Abstract
Plasmonic hot spots, generated by controlled 20-nm Au nanoparticle (NP) assembly, are shown to suppress fluorescent quenching effects of metal NPs, such that hair-pin FRET (Fluorescence resonance energy transfer) probes can achieve label-free ultra-sensitive quantification. The micron-sized assembly is a result of intense induced NP dipoles by focused electric fields through conic nanocapillaries. The efficient NP aggregate antenna and the voltage-tunable NP spacing for optimizing hot spot intensity endow ultra-sensitivity and large dynamic range (fM to pM). The large shear forces during assembly allow high selectivity (2-mismatch discrimination) and rapid detection (15 min) for a DNA mimic of microRNA.
Irregular expressions of a panel of microRNAs (miRNA) in blood and other physiological fluids may allow early diagnosis of many diseases, including cancer and cardiovascular diseases.1 However, quantifying all relevant miRNAs (out of 1000), with similar sequences over 22 bases2 and large variations in expression level (as much as 100 fold) at small copy numbers, requires a new molecular diagnostic platform with high-sensitivity, high-selectivity, and large dynamic range. Current techniques for miRNA profiling, such as Northern blotting,3 microarray-based hybridization,4 and real-time quantitative polymerase chain reaction5 are expensive and complex. A simple and rapid miRNA array would allow broad distribution of molecular diagnostic devices for cancer and chronic diseases, eventually into homes for frequent prescreening of many diseases.
At their low concentrations in untreated samples, optical sensing of miRNA is most promising. Plasmonically excited Raman scattering (SERS) and fluorescence sensors from metallic nanoparticles (NPs) or surfaces have enhanced the sensitivity of optical molecular sensors by orders of magnitude.6, 7, 8, 9 However, probe-less SERS sensing or fluorescent sensing of unlabeled targets are insufficiently specific for miRNA targets in heterogeneous samples. Plasmonic detection is also very compatible with FRET probes whose donor dye offers small light sources to excite fluorescently labelled targets upon hybridization.7, 10
A particular family of FRET reporters does offer label-free sensing: hairpin oligo probes whose end-tagged fluorophores are quenched by the Au NP to which they are functionalized.11 The fluorescent signal is only detected when the hairpin is broken by the hybridizing target nucleic acid or protein (for an aptamer probe), and the more rigid paired segment separates the end fluorophore from the quenching surface to produce a fluorescent signal. It is often hoped that plasmonics on the metal surface will enhance the intensity to overcome the quenching effect, if the linearized hairpin is within the NP plasmonic penetration length. However, since fluorescent quenching decays slowly (linearly) with fluorophore-metal spacing10 whereas the plasmonic intensity decays exponentially from a flat surface, careful experimentation shows that quenching dominates and the hairpin probe actually produces a larger intensity on non-metallic surfaces,10 on which it can not function as a label-free probe. Hence, only μM limit-of-detection (LOD) has been achieved with this technique on single NPs or on flat metal surfaces,12 with expensive laser excitation and confocal detection.
Plamonic hot spots formed between metal nanostructures and sharp nanocones can further amplify the plasmonic field.13, 14 The hot spot intensity decays algebraically with respect to the separation or cone tip distance and hence should dominate the linear decay of the metal quenching effect at some optimum separation.15 It is hence possible that plasmonic hot spots may allow much lower LOD with inexpensive optical instruments—ideally light-emitting diode light source and miniature camera. However, the dimension of the gaps, cones, and wedges needs to be at nanoscale, and the cost is now transferred to fabrication of such hot-spot substrates like bow-ties, double crescents, bull-eyes, etc.16 Low-cost wet-etching techniques for addressable nanocones that sustain converging plasmonic hot spots17 have been reported but the fabricated nanocones are often too non-uniform to allow precise quantification. NP monolayers have been shown to exhibit plasmonic hot spots and fluorescence enhancement.18, 19 However, the enhancement only occurs within a range of spacing between aggregated NPs, which is difficult to control and the location or even the existence of the hotspots are not known a priori.
Higher sensitivity is expected if a minimum number of NPs are used in an assembly at a known location and if the NP assembly can produce crystal-like aggregates with controllable NP spacing. Induced DC and AC NP dipoles (related to dielectrophoresis) have been used to assemble NP crystals by embedded micro-electrodes to provide the requisite high field.20, 21 The resulting NP crystals are ideal for plasmonic hot spots, since the spacing of the regimented NP crystal can be controlled by the applied voltage. Conic nanocapillaries22, 23 will be used here for such field-induced NP assembly because the submicron-tip can focus the electric field into sufficient high intensity for NP assembly without embedded-electrodes. Because the field is highest at the tip due to field focusing, the micron-sized crystal would be confined to a small volume, which will be shown to be less than typical confocal volumes, at a known location. So long as the hotspots are regimented, the quantification of target molecules is determined by the total fluorescent intensity and is hence insensitive to the exact geometry of the nanocapillary.
