Abstract
There is wide interest in understanding how genetic diversity is generated and maintained in parthenogenetic lineages, as it will help clarify the debate of the evolution and maintenance of sexual reproduction. There are three mechanisms that can be responsible for the generation of genetic diversity of parthenogenetic lineages: contagious parthenogenesis, repeated hybridization and microorganism infections (e.g. Wolbachia). Brine shrimps of the genus Artemia (Crustacea, Branchiopoda, Anostraca) are a good model system to investigate evolutionary transitions between reproductive systems as they include sexual species and lineages of obligate parthenogenetic populations of different ploidy level, which often co-occur. Diploid parthenogenetic lineages produce occasional fully functional rare males, interspecific hybridization is known to occur, but the mechanisms of origin of asexual lineages are not completely understood. Here we sequenced and analysed fragments of one mitochondrial and two nuclear genes from an extensive set of populations of diploid parthenogenetic Artemia and sexual species from Central and East Asia to investigate the evolutionary origin of diploid parthenogenetic Artemia, and geographic origin of the parental taxa. Our results indicate that there are at least two, possibly three independent and recent maternal origins of parthenogenetic lineages, related to A. urmiana and Artemia sp. from Kazakhstan, but that the nuclear genes are very closely related in all the sexual species and parthenogegetic lineages except for A. sinica, who presumable took no part on the origin of diploid parthenogenetic strains. Our data cannot rule out either hybridization between any of the very closely related Asiatic sexual species or rare events of contagious parthenogenesis via rare males as the contributing mechanisms to the generation of genetic diversity in diploid parthenogenetic Artemia lineages.
Introduction
There is wide interest in understanding how genetic diversity is generated and maintained in parthenogenetic lineages, as it will help clarify the debate of the evolution and maintenance of sexual reproduction. Many asexual species are genetically diverse and this genetic diversity can to some extent ameliorate the lack of meiotic recombination [1], [2]. Several different genetic mechanisms underlie transitions from sexual reproduction to asexuality, and these mechanisms influence in turn the genetic diversity of parthenogenetic lineages and their success and persistence [3], [4]. However, some mechanisms of origin of parthenogenetic lineages can be recurrent, resulting in many, repeated non-independent but polyphyletic origins.
One mechanism for the polyphyletic origin of parthenogenetic lineages diversity is contagious parthenogenesis [3], in which parthenogenetically produced functional rare males mate with sexual females and transmit parthenogenesis to their offspring. Some parthenogenetic lineages produce functional rare males or invest in male function [3], [5], [6]. In the presence of sexual females of related lineages or species, rare males could produce fertile hybrid offspring which would inherit the parthenogenesis-inducing alleles. This mechanism has been best studied in the water flea Daphnia pulex [4], [7]–[9], but is also known to occur in the aphid Myzus persicae [10] and in the parasitoid wasp Lisyphlebus fabarum [11]. The genetic consequence of the spread of asexuality via contagious mechanism is the recurrent origin of new parthenogenetic clones, which will capture some genetic diversity of the maternal sexual species but also maintain some common genomic background from their parthenogenetic ancestor.
A second mechanism is the recurrent generation of multiple parthenogenetic lineages through recent hybridization between related sexual species [3]. Parthenogenesis can result from hybridization between two co-occurring sexual species in vertebrates [12]–[14] and in invertebrates [3], [15], [16]. The repeated origin of hybrid asexuals might generate complex patterns of relationships between the parthenogenetic lineages [17].
A third mechanism of polyphyletic origin is through infection by vertically inherited microorganisms, such as Wolbachia [3]. Microorganisms associated with parthenogenesis can alter the reproduction of their host to favour their persistence in populations, for example by feminizing or killing males or inducing parthenogenesis [2], [18].
If parthenogenetic lineages arise repeatedly trough these mechanisms or a combination of them, their genetic diversity may be comparable to those of sexual populations [1], [19], [20]. Such repeated transitions between sexual and asexual lineages can generate many related but highly diverse asexual lineages which can potentially lead to confounding estimates of genetic diversity of parthenogenetic lineages, and conclusions of ancient asexuality [16].
Brine shrimps of the genus Artemia (Crustacea, Branchiopoda, Anostraca) are a good model system to investigate evolutionary transitions between reproductive systems as they include sexual species and lineages of obligate parthenogenetic populations of different ploidy level [21]. Parthenogenetic populations are found only in the Old World, where they co-occur with various sexual species, including A. salina (Linnaeus 1758) in the Mediterranean region and South Africa [22], A. urmiana (Günther 1899) in and around lake Urmia (Iran) and Crimean salt lakes [23], A. sinica in Central and Northern China [24], A. tibetiana in the Tibetan plateau [25], [26], and likely with a yet undescribed sexual species in Kazakhstan [27], [28]. Artemia species differ in genetic, morphometric, morphological, life history traits [23], [28], and show reproductive isolation, although this is weaker between Asiatic species [25].
Parthenogenetic diploid Artemia populations are automictic and most populations produce fully functional males in low proportions (from 1 to 17 per thousand individuals)[29]. These so called rare males can produce fertile offspring when mating with females of sexual Asiatic species [29]. Assessments of the mitochondrial genetic diversity of Mediterranean parthenogenetic Artemia populations suggested that there were at least two maternal origins of diploid parthenogenesis from a group of closely related Central Asiatic sexual species [30]: one of the mitochondrial lineages – largely responsible for the recent expansion of diploid parthenogenetic Artemia in the Mediterranean – is closely related to those of a sexual undescribed species from Kazakhstan, and the other, rarer lineage, which is closely related to haplotypes of Iranian A. urmiana. The occurrence of two diploid parthenogenetic lineages, and the origin of triploid strains from the common parthenogenetic lineage was also supported by a study of microsatellite and mtDNA sequence diversity of parthenogenetic populations [31]. Nuclear gene sequence variation such as ITS1 [32], also indicated that there were multiple origins of parthenogenesis amongst the sexual species from Asia including A. urmiana, A. tibetiana and A. sinica, but as the ploidy of the samples was not identified, conclusions could not be drawn regarding the origin of diploid parthenogenetic Artemia. However, A. salina and the two American species, are only distantly related to parthenogenetic lineages [32].
Although diploid parthenogenetic Artemia can be identified by their morphology, a genetic marker to characterise would be very useful. In this respect, a study by Manaffar et al. [33] revealed that the digestion of the fragment of exon-7 of Na+/K+ ATPase by Tru1I restriction enzyme showed a polymorphism that allowed discriminating between sexual species and parthenogenetic populations. The sexuals resulted to be homozygote whereas the parthenogens were heterozygote in this position.
Little is known about the mechanisms of origin of parthenogenetic lineages from the ancestral sexual condition, although the possibility of an infectious origin of parthenogenetic Artemia lineages through Wolbachia parasites has been ruled out [34]. Given the functionality of rare males when crossed with Asiatic sexual females, Maccari et al. [29] suggested that they may have an evolutionary role through genetic exchange between parthenogenetic lineages and Asiatic related sexual species. Another possibility would be a hybrid origin between two related sexual species which could give rise to parthenogenetic lineages, especially given the evidence for interspecific hybridization in Artemia in natural populations [35] and in the laboratory [25]. The limited analysis of Asiatic diploid parthenogenetic populations, where the coexistence with closely related sexual species is more likely, has also hampered our understanding of the origin of parthenogenetic lineages.
Here we obtained and analysed sequences from one mitochondrial and two nuclear genes (including the putatively diagnostic marker Na+/K+ ATPase) from an extensive set of populations of diploid parthenogenetic Artemia and sexual species with emphasis on Central and East Asia in order to gain insights into the evolutionary origin of diploid parthenogenetic Artemia, its mode of origin and geographic origin of the parental taxa.
