv IR-50 overexpressing Vigna aconitifolia Δ1-pyrroline-5-carboxylate synthetase cDNA shows tolerance to high salt. J Plant Biochem Biotechnol 12: 109–116. [Google Scholar]
- 41. Su J, Wu R (2004) Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis, Plant Sci. 166: 941–948. [Google Scholar]
- 42. Vendruscolo EC, Schuster I, Pileggi M, Scapim CA, Molinari HB, et al. (2007) Stress-induced synthesis of proline confers tolerance to water deficit in transgenic wheat. J Plant Physiol 164: 1367–1376. [DOI] [PubMed] [Google Scholar]
- 43. Delauney AJ, Verma DPS (1990) A soybean gene encoding △1-pyrroline-5-carboxylate reductase was isolated by functional complementation in Escherichia coli and is found to be osmoregulated. Mol Gen Genet 221: 299–305. [DOI] [PubMed] [Google Scholar]
- 44. Hu CAA, Delauney AJ, Verma DPS (1992) A bifunctional enzyme (△1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Nat Acad Sci U S A 89: 9354–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu W, Zabirnyk O, Wang H, Shiao YH, Nickerson ML, et al. (2010) miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene 29: 4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]