Abstract
Coat color dilution turns black coat color to blue and red color to cream and is a characteristic in many mammalian species. Matings among Netherland Dwarf, Loh, and Lionhead Dwarf rabbits over two generations gave evidence for a monogenic autosomal recessive inheritance of coat colour dilution. Histological analyses showed non-uniformly distributed, large, agglomerating melanin granules in the hair bulbs of coat color diluted rabbits. We sequenced the cDNA of MLPH in two dilute and one black rabbit for polymorphism detection. In both color diluted rabbits, skipping of exons 3 and 4 was present resulting in altered amino acids at p.QGL[37-39]QWA and a premature stop codon at p.K40*. Sequencing of genomic DNA revealed a c.111-5C>A splice acceptor mutation within the polypyrimidine tract of intron 2 within MLPH. This mutation presumably causes skipping of exons 3 and 4. In 14/15 dilute rabbits, the c.111-5C>A mutation was homozygous and in a further dilute rabbit, heterozygous and in combination with a homozygous frame shift mutation within exon 6 (c.585delG). In conclusion, our results demonstrated a colour dilution associated MLPH splice variant causing a strongly truncated protein (p.Q37QfsX4). An involvement of further MLPH-associated mutations needs further investigations.
Introduction
Coat color dilution in rabbits affects eumelanin as well as pheomelanin. A dilution of eumelanin in black and brown coat color leads to blue and cream-brown, respectively, while a dilution of pheomelanin in yellow coat color leads to cream-yellow [1]. The rabbit dilution allele (d) is a monogenic, autosomal recessive trait [1]. Besides the rabbit, color dilution of the same phenotype is known and often favoured by selective breeding in many different animal species including cats [2], chicken [3], quails [4], mice [5], foxes [6], and minks [7]. Dilution of the coat color is also seen in dogs where color-dilution can be accompanied by alopecia [8-10]. This defect causes poor quality of the hair coat and hair re-growth to the point of progressive and extensive hair loss as well as comedo formation [9]. It was not reported for the dilution phenotype in any of the other species mentioned above. In humans, Griscelli syndromes are characterized by pigmentary dilution due to large agglutinations of pigment in the hair shafts and accumulation of melanosomes in melanocytes [11]. Similar histological findings including uneven granule-distribution and pigment agglutinations especially around the nucleus were found in dilute cats [2], foxes [6] and mice [5,12]. In diluted dogs, hair shafts, follicles, and bulbs showed a massive, abnormal clumping of melanin in the epidermis, dermis, and hair follicles [9,10,13]. For the coordination of melanosome capture, transport, and distribution, a tripartite protein complex of Rab27A, melanophilin, and Myosin Va was shown to be essential [14]. In this complex, Rab27A binds to melanosomes and captures melanophilin, which subsequently captures myosin Va. Myosin Va again binds to actin filaments and thus facilitates melanosome transport [14-17]. In human, mutations within any of the genes RAB27A (RAB27A, member RAS oncogene family), MLPH (melanophilin), and MYO5A (myosin VA) encoding for the proteins of the tripartite complex can cause Griscelli syndromes. However, dilution of the hair is commonly accompanied by further symptoms depending on the defective gene [18]. Griscelli syndrome type 1 (GS1) is caused by mutations of MYO5A and hypomelanosis is accompanied by neurologic deficits [19], while Griscelli syndrome type 2 (GS2) is caused by RAB27A mutations and patients show immune impairment in combination with the hair color dilution [20]. Griscelli syndrome type 3 (GS3) is caused by mutations within MLPH or MYO5A [21] and only MLPH defects are usually not accompanied by severe clinical diseases [22]. The human Griscelli phenotypes GS1, GS2, and GS3 correspond to the dilute, ashen, and leaden phenotypes in the mouse, respectively [23] and the human Griscelli phenotype GS1 corresponds to the lavender foal syndrome in the horse [24].
In this study, we analysed the dilute coat color in rabbits. MLPH was chosen as the most likely candidate gene due to histological analyses and phenotypic examination. We sequenced the cDNA of MLPH in black and dilute rabbits and performed mutation screening for the dilution phenotype.