Materials and Methods
Samples
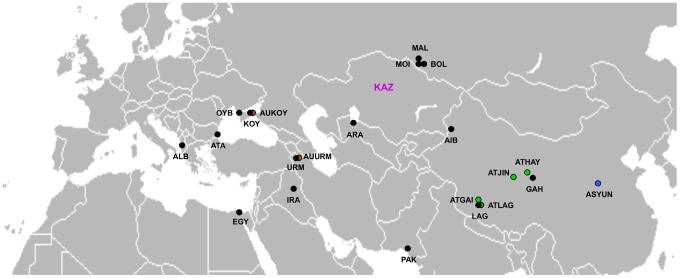
Cyst samples from 15 Eurasian populations of diploid parthenogenetic Artemia (from here onwards, we will use ‘parthenogenetic Artemia’ or ‘parthenogens’ to refer to diploid parthenogenetic Artemia for simplicity) were obtained from the cyst bank collection of the Instituto de Acuicultura de Torre de la Sal (IATS-CSIC) (Figure 1). Laboratory populations were reared from these cyst samples. We assessed the reproductive mode of each population using a sex ratio criterion [29] and whenever the original cyst samples contained an additional sexual species (see Table 1), we obtained pure laboratory parthenogenetic populations using morphometric methods (for culture conditions and other details see [29]). Cyst samples from Asiatic sexual species were also obtained from the same cyst bank collection, including A. urmiana from Urmia lake and from Koyashskoe lake, A. tibetiana from four lakes of the Tibetan plateau (Lagkor Co, Gaize, Hayan, Jingyu), an undescribed sexual Artemia population from Kazakhstan (originally Artemia Reference Center code - ARC 1039, unknown locality) and A. sinica from Yuncheng (China) (Figure 1).
Figure 1. Map of geographic distribution of Artemia populations sampled.
Black circles represent diploid parthenogenetic populations and coloured ones sexual species. Note that due to its unknown locality, Artemia sp. Kazakhstan is represented without circle. See Table 1 for population codes.
Table 1. Detailed information on Artemia samples: population name, population codes, location details and additional co-occurring species found in the sample.
| Population | Codes | Coordinates | Other species | |
| Diploid parthenogens | Narte saltern, Albania | ALB | 40°34′46″N-19°28′16″E | |
| Atanasovko Lake,Bulgaria | ATA | 42°34′25″N-27°28′09″E | ||
| Oybuskoye Lake, Ukraine | OYB | 45°16′15″N-33°04′40″E | ||
| Koyashskoe Lake,Ukraine | KOY | 45°02′09″N-36°12′00″E | A. urmiana | |
| Alexandria saltern,Egypt | EGY | 31°04′13″N-29°46′57″E | ||
| Bagdad saltern, Iraq | IRA | 33°20′19″N-44°29′32″E | ||
| Urmia Lake, Iran | URM | 37°20′00″N-45°40′00″E | A. urmiana | |
| Aral Sea, Uzbekistan | ARA | 45°00′00″N-59°56′00″E | ||
| Maloje Jarovoe Lake, W.Altai | MAL | 52°47′31″N-79°33′39″E | ||
| Bolshoe Jarovoe Lake, W.Altai | BOL | 52°50′N-79°45′E | ||
| Moimishanskoe Lake, W.Altai | MOI | 52°50′N-79°45′E | ||
| Korangi Creek saltern, Pakistan | PAK | 24°47′25″N-67°09′33″E | ||
| Aibi Lake, China | AIB | 44°45′42″N-82°51′54″E | ||
| Lagkor Co Lake, Tibet | LAG | 32°03′N-84°13′E | A. tibetiana | |
| Gahai Lake, China | GAH | 36°58′18″N-98°09′53″E | ||
| Sexuals | ||||
| A. urmiana | Koyashskoe Lake, Ukraine | AUKOY | 45°02′09″N-36°12′00″E | diploid parthenogenetic |
| A. urmiana | Urmia Lake, Iran | AUURM | 37°20′00″N-45°40′00″E | diploid parthenogenetic |
| Kazakhstan sp. | unknown, Kazakhstan | KAZ | ? | |
| A. tibetiana | Lagkor Co Lake, Tibet | ATLAG | 32°03′N-84°13′E | diploid parthenogenetic |
| A. tibetiana | Gaize Lake, Tibet | ATGAI | 32°20′N- 84°10′E | |
| A. tibetiana | Jingyu Lake, Tibet | ATJIN | 36°03′N-89°09′E | |
| A. tibetiana | Hayan Lake, Tibet | ATHAY | 36°03′N-100°11′E | |
| A. sinica | Yuncheng saltern, China | ASYUN | 35°00′N-111°00′E |
DNA isolation, polymerase chain reaction, and sequencing
Total DNA was extracted from cysts using a modified HotSHOT protocol [36]. We amplified fragments of one mitochondrial (cytochrome c oxidase subunit I, COI) and two nuclear genes (internal transcribed spacer 1, ITS1, and Na+/K+ ATPase).
The COI fragment was amplified using the primers HCO2198 and LCOI490 [37]. PCR was carried out in a total volume of 50 µl containing 5 µl of template DNA, 0.2 mM of each nucleotide, 0.2 µM of each primer, 0.05 U of Taq polymerase (Bioline) and 10×Bioline buffer (producing a MgCl2 final concentration of 2 mM). The cycling profile consisted of one cycle of 3 min at 95°C, followed by 40 cycles of 15 s at 95°C, 20 s at 50°C, and 30 s at 72°C, with a final step of 5 min at 72°C.
PCR of the ITS1 region was performed using primers PTF and PTR [38] in a total volume of 30 µl consisting of 3 µl of template DNA, 0.2 mM of each nucleotide, 0.2 µM of each primer, 0.03 U of Taq polymerase (Bioline) and 10×Bioline buffer (producing a MgCl2 final concentration of 1.5 mM) using the following conditions: a cycle of 3 min at 95°C, followed by 35 cycles of 60 s at 95°C, 50 s at 59°C, and 90 s at 72°C, and a final step of 7 min at 72°C.
A fragment of 280-bp, representing exon-7 of Na+/K+ ATPase, was amplified using the primers designed by [33]. PCR was performed in a total volume of 20 µl, containing 3 µl of template DNA, 0.2 mM of each nucleotide, 0.2 µM of each primer, 0.02 U of Taq polymerase (Bioline) and 10×Bioline buffer (producing a MgCl2 final concentration of 2 mM) using the following program: 94°C for 2 min, 32 cycles at 94°C for 25 s followed by 56°C for 25 s and 72°C for 1 min, and a final extension at 72°for 3 min.
All amplifications were performed on a Verity 96 well thermal cycler (Applied Biosystems). PCR products were purified and sequenced by Macrogen Europe Inc. (Amsterdam, The Netherlands). The electrophoregrams were checked by eye using CodonCode Aligner v. 3.5 (CodonCode Corporation, Dedham, MA). COI and ITS1 sequences generated were deposited in GenBank (for Accession Numbers see Tables 2 and 3) and all alignments are available in Dryad (http://doi.org/10.5061/dryad.kd0k4).