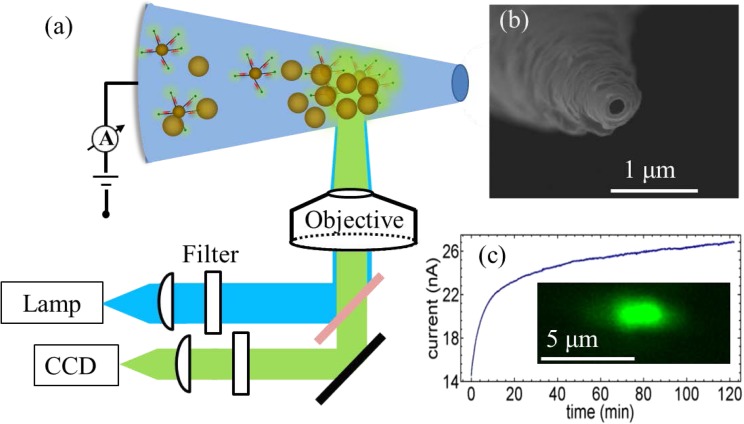
Fluorescent microscope equipped with tungsten lamp light source and normal CCD camera from Q Imaging were used for simultaneous optical and ion current measurements, as shown in Fig. 1a. The nanocapillaries were pulled from commercial glass capillaries using laser-assisted capillary puller. SEM image of a typical pulled glass nanocapillary in Fig. 1b shows an inner diameter of 111 nm and cone angle of 7.3°. The capillary was inserted into a Polydimethylsiloxane chip with two reservoirs. The 20 nm Au NPs, functionalized with fluorescently labelled dsDNA, were injected into the base reservoir. With SEM imaging (Fig. S3 in the supplementary material24), the functionalized DNA is found to prevent NP aggregation even in high ionic-strength Phosphate buffered saline buffer. The NP solution is then driven into the capillary through the tip by applying a positive voltage. Fig. 1c shows the ion current evolution over 2 h at +1 V packing voltage. The ion current increases rapidly in the first 10 min, then at a much slower rate. The rise of current indicates assembly of conductive Au NP assembly at the tip. This was confirmed by the strong fluorescence signal at the tip region during the packing process (inset of Fig. 1c). The one-micron region (corresponding to roughly an aggregate volume of one attoliter) near the capillary tip shows a fluorescence signal after 1 min and also appeared as a dark spot in the transmission image (supplementary material, Fig. S124). This spot darkens with longer packing time but does not grow in size, consistent with the monotonically increasing ion current with increased packing density of the NP assembly. As contrast, a strong fluorescence appeared after only 1 min of packing, but the signal became weaker after 15 min (supplementary material, Fig. S124). This reduction in fluorescence is not due to bleaching of fluorophores because we took 2 images in 15 min at 5 s exposure each and control experiments show significant bleaching only beyond an exposure time of 100 s (see supplementary material).24 Instead, the non-monotonic dependence of the fluorescence intensity with respect to time is because of the optimal hotspot spacing for highest plasmonic intensity at about 5–20 nm,25, 26, 27 which is reached at about 10 min.
Figure 1.
Plasmonic hotspots generated at the tip of a nano-capillary. (a) Schematic of the experimental set up. (b) SEM image of glass nanocapillary shows opening at the tip with a diameter of 111 nm. (c) Current evolution during packing of fluorescently labeled gold particles at +1 V. Inset shows strong fluorescence only after 1 min of packing.