Results
Phenotypes and Histological Analyses
In total 23 Netherland Dwarf, Loh, and Lionhead Dwarf rabbits and their crosses of our breeding trial, including six individuals showing coat color dilution, were clinically examined for defects accompanying the dilute color. No abnormal clinical findings were detected associated with this phenotype, neither serious clinical defects nor less severe signs like color dilution alopecia or comedo formations. The oldest rabbit showing the dilution phenotype was more than three years of age at examination. The phenotypes of a black and a blue colored rabbit are shown in Figure 1.
Figure 1. Phenotypic appearance of black (wildtype) and blue (dilute) rabbits.
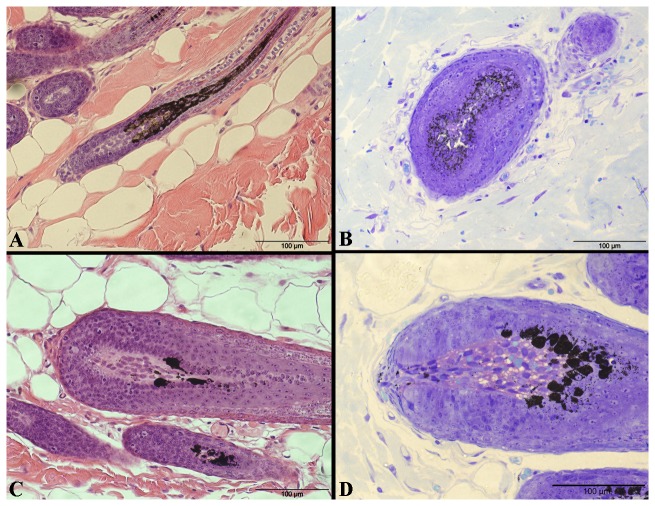
Subsequently, histological analyses were performed using skin samples of black rabbits (n=6) and blue rabbits (n=4). Cells of the hair bulb matrix from black rabbits contained large numbers of fine melanin granules which are moderately and uniformly distributed in the cytoplasm. Nuclei in the horizontally sectioned hair bulb of black rabbits were free of melanin (Figure 2). The melanin granules in the hair bulbs from blue rabbits differ in several aspects from those in black ones. The majority of the melanin granules seem somewhat larger and form aggregates (melanin clumps). The pattern of melanin granules in hair matrix cells shows significant differences (P-value<0.01) between black and blue rabbit hair bulbs (Figure 3). These results are based on a qualitative scoring system, which was then semi-quantitatively analysed. It is important to note that blue rabbits only form these melanin clumps, yet in the blue rabbit hair bulbs, >90% of the hair matrix cells contain melanin clumps but no melanin granules. In contrast, approximately 50% of hair matrix cells of black rabbits contain either few or several melanin granules. Furthermore, the pattern of melanin granule distribution differs from dilute individuals as granule containing cells are uniformly distributed throughout the hair bulb matrix in black rabbits (Figures 2 and 3).
Figure 2. Representative light microscopic comparison of black (A, B) and blue (C, D) rabbit skin samples.
HE-staining (A, C) and Toluidine blue O staining (B, D) was performed. Sagittal sections through hair bulbs are shown in A, C, D, and horizontal section through hair bulb is shown in B. Note the uneven distribution pattern of melanin granules and melanin clumps in hair matrix cells of hair bulbs of blue rabbit skin samples. Scale bars = 100 µm.