Table 2. COI samples and haplotypes: sample size; number of haplotypes per population; πJC, corrected nucleotide diversity; Hd, gene diversity.
| Population code | Sample size | Number of haplotypes | Haplotypes and sample size | πJC | Hd | Acc.Num |
| Diploid parthenogens | ||||||
| URM | 20 | 2 | APD02(17), APD05(3) | 0.0009 | 0.2684 | KF707710-19, KF707765-74 |
| KOY | 15 | 1 | APD02(15) | 0.0000 | 0.0000 | KF707700-09, KF707805-09 |
| ATA | 12 | 3 | APD02(10), APD07(1), APD12(1) | 0.0071 | 0.3182 | KF707720-26, KF707800-04 |
| IRA | 19 | 1 | APD02(19) | 0.0000 | 0.0000 | KF707727-45 |
| EGY | 5 | 2 | APD02(3), APD05(2) | 0.0020 | 0.6000 | KF707785-89 |
| ALB | 10 | 2 | APD02(2), APD05(8) | 0.0012 | 0.3556 | KF707790-99 |
| PAK | 10 | 1 | APD02(10) | 0.0000 | 0.0000 | KF707775-84 |
| OYB | 10 | 2 | APD10(3), APD08(7) | 0.0008 | 0.4667 | KF707810-19 |
| ARA | 6 | 4 | APD02(2), APD11(2), APD13(1),APD14(1) | 0.0021 | 0.8667 | KF707820-25 |
| MAL | 10 | 3 | APD02(3), APD15(5),APD16(2) | 0.0015 | 0.6889 | KF707826-35 |
| BOL | 9 | 3 | APD02(7), APD15(1),APD16(1) | 0.0007 | 0.4167 | KF707836-44 |
| MOI | 10 | 3 | APD02(2),APD18(7), APD19(1) | 0.0026 | 0.5111 | KF707865-74 |
| AIB | 9 | 3 | APD02(5), APD09(1),APD10(3) | 0.0136 | 0.6389 | KF707746-54 |
| GAH | 10 | 1 | APD11(10) | 0.0000 | 0.0000 | KF707755-64 |
| LAG | 10 | 3 | APD02(4), APD05(1),APD17(5) | 0.0145 | 0.6444 | KF707845-54 |
| Sexuals | ||||||
| KAZ | 10 | 4 | KAZSEX06(2), KAZSEX05(2), KAZSEX03(4), KAZSEX08(2) | 0.0038 | 0.8000 | KF707671-80 |
| AUURM | 20 | 12 | AUURM01(1), AUURM02(1), AUURM03(1), AUURM04(7), AUURM05(1), AUURM06(1), AUURM07(1), AUURM08(1), AUURM09(1), AUURM10(2), AUURM11(2), AUURM12(1) | 0.0074 | 0.8790 | KF707681-90, KF707875-84 |
| AUKOY | 9 | 2 | AUKOY01(5),AUKOY02(4) | 0.0027 | 0.5556 | KF707691-99 |
| ATLAG | 20 | 4 | AT01(17), AT08(1), AT09(1),AT10(1) | 0.0007 | 0.2842 | KF707855-64, KF707919-28 |
| ATGAI | 5 | 1 | AT01(5) | 0.0000 | 0.0000 | KF707895-99 |
| ATHAY | 9 | 4 | AT02(3),AT03(4), AT04(1), AT05(1) | 0.0036 | 0.7500 | KF707900-08 |
| ATJIN | 10 | 3 | AT05(4), AT06(1), AT06(5) | 0.0015 | 0.6444 | KF707909-18 |
| ASYUN | 10 | 2 | AS01(6), AS02(4) | 0.0017 | 0.5333 | KF707885-90 |
Table 3. Nuclear loci summary of polymorphic sites in each Artemia population. A dash means that heterozygote individuals were found, a forward slash indicate that the position is polymorphic in the population, with both homozygote and heterozygotes found.
| ITS | NA+/K+ ATPase | ||||||||||||
| Sample size | 522bp | 721bp | 695bp | Acc. Num. | Sample size | 26bp | 56bp | 80bp | 95bp | 140bp | 152bp | ||
| Diploid parthenogens | ALB | 2 | C | C | T | KF736274,75 | 2 | C | T | T | A | G-T | T |
| ATA | 2 | A | C | T | KF736258,59 | 2 | C | T | T | A | G-T | T | |
| OYB | 2 | A | C | T | KF736276,77 | 3 | C | T | T | A | G-T | T | |
| KOY | 2 | C-A/A | C | T | KF736255-57 | 2 | C | T-C | T | A | G-T | T | |
| EGY | 2 | C | C | T-A | KF736266-69 | 2 | C | T | T | A | G-T | T | |
| IRA | 2 | A | C | T | KF736264,65 | 4 | C | T | T | A | G-T | T | |
| URM | 2 | C | C | T | KF736253,54 | 2 | C | T-C/T | T-A/T | A | G-T | T | |
| ARA | 2 | C/A | C | T | KF736278,79 | 2 | C | T-C | T | A | G-T | T | |
| MAL | 2 | C | C | T | KF736280,81 | 2 | C | T-C | T | A | G-T | T | |
| BOL | 2 | C | C | T | KF736282,83 | 2 | C | T | T | A | G-T | T | |
| MOI | 2 | C | C | T | KF736284,85 | 3 | C | T-C | T-A | A | T/G-T | T | |
| PAK | 2 | A | C-T | T | KF736270-73 | 2 | C | T | T | A | G-T | T | |
| AIB | 2 | C | C | T | KF736260,61 | 2 | C | T | T | A | G-T | T | |
| LAG | 2 | C-A | C | T | KF736286-89 | 4 | C | T | T | A | G-T | T | |
| GAH | 2 | C | C | T | KF736262,63 | 2 | C | T-C | T-A | T-A | T | T | |
| Sexuals | AUKOY | 2 | C | C | T | KF736251,52 | 5 | C | T | T | A | T | T |
| AUURM | 2 | C | C | T | KF736249,50 | 4 | C | T | T | A | T | T | |
| KAZ | 2 | C | C | T | KF736247,48 | 6 | C | T-C | T-A | A | T | T | |
| ATLAG | 2 | C | C | T | KF736290,91 | 3 | C | T | T | A | T | T | |
| ATGAI | 2 | C | C | T | KF736294,95 | 4 | C | T | T | A | T | T | |
| ATJIN | 2 | C | C | T | KF736291,92 | 3 | C | T | T | A | T | T | |
| ASYUN | 2 | C | T | T | KF736296,97 | 2 | T | T | T | A | T | C-T | |
Sequence alignment and phylogenetic analyses
The COI fragment was sequenced in 258 individuals, 165 of which were diploid parthenogens (see Table 2). For the nuclear markers we sequenced a subset of these individuals, 44 for the ITS1 region (two for each population sampled) and 63 for the Na+/K+ ATPase fragment (Table 3).
To the COI marker alignment we also added 55 published available sequences from GenBank (parthenogenetic rare males and females KC193638-KC193677, parthenogenetic haplotypes DQ426824-DQ426826, haplotypes from parthenogenetic populations and from Artemia sp. Kazakhstan GU591380-GU591389 and A.tibetiana EF615588-89). Sequences were aligned using ClustalW in MEGA5 [39] using the default settings and checked by eye. The number of polymorphic and parsimony informative sites was computed with MEGA5. Patterns of nucleotide diversity, synonymous and non-synonymous substitutions, population haplotype and nucleotide diversity were computed using DnaSP5 [40].
Before phylogenetic reconstruction, sequences were collapsed into haplotypes using FaBox v.1.40 [41]. For both COI and ITS1 markers, phylogenetic analysis was implemented using Maximum Likelihood (ML) approaches in MEGA5 and Bayesian approaches in MrBayes v 3.2.2 [42] on the Cipres Science Gateway portal [43]. We estimated the best-scoring ML tree using the model selected by the inbuilt model generator in MEGA5. The robustness of the branches was assessed with 1000 bootstrap pseudo-replicates. For Bayesian analysis we used the default parameters on the Cipres gateway. In two simultaneous runs, four Markov chains (one cold and three heated) were started from a random tree and run for 1,000,000 generations with sampling frequency every 100 generations. The first 2500 trees were discarded as burn-in.
In addition, we constructed a statistical parsimony haplotype network for COI using TCS v. 1.21 [44] to visualize the genealogical relationships between the mitochondrial haplotypes. For this analysis we used all the COI sequences generated here, two A. tibetiana sequences from GenBank (EF615587-8), the sequences from Maccari et al. [29] and Muñoz et al. [30]. For sequences from the latter paper, including Mediterranean populations of diploid parthenogenetic Artemia, we reconstructed the sequence of each individual from the paper haplotype information.