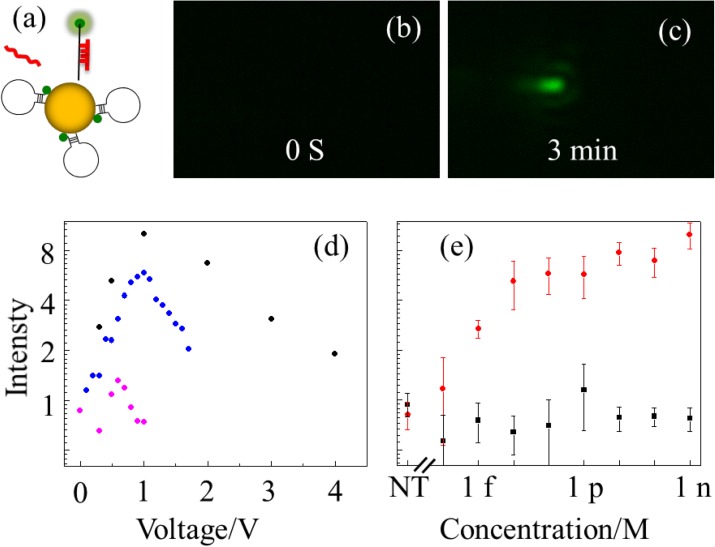
The FRET probe is designed to exploit the plasmonic hotspot.24 We first electrophoretically drove the target molecules in the tip side reservoir into the nano-capillary by applying a negative voltage of −1 V. During this process, the targets are trapped within the capillary and hybridize with the hairpin probes on the Au NP in the nanocapillary. Fluorescence of the unquenched hybridized probes is too weak to be detected by our detector as shown in Fig. 2b. A reverse positive voltage of +1 V was then applied to the capillary to pack the Au NPs to the tip. Due to plasmonic hot spots of aggregated gold nanoparticles, the fluorescence signal is significantly enhanced at the tip and can be detected by our CCD camera, as shown in Fig. 2c.
Figure 2.
(a) Schematics of designed hairpin probe on gold particle. (b) Before packing gold particles, probe fluorescence signal was too weak to be detect. (c) After packing for 3 minutes, a strong fluorescence signal appears at the NP aggregate. (d) Normalized intensity (average of all pixels above a threshold (15 au) normalized with respect to the average over all pixels (with 0-250 au)) as a function of packing voltage for different samples. Black, 1 nM target ; blue, 10 pM target; purple, 10 nM 2-mismatch non-target. (e) Intensity dependence on target concentration. Measured normalized intensity before packing (black) and after packing (red), for three independent experiments with different nano-capillaries at each concentration. NT stands for non-target at 10 nM as a reference.
For the same packing time, the fluorescence intensity increases initially but saturates after 10 min time of trapping (supplementary material, Fig. S2(a)24). Over 10 min of trapping with a negative voltage, we found the fluorescence intensity exhibits a maximum at a packing time of 3 min (supplementary material, Fig. S2(b)24). In later experiments, we used 10 min trapping time and 3 min packing time as standards.
Fig. 2d shows the fluorescence intensity is sensitive to the positive packing voltage at different concentration of target and non-target molecules. For target samples (1 nM and 10pM), the optimal voltage is about 1 V. We suspect that with larger voltage, the NPs are packed too tightly such that the NP spacing is smaller than the optimal distance for plasmonic hotspots. The fluorescence intensity for a nontarget with two mismatches is 7 times lower than the target even with a 10 times higher concentration (10 nM). Moreover, the optimal voltage for the non-target miRNA is reduced to 0.5 V instead 1 V for the target miRNA. Strong shear during electrophoretic packing has probably endowed this high selectivity.20
Using the protocol above, the LOD and dynamic range of the target was determined (Fig. 2e). The intensity at each concentration is measured with three independent experiments with different nanocapillaries to verify insensitivity with respect to the nanocapillary. The intensity increases monotonically with respect to the concentration from 1fM to 1pM. Beyond 1pM, the fluorescence signal saturates, presumably because all hairpin probes at the tip have been hybridized. At 1 fM, the fluorescent intensity is still well above the background measured from the non-target sample. Note both auto-fluorescence of gold nanoparticles and free diffusing non-target DNA molecules contribute to the background. Given the volume of tip side reservoir (∼50 μl), there are about 30 000 target molecules in the reservoir at 1 fM. However, with a short 10 min trapping time, we estimate only a small fraction of these molecules, less than 100, have been transferred from the tip reservoir into the nanocapillary.
Acknowledgments
This work was supported by NSF CBET 1065652.