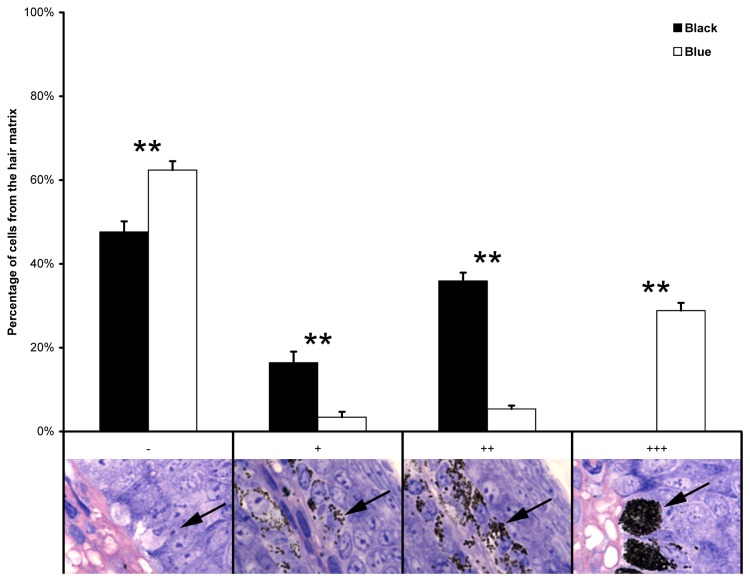
Figure 3. Proportions of cells with melanin granules in the hair matrix layer of hair bulbs for black and blue rabbits using a semi-quantitative analysis.
The scoring system is: “-” = no melanin granules, “+” = few melanin granules, “++” = several melanin granules, “+++” = many melanin granules (several appear as melanin clumps). Scoring is represented in semi-thin sections (Technovit®) below the graph. Arrows point to characteristic cells of each category/score. Original magnification of insets is 63x. Statistical analysis revealed each single scores as significantly different (**, P-value<0.01) between black and blue rabbits.
Sequencing MLPH and Mutation Analysis
The oryctolagus cuniculus (OryCun2.0) map viewer of NCBI (http://www.ncbi.nlm.nih.gov/projects/mapview) locates rabbit MLPH on an unknown chromosome. MLPH is designated LOC100343360, which contains 35,141 bp genomic sequence. We ascertained the complete coding sequence of MLPH in one black (DD) and two dilute (dd) rabbits, including all exon-exon boundaries and 51 bp of the 5’ untranslated region (UTR) and 60 bp of the 3’ UTR (Figure 4). We identified a 1,689 bp open reading frame (ORF) in the cDNA of a black rabbit, which was translated into a protein of 563 amino acids. A total of 15 exons were detected in the DD-rabbits. In the cDNA of both dd-rabbits, however, exons 3 and 4 were not present and exon 2 was directly spliced to exon 5 (Figure 5). This exon skipping caused a frameshift resulting in a change of two amino acids (p.QGL[37-39]QWA) followed by a premature stop codon (p.K40*) (Figure S1). In the next step, we sequenced 570 bp genomic DNA containing 51 bp of intron 2, exon 3 (222 bp), intron 3 (92 bp), exon 4 (113 bp), and 92 bp of intron 4 in two black and two dilute rabbits. In total, 32 polymorphisms were detected within the exons of MLPH and two polymorphisms were detected within intron 2 and 3, respectively (Table 1). Of all exonic polymorphisms, twelve caused amino acid exchanges and were analysed for a potentially damaging effect. Two polymorphisms within exon 3 were classified as probably damaging (c.214CT, c.262AG). A deletion of one base pair within exon 6 (c.585delG), may be regarded as probably damaging as it causes a frameshift leading to a premature stop codon. This stop codon is located 166 triplets downstream of c.585delG in both dilute rabbits sequenced. In the case of the presence of the wild type allele at c.953TC, the stop codon is 123 triplets downstream of the deletion (Figure S1). Analysing the genomic DNA containing exon 3 and 4, no polymorphisms affecting a branch site or an invariant base of a splice site were found. However, we detected a SNP (c.111-5C>A) five base pairs upstream of the 5’end of exon 3 within the polypyrimidine tract. The pyrimidine base seems to be conserved at this position after comparing 35 species (Table S1). Only in chicken, a guanine is found at this location in some sequences. Another polymorphism located within the 5’ UTR of MLPH (c.1-10A>G) also seems to be located at a position conserved in mammals and marsupials (Table S1).
Figure 4. Structure of the melanophilin (MLPH) gene.