Results
Cytochrome oxidase subunit I
The sequence alignment was trimmed to 614 bp long, with all the 313 sequences of the same length. No insertions, deletions or stop codons were present. The COI alignment consisted of 143 variable sites and 133 parsimony informative sites with a total of 144 synonymous and 10 nonsynonymous substitutions.
The sequences generated here collapsed into 45 haplotypes (see Table 2). No haplotype was shared between parthenogens and sexuals, despite both parthenogens and sexuals coexisting in three of the sampled populations. Diploid parthenogenetic populations had a total of 15 haplotypes, 11 of them newly found in this study. APD02, the most common and widespread haplotype, was found in 99 individuals from 13 out of the 15 diploid parthenogenetic populations sampled. The next most common haplotype, APD05 was found in four populations (URM, EGY, ALB and LAG), APD10 in two populations (OYB and AIB), as APD11 (ARA and GAH). Haplotypes APD15, APD16 were found in both populations from the Altai (MAL and BOL). The remaining nine haplotypes were found in single populations.
The sexual populations sequenced here had 30 COI haplotypes. We found four exclusive haplotypes in the undescribed sexual species from Kazahkstan, 12 in A. urmiana from Urmia Lake, and two in A. urmiana from Koyashskoe Lake, with no shared haplotypes between these A. urmiana populations. The populations of A. tibetiana had 11 haplotypes. The population of A. sinica was characterized by two haplotypes. The highest haplotype diversity (Hd) was found in A. urmiana from lake Urmia (0.88) and in the parthenogenetic population from Aral Sea (0.87) (Table 2). Populations from Koyashskoe Lake, Bagdad saltern, Korangi Creek saltern and Gahai Lake amongst the parthenogens and A. tibetiana from Gaize Lake among the sexuals were characterized by a single haplotype.
The nucleotide diversity values (π-values) ranged from 0.0000 to 0.0145 (Table 2). The highest value was found in two parthenogenetic populations from Lagkor Co and Aibi Lake, but the sexual populations from Urmia Lake, Kazakhstan and Hayan Lake and the parthenogenetic population from Atanosovko Lake also showed high π-values compared with the rest of the populations.
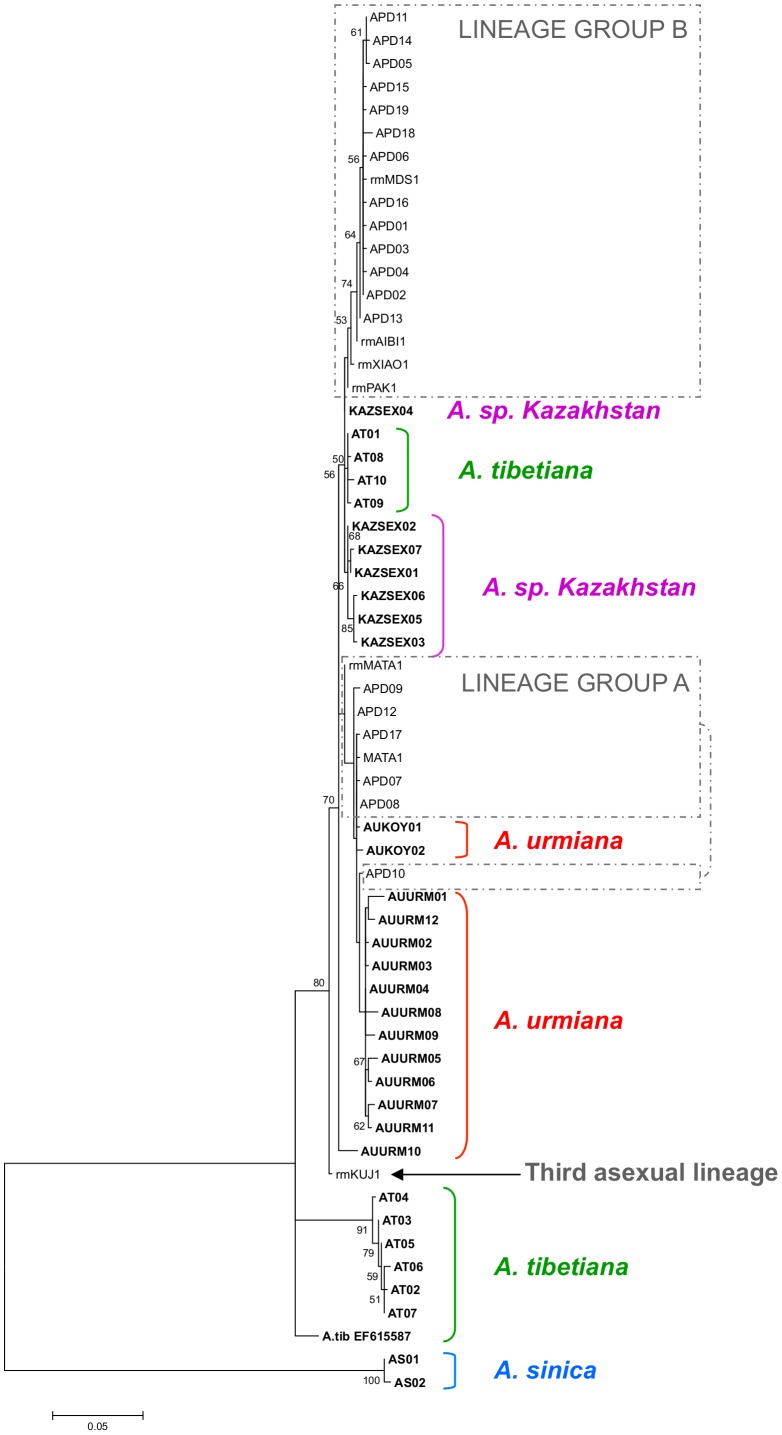
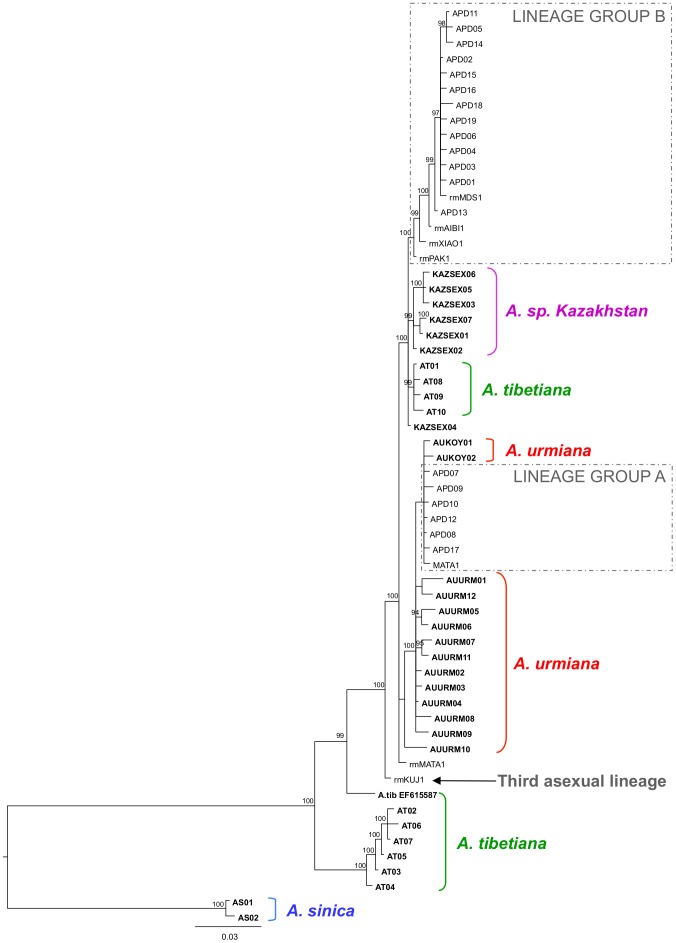
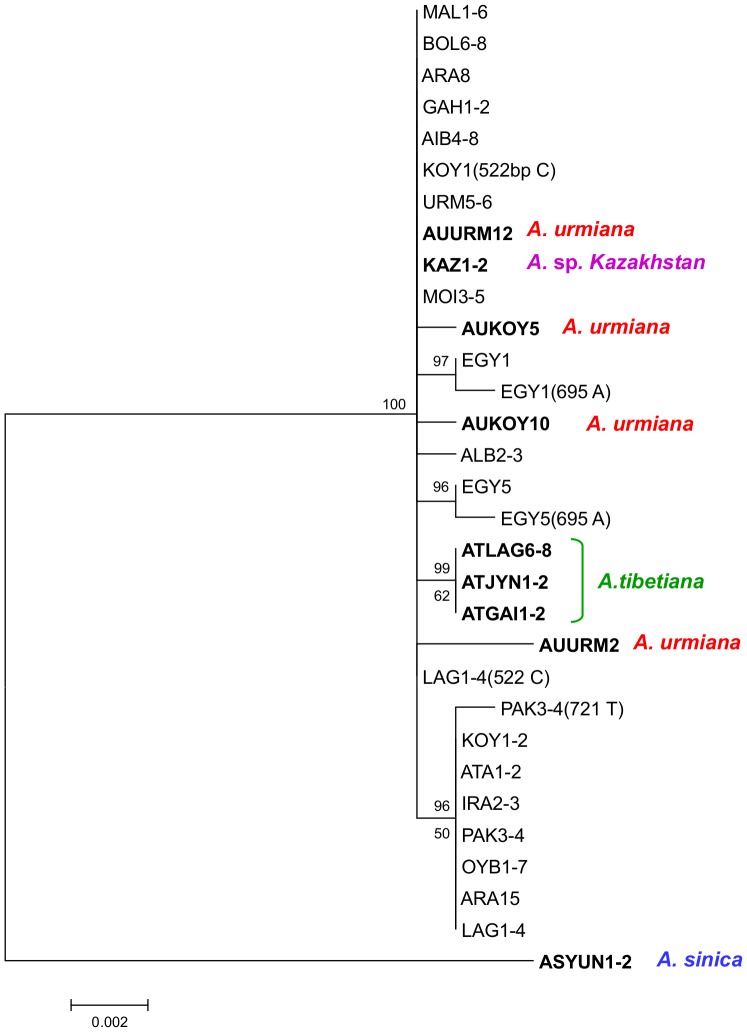
The ML tree (Figure 2) was obtained using the Tamura-3 parameter (T92) plus gamma model, the one selected by the inbuilt model generator in MEGA5. The tree showed that all diploid parthenogenetic Artemia haplotypes, plus the haplotypes of A. urmiana populations, Artemia sp. Kazakhstan and the haplotypes of A. tibetiana from Lagkor Co and Gaize Lake formed a highly supported monophyletic lineage. A group of diploid parthenogenetic