References
- Croce C. M., Nat. Rev. Genet. 10, 704 (2009). 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., Cell 136, 215 (2009). 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G. S., Codony-Servat C., Byrne J., Ritchie L., and Hamilton A., Nucleic Acids Res. 35, e60 (2007). 10.1093/nar/gkm112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. M., Parker J., Perou C. M., and Hammond S. M., Nat. Methods 1, 47 (2004). 10.1038/nmeth704 [DOI] [PubMed] [Google Scholar]
- Raymond C. K., Roberts B. S., Garrett-Engele P., Lim L. P., and Johnson J. M., RNA 11, 1737 (2005). 10.1261/rna.2148705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ru E. C. and Etchegoin P. G., Annu. Rev. Phys. Chem. 63, 65 (2012). 10.1146/annurev-physchem-032511-143757 [DOI] [PubMed] [Google Scholar]
- Etchegoin P. G. and Le Ru E. C., Phys. Chem. Chem. Phys. 10, 6079 (2008). 10.1039/b809196j [DOI] [PubMed] [Google Scholar]
- Sagle L. B., Ruvuna L. K., Ruemmele J. A., and Van Duyne R. P., Nanomedicine 6, 1447 (2011). 10.2217/nnm.11.117 [DOI] [PubMed] [Google Scholar]
- Bhatnagar K., Pathak A., Menke D., Cornish P. V., Gangopadhyay K., Korampally V., and Gangopadhyay S., Nanotechnology 23, 495201 (2012). 10.1088/0957-4484/23/49/495201 [DOI] [PubMed] [Google Scholar]
- Ray P. C., Darbha G. K., Ray A., Walker J., and Hardy W., Plasmonics 2, 173 (2007). 10.1007/s11468-007-9036-9 [DOI] [Google Scholar]
- Tu Y., Wu P., Zhang H., and Cai C., Chem. Commun. 48, 10718 (2012). 10.1039/c2cc35564g [DOI] [PubMed] [Google Scholar]
- Du H., Strohsahl C. M., Camera J., Miller B. L., and Krauss T. D., J. Am. Chem. Soc. 127, 7932 (2005). 10.1021/ja042482a [DOI] [PubMed] [Google Scholar]
- Farahani J. N., Pohl D. W., Eisler H. J., and Hecht B., Phys. Rev. Lett. 95, 017402 (2005). 10.1103/PhysRevLett.95.017402 [DOI] [PubMed] [Google Scholar]
- Zhang J., Fu Y., Chowdhury M. H., and Lakowicz J. R., Nano Lett. 7, 2101 (2007). 10.1021/nl071084d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao E. and Schatz G. C., J. Chem. Phys. 120, 357 (2004). 10.1063/1.1629280 [DOI] [PubMed] [Google Scholar]
- De Angelis F., Patrini M., Das G., Maksymov I., Galli M., Businaro L., Andreani L. C., and Di Fabrizio E., Nano Lett. 8, 2321 (2008). 10.1021/nl801112e [DOI] [PubMed] [Google Scholar]
- Wang Y., Plouraboue F., and Chang H.-C., Opt. Express 21, 6609 (2013). 10.1364/OE.21.006609 [DOI] [PubMed] [Google Scholar]
- Dodson S., Haggui M., Bachelot R., Plain J., Li S., and Xiong Q., J. Phys. Chem. Lett. 4, 496 (2013). 10.1021/jz302018x [DOI] [PubMed] [Google Scholar]
- Gill R., Tian L. J., Somerville W. R. C., Le Ru E. C., van Amerongen H., and Subramaniam V., J. Phys. Chem. C 116, 16687 (2012). 10.1021/jp305720q [DOI] [Google Scholar]
- Cheng I. F., Senapati S., Cheng X., Basuray S., and Chang H.-C., Lab Chip 10, 828 (2010). 10.1039/b925854j [DOI] [PubMed] [Google Scholar]
- Hoffman P. D., Sarangapani P. S., and Zhu Y., Langmuir 24, 12164 (2008). 10.1021/la8013392 [DOI] [PubMed] [Google Scholar]
- Karhanek M., Kemp J. T., Pourmand N., Davis R. W., and Webb C. D., Nano Lett. 5, 403 (2005). 10.1021/nl0480464 [DOI] [PubMed] [Google Scholar]
- Thacker V. V., Ghosal S., Hernandez-Ainsa S., Bell N. A. W., and Keyser U. F., Appl. Phys. Lett. 101, 223704 (2012). 10.1063/1.4768929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4832095 for methods and estimation of plasmonic hotspots of dynamically assembled nanoparticles in nanocapillaries.
- Zhang J., Matveeva E., Gryczynski I., Leonenko Z., and Lakowicz J. R., J. Phys. Chem. B 109, 7969 (2005). 10.1021/jp0456842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D. and Xu Q.-H., Chem. Commun. 2007, 248. 10.1039/b612401a [DOI] [PubMed] [Google Scholar]
- Anger P., Bharadwaj P., and Novotny L., Phys. Rev. Lett. 96, 113002 (2006). 10.1103/PhysRevLett.96.113002 [DOI] [PubMed] [Google Scholar]