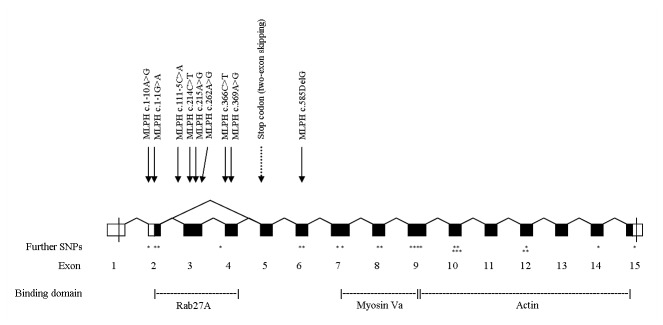
Coding regions of exons are painted black, the 5’ and 3’ untranslated regions are white. The positions of the nine SNPs chosen for further analyses are indicated by arrows. Further SNPs detected within MLPH are indicated by asterisks. The premature stop codon caused by the skipping of exons 3 and 4 in dilute rabbits is indicated by a dashed arrow. The functional protein binding domains Rab27A, Myosin Va and Actin are given.
Figure 5. Comparisons of the cDNA sequences for melanophilin among a colour dilute rabbit and a black wild type rabbit.
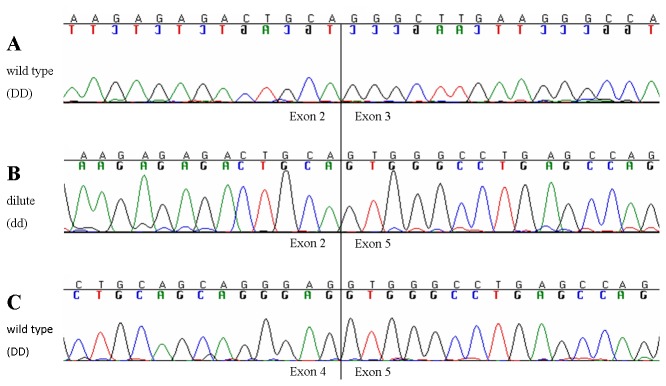
The sequences show the boundary of exon 2 to exon 3 (A) as well as of exon 4 to exon 5 (C) in the wild type rabbit in comparison with the sequence of a dilute rabbit (B), where exon 2 is spliced to exon 5. The vertical line marks the boundaries.
Table 1. SNPs detected within melanophilin (MLPH), their location, predicted effects on the protein sequence and protein function are given.
| Polymorphism | Location | Amino acid | Predicted effect |
|---|---|---|---|
| within | exchange | ||
| MLPH | |||
| c.1-11C>T | exon 2 | - | |
| c.1-10A>G | exon 2 | - | |
| c.1-1G>A | exon 2 | - | |
| c.48AG | exon 2 | - | |
| c.52GA | exon 2 | p.V18I | Benign |
| c.111-5C>A | intron 2 | (p.Q37QfsX4) | Truncated protein |
| c.214CT | exon 3 | p.Q72W | Probably damaging |
| c.215AG | exon 3 | p.Q72R | Benign |
| c.262AG | exon 3 | p.T88A | Probably damaging |
| c.333-17insGC | intron 3 | - | |
| c.366CT | exon 4 | - | |
| c.369AG | exon 4 | - | |
| c.585delG | exon 6 | p.L195LfsX123* | Truncated protein |
| p.L195LfsX166* | |||
| c.610TC | exon 6 | p.R204W | Benign |
| c.642CG | exon 6 | - | |
| c.718AG | exon 7 | p.T240A | Benign |
| c.774AT | exon 7 | - | |
| c.941GC | exon 8 | p.G314A | Possibly damaging |
| c.953TC | exon 8 | p.V318A | Benign |
| c.1029AG | exon 9 | - | |
| c.1041GA | exon 9 | - | |
| c.1092CT | exon 9 | - | |
| c.1143TC | exon 9 | - | |
| c.1197TC | exon 10 | - | |
| c.1198AG | exon 10 | p.G400S | Benign |
| c.1206AG | exon 10 | - | |
| c.1209TC | exon 10 | - | |
| c.1284AG | exon 10 | - | |
| c.1440CG | exon 12 | - | |
| c.1462GA | exon 12 | p.A488D | Possibly damaging |
| c.1463AG | exon 12 | p.A488D | Possibly damaging |
| c.1482AG | exon 12 | - | |
| c.1602CT | exon 14 | - | |
| c.1689+45GC | exon 15 | - |
Predicted effects on protein function were obtained using PolyPhen2 software. The c.111-5C>A mutation presumably causes dilution in rabbits due to two-exon skipping.