Artemia haplotypes formed a polyphyletic, not well supported assemblage amongst haplotypes from both A. urmiana populations (lineage group A). A second group of haplotypes, including the most common APD02 haplotype, formed a monophyletic, but not highly supported lineage, closely related to Artemia sp. Kazahkstan and to the lineage of A. tibetiana (which we called lineage group B). The haplotype from Kujalnik (rmKUJ1), obtained in two rare males [29] formed a well supported sister branch to those containing all other parthenogenetic. The mtDNA lineages of the other two A. tibetiana populations (Hayan and Jingyu Lake) and A. sinica were only distantly related to those of diploid parthenogenetic Artemia. The Bayesian consensus tree (Figure 3) showed a similar topology, although it resolves the relationships of two A. tibetiana lineages. A. tibetiana from GenBank (EF615587) forms a highly supported branch with all diploid parthenogens, A. urmiana, Artemia sp. Kazakhstan and the haplotypes of A. tibetiana from Lagkor Co and Gaize Lake. Lineage group A, with the exception of rmMATA1, together with all A. urmiana haplotypes forms a well supported lineage. Lineage group B forms a well supported monophyletic lineage and its relationship with Artemia sp. Kazakhstan and the haplotypes of A. tibetiana from Lagkor Co and Gaize Lake was also highly supported. Further differences with the ML analysis are represented by the position of AURM010, which in the Bayesian analysis falls at the base of the rest of A. urmiana haplotypes and Lineage group A, and by the position of rmMATA1 which forms a polytomy more basal in the tree, instead of belonging to lineage group A.
Figure 2. Maximum Likelihood (ML) phylogenetic tree of diploid parthenogenetic Artemia and Asiatic sexual species based on COI haplotypes.
Sequence evolution is based on the T92 + G model. One thousand pseudoreplications of bootstrapping were used. For haplotypes from GenBank, the code for each haplotype shown corresponds to the code for the first individual in the alignment with that haplotype (see text, Table 2 and Figure 4 for the individuals included in each haplotype). Sexual species are shown in bold. Rare males are noted by rm followed by the population code as reported en GenBank.
Figure 3. Bayesian inference of phylogenetic relationships of diploid parthenogenetic Artemia and Asiatic sexual species based on COI haplotypes.
Support values higher than 0.90 are shown. For haplotypes from GenBank, the code for each haplotype shown corresponds to the code for the first individual in the alignment with that haplotype (see text, Table 2 and Figure 4 for the individuals included in each haplotype). Sexual species are shown in bold. Rare males are noted by rm followed by the population code as reported en GenBank.
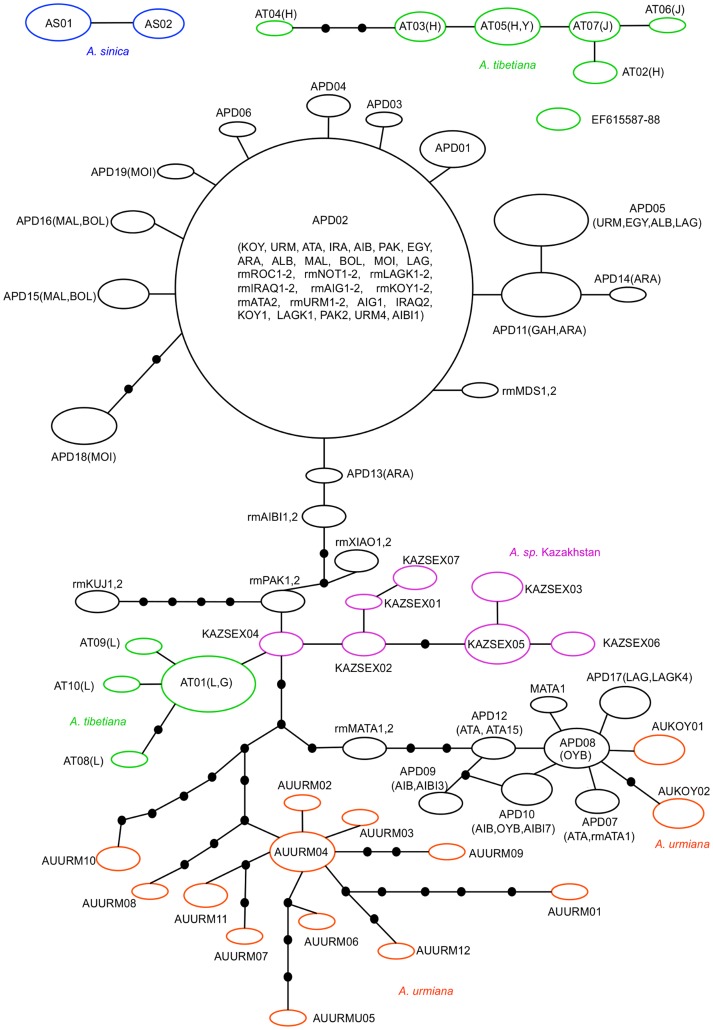
The statistical parsimony network shows the relationship between the mitochondrial haplotypes of parthenogenetic and related sexual species more clearly (Figure 4). There were four unlinked networks. The two haplotypes from A. sinica formed a network, the two A. tibetiana populations from Hayan and Jinyu Lake resulted in a second haplotype network, and the two A. tibetiana sequences from GenBank (EF615587-88) formed a third network. The remaining haplotypes including all diploid parthenogenetic samples, A. urmiana, Artemia sp. from Kazakhstan and the A. tibetiana populations of Lagkor Co and Gaize Lake were joined in a single network. Haplotypes of diploid parthenogenetic Artemia formed three distinct mitochondrial lineage groups as in the phylogenetic reconstructions. Lineage group A, with eight haplotypes, is nested within the diversity of A. urmiana haplotypes and most closely related to haplotypes from Koyashkoe Lake population. This is a relatively rare parthenogenetic lineage, but found at very geographically widespread populations (Atanosovsko Lake, Oybuskoye Lake, Lagkor Co Lake, la Mata Lagoon and Aibi Lake parthenogenetic populations). Lineage group B is more common and widespread, and is formed by the common haplotype APD02 and a number of closely related ones forming a star-like network. Lineage group B is closely related to haplotypes from A. tibetiana from Lagkor Co and Gaize Lake (AT01, AT08, AT09 and AT10) and Artemia sp. from Kazakhstan (KAZSEX01-07), which are also closely related between them. There is no geographic association of the two lineages with a well-defined region because both diploid parthenogenetic haplotype lineage groups coexist in Atanosovsko Lake (ATA), Aibi Lake (AIB) and Lagkor Co Lake (LAG) populations. Some haplotypes found exclusively in rare males from diploid parthenogenetic populations of diverse origins (rmPAK from Korangi Creek in Pakistan; rmXIAO from Xiaotan in China; rmMATA from La Mata in Spain) appeared in the center of the network, and were more closely related to haplotypes of sexual populations. The haplotype from rare males of Kujalnik (rmKUJ from Kujalnik in Ukraine) formed a separate branch to the rest, and would be a third group of parthenogenetic lineages.
Figure 4. Statistical Parsimony networks showing the nested relationships of diploid parthenogenetic Artemia haplotypes and Asiatic sexual species.
Black circles represent diploid parthenogenetic Artemia haplotypes and coloured circles represent Asiatic sexual species. Circle diameter is proportional to the relative haplotype frequency. Connecting lines indicate single substitutions and small black circles represent putative missing haplotypes. The haplotypes codes correspond to those listed in Table 2 or those from GenBank. Rare males are noted by rm followed by the population code as reported en GenBank.
ITS-1
The ITS1 sequences, excluding gaps in the alignment, ranged from 991 (A. tibetiana, Artemia sp. from Kazakhstan, A. urmiana from Koyashskoe lake and all the parthenogens) to 1000 bp (A. sinica), including the sequences of A. urmiana from Urmia lake which have a variable length (994–999 bp). The final ITS1 alignment was 1002 bp long, with 34 variable sites and 28 parsimony informative sites and collapsed into 14 haplotypes. Evidence of heterozygosity was found in 5 parthenogenetic populations and allele identification in these was straightforward (Table 3).
Prior to the phylogenetic analysis, we collapsed identical haplotypes for each population. Both phylogenetic reconstructions (Maximum Likelihood and Bayesian analysis) had a virtually identical topology and branch support (Figure 5). The ML tree was obtained using the Hasegawa-Kishino-Yano model, the one selected by the inbuilt model generator in MEGA5. It showed A. sinica as the most divergent species. The remaining samples were very closely related. The parthenogenetic samples had a total of nine very closely related haplotypes, one of them found in nine populations, was shared with both Artemia sp. from Kazakhstan and one of the haplotypes from the Iranian A. urmiana, although this latter haplotype contained an indel. The populations of A. urmiana from Koyashskoe Lake and A. tibetiana present different haplotypes, although still closely related to the parthenogenetic ones.
Figure 5. Phylogenetic relationships of diploid parthenogenetic Artemia and Asiatic sexual species based on ITS-1 sequences.
The topology inferred by Maximum Likelihood (ML) method using HKY model is shown. Bayesian (BA) phylogenetic reconstruction showed a very similar topology. The ML bootstrap values higher than 50 are shown below the branch, and the Bayesian support values over 90% are shown above the branch. Haplotypes found in each population are shown, with population codes corresponding to those listed in Table 3. Sequences corresponding to heterozygous individuals are noted with the polymorphic site in parenthesis.
Na+/K+ ATPase
The Na+/K+ ATPase alignment was 160 bp long and consisted of sequences of 63 individuals. The alignment did not contain indels and had nine polymorphic sites (Table 3). Evidence of heterozygosity was found in all parthenogenetic populations and in only the sexual population from Kazakhstan. The populations from Moimishanskoe Lake (Altai), Gahai Lake (China) and Urmia Lake (Iran) share the same alleles at all polymorphic sites with the sexual population from Kazakhstan (see Table 3).
Discussion
In order to shed light on the origin and evolution of parthenogenesis in Artemia, we explored the genetic variability of nuclear and mitochondrial DNA of diploid parthenogenetic strains and sexual species, with an emphasis on Asia, the region considered to be the most likely centre of origin of asexual lineages [29]–[31]. Our analyses confirmed the existence of at least two and possibly three maternal clades of diversity, two of them most related to two different sexual Artemia species, A. urmiana and Artemia sp. Kazakhstan in agreement with Muñoz et al. [30], but also revealed a possibly new lineage of parthenogenetic lineages represented by KUJ [29]. Overall, nuclear genes indicate that diploid parthenogenetic Artemia is very closely related to A. urmiana, Artemia sp. Kazakhstan and A. tibetiana, with the exclusion of A. sinica. Both nuclear and mitochondrial data for A. sinica are very divergent to those of diploid parthenogens, suggesting that this species did not contribute to the genetic diversity of diploid parthenogenetic Artemia. Our survey substantially expands our knowledge of its genetic diversity in Eurasia.
Our geographically wider number of Artemia populations sampled, inclusion of rare males and samples of a recently found population of A. urmiana not sequenced before revealed that the lineages in Muñoz et al [45] are not highly supported phylogenetically, as we found further intermediate haplotypes and also identified the key role of the new A. urmiana population from Koyashskoe Lake. Furthermore, we found that the less common mitochondrial group (A) is closely related to haplotypes newly sequenced here from A. urmiana from Koyashskoe Lake, but occupies a non-monophyletic position in the network between both A. urmiana populations, which appears incompatible with a mutational origin, and points to a possible event of contagious parthenogenesis. In contrast, the most common lineage (B), is monophyletic and closely related both to the haplotypes of Artemia sp. from Kazakhstan, and to those of two A. tibetiana populations from Lagkor Co and Gaize lakes, which represent a new lineage of A. tibetiana (see below). Our analysis also revealed a possibly further lineage, so far only found in rare males from Kujalnik population, indicating that they might be present in some populations at low frequencies, maybe resulting from the emergence of new parthenogenetic lineages [29].
In agreement with previous work [30], [38], our results support that the Asiatic sexual species A. urmiana, A. tibetiana and the undescribed species from Kazakhstan, are closely related such that they might be considered a species complex, despite clear morphological differences [29], [46]. This is further supported by experimental crosses showing that, under laboratory conditions there is a proportion of fertile interspecific crosses between these sexual species, indicating weak post mating isolating barriers to gene flow [25].
A. tibetiana contains several divergent, polyphyletic mtDNA lineages, but, in contrast, its nuclear diversity is very homogeneous (monomorphic ITS1 and ATP) and shows little or no differentiation to A. urmiana and Artemia sp. Kazakhstan. A possibility to explain this pattern is that introgression from other species, in particular from females of Artemia sp. Kazakhstan, has resulted on capture of mitochondrial lineages. The genetic diversity of this species needs to be explored further and its taxonomic status might have to be re-evaluated. Given that we have a limited number of samples from A. tibetiana, and the richness of hypersaline habitats in Tibet is high [47], [48], it is likely that the level of diversity within A. tibetiana might still be underestimated. The mitochondrial lineages of A. tibetiana are diverse and the genetic diversity of the rest of the Asiatic species appears to be a subset of it, therefore, A. tibetiana might have a key role in the origin of the species complex and the origin of parthenogenetic lineages.
Although mitochondrial markers have allowed us to identify the minimum number of maternal origins of each diploid Artemia parthenogenetic lineage, nuclear markers should provide information on both parental species and therefore, shed some light on their modes of origin. For example, diploid parthenogenetic lineages resulting from hybridization between conspecific or interspecific sexuals are expected to have a characteristic signature of high heterozygosity, with diploid asexual lineages containing alleles typical of both parental species [49]. If asexuality arises by contagious parthenogenesis through rare males, we could expect a different maternal origin and possibly distinctive genomic component of parthenogenetic lineages. However, repeated gene flow through contagious parthenogenesis should result in a regular emergence of asexual strains and the genetic differentiation between asexuals and sexuals relatives should be low. Our nuclear analysis shows that ITS-1 from parthenogens is closely related to Artemia sp. from Kazakhstan, A. tibetiana and A. urmiana. Some parthenogens and Artemia sp. from Kazakhstan share the same haplotype, whereas A. sinica is very divergent. Baxevanis et al. [32] found four parthenogenetic Artemia lineages, three of which clustering with A. urmiana and A. tibetiana and another one more closely related to A. sinica. The closely related nature of the sexual species from Asia and the lack of divergence between the investigated nuclear genes, however, make it difficult to assess the mechanism or mechanisms of origin of parthenogenesis. However, our mitochondrial phylogenies do not provide clear evidence of rampant contagious parthenogenesis, as it would result in repeated occurrences of new asexual strains and higher mitochondrial diversity. Moreover, parthenogenetic populations coexisting with the known populations of A. urmiana do not have a local origin, as they do not share any haplotypes with the local sexual population. On the contrary, only three mtDNA lineages are found, one of them a minor lineage identified in rare males. That might indicate either that some occasional contagious parthenogenesis does occur or that these are low frequency parthenogenetic lineages with a higher propensity to produce rare males, and have persisted in populations at low frequency. These events would increase the diversity of parthenogenetic strains but playing little role on the geographical expansion and success of parthenogenetic lineages.
The three mtDNA lineages in diploid parthenogenetic Artemia are not differentiated in their nuclear DNA. Although this pattern could result both from repeated hybridization between two similar lineages or from a contagious event between one lineage group and another, the possible existence of contagious parthenogenesis is also supported by microsatellite data. The set of microsatellite loci developed for diploid parthenogenetic Artemia [45] did not amplify consistently in all the sexual species from Asia [29], [31], suggesting that parthenogenetic strains have enough nuclear distinctiveness, and this may be more consistent with contagious parthenogenesis than with a hybrid origin, although it is possible that different mechanisms underlie the origin of each lineage group.
As we used Manaffar et al.'s [33] primers to amplify and sequence a fragment presumably containing a diagnostic SNP between parthenogenetic and sexual strains, we were able to test their finding on a wider array of samples. Our results indicate that, although most samples from a wide range of parthenogenetic populations do meet this criterion (position 140 in our alignment, see Table 3), we identified some parthenogenetic populations that were homozygous for this position (GAH and MOI) and do not confirm the universality of the polymorphism at this site to distinguish parthenogenetic and sexual populations.
Our data cannot rule out either hybridization between any of the very closely related Asiatic sexual species, or rare events of contagious parthenogenesis via rare males as the contributing mechanisms to the generation of genetic diversity in diploid parthenogenetic Artemia lineages. Although our work has provided information on the origin of diploid parthenogenetic Artemia, much is still unknown, and the close relationship of sexual species has hampered this, therefore, more research possibly using genomic approaches is needed to disentangle the evolutionary origin of diploid parthenogenetic Artemia.
Acknowledgments
The authors are especially grateful to all those colleagues and institutions that kindly provided Artemia cyst samples during over three decades. We would like to thank two reviewers and the editor for their constructive comments on the manuscript.
Funding Statement
This study has been funded by the Plan Nacional CGL2008-03277 project to Francisco Amat, sponsored by Spanish Government MICIN. Africa Gomez was supported by a National Environment Research Council (NERC) Advanced Fellowship (NE/B501298/1). Marta Maccari was supported by a fellowship of the JAE Program from CSIC and European Social Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schön I, Gandolfi A, Di Masso E, Rossi V, Griffiths HI, et al. (2000) Persistence of asexuality through mixed reproduction in Eucypris virens (Crustacea, Ostracoda). Heredity 84 (Pt2): 161–169. [DOI] [PubMed] [Google Scholar]
- 2.Shoen I, Martens K, Van Dijk P (2009) Lost Sex: The evolutionary biology of parthenogenesis. Shoen I, Martens K, van Dijk P, editors Dordrecht: Springer.
- 3. Simon J-C, Delmotte F, Rispe C, Crease T (2003) Phylogenetic relationships between parthenogens and their sexual relatives: the possible routes to parthenogenesis in animals. Biological Journal of the Linnean Society 79: 151–163. [Google Scholar]
- 4. Xu S, Innes DJ, Lynch M, Cristescu ME (2013) The role of hybridization in the origin and spread of asexuality in Daphnia . Molecular Ecology 22: 4549–4561 10.1111/mec.12407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D'Souza TG, Michiels NK (2010) The costs and benefits of occasional sex: theoretical predictions and a case study. The Journal of Heredity 101 SupplS34–41 10.1093/jhered/esq005 [DOI] [PubMed] [Google Scholar]
- 6. Neiman M, Larkin K, Thompson AR, Wilton P (2012) Male offspring production by asexual Potamopyrgus antipodarum, a New Zealand snail. Heredity 109: 57–62 10.1038/hdy.2012.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Innes DJ, Hebert PDN (1988) The origin and genetic basis of obligate parthenogenesis in Daphnia pulex . Evolution (N Y) 42: 1024–1035. [DOI] [PubMed] [Google Scholar]
- 8. Lynch M, Seyfert A, Eads BD, Williams E (2008) Localization of the genetic determinants of meiosis suppression in Daphnia pulex . Genetics 180: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eads BD, Tsuchiya D, Andrews J, Lynch M, Zolan ME (2012) The spread of a transposon insertion in Rec8 is associated with obligate asexuality in Daphnia. Proceedings of the National Academy of Sciences of the United States of America: 1–6. doi: 10.1073/pnas.1119667109. [DOI] [PMC free article] [PubMed]
- 10. Blackman RL (1972) The inheritance of life-cycle differences in Myzus persicae (Sulz.) (Hem., Aphididae). . Bulletin of Entomological Research 62: 281–294. [Google Scholar]
- 11. Sandrock C, Vorburger C (2011) Single-locus recessive inheritance of asexual reproduction in a parasitoid wasp. Current Biology 21: 433–437 10.1016/j.cub.2011.01.070 [DOI] [PubMed] [Google Scholar]
- 12. Moritz C (1983) Parthenogenesis in the endemic Australian lizard Heteronotia binoei (Gekkonidae). Science 220: 735–737. [DOI] [PubMed] [Google Scholar]
- 13.Lutes AA, Baumann DP, Neaves WB, Baumann P (2011) Laboratory synthesis of an independently reproducing vertebrate species. Proceedings of the National Academy of Sciences: 1–6. doi: 10.1073/pnas.1102811108. [DOI] [PMC free article] [PubMed]
- 14.Kearney M, Fujita M, Ridenour J (2009) Lost sex in the reptiles: constraints and correlations. Lost Sex.
- 15. Morgan-Richards M, Trewick SA (2005) Hybrid origin of a parthenogenetic genus? Molecular ecology 14: 2133–2142 10.1111/j.1365-294X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- 16. Lunt DH (2008) Genetic tests of ancient asexuality in root knot nematodes reveal recent hybrid origins. BMC Evolutionary Biology 8: 194 10.1186/1471-2148-8-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtsson B (2009) Asex and evolution: a very large-scale overview. Lost Sex.
- 18. Stouthamer R, Breeuwer JAJ, Hurst GDD (1999) Wolbachia pipientis: Microbial Manipulator of Arthropod Reproduction. Annual Review of Microbiology 53: 71–102. [DOI] [PubMed] [Google Scholar]
- 19. Delmotte F, Leterme N, Bonhomme J, Rispe C, Simon J-C (2001) Multiple routes to asexuality in an aphid species. Proceedings of the Royal Society B: Biological Sciences 268: 2291–2299 10.1098/rspb.2001.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwander T, Henry L, Crespi BJ (2011) Molecular evidence for ancient asexuality in timema stick insects. Current biology: CB 21: 1129–1134 10.1016/j.cub.2011.05.026 [DOI] [PubMed] [Google Scholar]
- 21.Abatzopoulos TJ (2002) Artemia: basic and applied biology. Dordretch, The Netherlands: Kluwer Academic Publishers.
- 22. Amat F, Barata C, Hontoria F (1995) A Mediterranean origin for the Veldrif (South Africa) Artemia Leach population. Journal of Biogeography 22: 49–59. [Google Scholar]
- 23. Abatzopoulos TJ, Amat F, Baxevanis AD, Belmonte G, Hontoria F, et al. (2009) Updating Geographic Distribution of Artemia urmiana Günther, 1890 (Branchiopoda: Anostraca) in Europe: An Integrated and Interdisciplinary Approach. International Review of Hydrobiology 94: 560–579 10.1002/iroh.200911147 [DOI] [Google Scholar]
- 24. Cai Y (1989) A redescription of the brine shrimp (Artemia sinica). The Wasman Journal of Biology 47: 105–110. [Google Scholar]
- 25. Abatzopoulos TJ, Kappas I, Bossier P, Sorgeloos P, Beardmore JA (2002) Genetic characterisation of Artemia tibetiana (Crustacea: Anostraca). Biological Journal of the Linnean Society 75: 333–344. [Google Scholar]
- 26. Van Stappen G, Sui L, Xin N, Sorgeloos P (2003) Characterisation of high-altitude Artemia populations from the Qinghai-Tibet Plateau, PR China. Hydrobiologia 500: 179–192. [Google Scholar]
- 27. Pilla EJS, Beardmore JA (1994) Genetic and morphometric differentiation in Old World bisexual species of Artemia (the brine shrimp). Heredity 73: 47–56. [Google Scholar]
- 28. Litvinenko LI, Boyko E (2008) The morphological characteristics of Artemia shrimps from Siberian populations. Inland Water Biology 1: 37–45 10.1134/S1995082908010070 [DOI] [Google Scholar]
- 29. Maccari M, Gomez A, Hontoria F, Amat F (2013) Functional rare males in diploid parthenogenetic Artemia . Journal of Evolutionary Biology 26: 1934–1948 10.1111/jeb.12191 [DOI] [PubMed] [Google Scholar]
- 30. Muñoz J, Gómez A, Green AJ, Figuerola J, Amat F, et al. (2010) Evolutionary origin and phylogeography of the diploid obligate parthenogen Artemia parthenogenetica (Branchiopoda: Anostraca). PLoS One 5: e11932 10.1371/journal.pone.0011932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maniatsi S, Baxevanis AD, Kappas I, Deligiannidis P, Triantafyllidis A, et al. (2011) Is polyploidy a persevering accident or an adaptive evolutionary pattern? The case of the brine shrimp Artemia . Molecular Phylogenetics and Evolution 58: 353–364 10.1016/j.ympev.2010.11.029 [DOI] [PubMed] [Google Scholar]
- 32. Baxevanis AD, Kappas I, Abatzopoulos TJ (2006) Molecular phylogenetics and asexuality in the brine shrimp Artemia . Molecular Phylogenetics and Evolution 40: 724–738 10.1016/j.ympev.2006.04.010 [DOI] [PubMed] [Google Scholar]
- 33. Manaffar R, Zare S, Agh N, Abdolahzadeh N, Soltanian S, et al. (2011) SNP detection in Na/K ATP-ase gene α1 subunit of bisexual and parthenogenetic Artemia strains by RFLP screening. Molecular Ecology Resources 11: 211–214 10.1111/j.1755-0998.2010.02908.x [DOI] [PubMed] [Google Scholar]
- 34. Maniatsi S, Bourtzis K, Abatzopoulos TJ (2010) May parthenogenesis in Artemia be attributed to Wolbachia? Hydrobiologia 651: 317–322 10.1007/s10750-010-0306-8 [DOI] [Google Scholar]
- 35. Kappas I, Baxevanis AD, Maniatsi S, Abatzopoulos TJ (2009) Porous genomes and species integrity in the branchiopod Artemia . Molecular Phylogenetics and Evolution 52: 192–204 10.1016/j.ympev.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 36. Montero-Pau J, Gómez A, Muñoz J (2008) Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnology and Oceanography- Methods 6: 218–222. [Google Scholar]
- 37. Folmer O, Black M, Hoeh WR, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299. [PubMed] [Google Scholar]
- 38. Hou L, Bi X, Zou X, He C, Yang L, et al. (2006) Molecular systematics of bisexual Artemia populations. Aquaculture Research 37: 671–680 10.1111/j.1365-2109.2006.01480.x [DOI] [Google Scholar]
- 39. Tamura K, Peterson D, Peterson N (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. [DOI] [PubMed]
- 41. Villesen P (2007) FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes 7: 965–968 10.1111/j.1471-8286.2007.01821.x [DOI] [Google Scholar]
- 42. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Systematic Biology 61(3): 539–42 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller M, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans.
- 44. Clement M, Posada D, Crandall K a (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- 45.Muñoz J, Gómez A, Green AJ, Figuerola J, Amat F, et al. (2008) Phylogeography and local endemism of the native Mediterranean brine shrimp Artemia salina (Branchiopoda: Anostraca). Molecular Ecology: 3160–3177. doi: 10.1111/j.1365-294X.2008.03818.x. [DOI] [PubMed]
- 46. Triantaphyllidis GV, Criel GRJ, Abatzopoulos TJ, Thomas KM, Peleman J, et al. (1997) International study on Artemia. LVII. Morphological and molecular characters suggest conspecificity of all bisexual European and North African Artemia populations. Marine Biology 129: 477–487 10.1007/s002270050188 [DOI] [Google Scholar]
- 47.Williams W (1991) Chinese and Mongolian saline lakes: a limnological overview. Hydrobiologia: 39–66.
- 48. Wen Z, Mian-ping Z, Xian-zhong X, Xi-fang L, Gan-lin G, et al. (2005) Biological and ecological features of saline lakes in northern Tibet, China. Hydrobiologia 541: 189–203 10.1007/s10750-004-5707-0 [DOI] [Google Scholar]
- 49. Kearney M (2005) Hybridization, glaciation and geographical parthenogenesis. Trends in Ecology & Evolution 20: 495–502 10.1016/j.tree.2005.06.005 [DOI] [PubMed] [Google Scholar]