* Depending on the c.953TC genotype
To exclude the presence of an inversion, which might include exon 3 and 4 and therefore cause the two-exon skipping, we performed a long range PCR with the forward primer located within exon 3 and the reverse primer located within exon 5. In the two black as well as in the two dilute rabbits, we obtained PCR products of the appropriate size. Therefore, an inversion affecting this region could be excluded.
All polymorphisms with predicted probably damaging effects (c.214CT, c.262AG, c.585delG) as well as the two polymorphisms located within conserved, non-coding regions (c.1-10A>G, c.111-5C>A) and four further polymorphisms located within exon 2 (c.1-1G>A), exon 3 (c.215AG) or exon 4 (c.366CT, c.369AG) were analysed in all 23 rabbits of the breeding trial. The SNPs c.1-1G>A, c.111-5C>A, c.366CT, c.369AG, and c.585delG showed associations with the dilute phenotype and were subsequently genotyped in further 30 rabbits of several breeds, which were not related with those of the breeding trial. Of these rabbits, seven showed the dilution phenotype, two rabbits showed a coat color which was classified as dark-dilute and 21 rabbits were wild type colored. The polymorphisms c.1-1G>A, c.111-5C>A, c.366CT, and c.369AG were highly correlated with each other at r2>0.9 (Figure S2).
Association Analysis
We performed a case-control analysis for the SNPs c.1-1G>A, c.111-5C>A, c.366CT, c.369AG, and c.585delG with coat colour dilution (Table S2). The P-values for association with the dilution phenotype were lowest for c.111-5C>A (1.5x10-10 to 6.8x10-7) and c.585delG (1.3x10-10 to 1.8x10-7). The c.111-5C>A mutation was homozygous in 14/15 colour diluted rabbits and the c.585delG deletion in 13/15 colour diluted rabbits (Table 2). Out of the 15 colour diluted rabbits, twelve individuals were homozygous for the mutations, c.111-5C>A and c.585delG. One colour diluted individual homozygous for the A/A genotype at c.111-5C>A, carried the wild type alleles (wt/wt) at c.585delG. A further at c.111-5C>A homozygous A/A rabbit was heterozygous at c.585delG. This rabbit showed a darker blue coat color and was classified as dark-dilute. The same dark-dilute phenotype was seen in a rabbit being heterozygous A/C at c.111-5C>A, but homozygous del/del at c.585delG. The c.953TC mutation was not conclusive for distinguishing dilute and dark-dilute phenotypes (Table S3).
Table 2. Distribution of the melanophilin (MLPH) c.111-5C>A and c.585delG mutations in rabbits of different breeds.
| Rabbit breed | Fur color | Dilute genotype | Number of animals | SNP c.111-5C>A | SNP c.585delG | ||||
|---|---|---|---|---|---|---|---|---|---|
| C/C | A/C | A/A | w/w | w/del | del/del | ||||
| Netherland | Wild Type | D- | 11 | x | x | ||||
| Dwarf | Wild Type | D- | 2 | x | x | ||||
| Wild Type | D- | 2 | x | x | |||||
| Dilute | dd | 5 | x | x | |||||
| Lionhead | Wild Type | D- | 1 | x | x | ||||
| Dwarf | Wild Type | D- | 2 | x | x | ||||
| Loh | Wild Type | D- | 2 | x | x | ||||
| Netherland | Wild Type | D- | 5 | x | x | ||||
| Dwarf x Loh | Wild type | Dd | 5 | x | x | ||||
| Dilute | dd | 4 | x | x | |||||
| Vienna Blue | Dilute | dd | 2 | x | x | ||||
| Dwarf Lop | Wild Type | D- | 2 | x | x | ||||
| Wild Type | D- | 2 | x | x | |||||
| Dilute | dd | 1 | x | x | |||||
| Giant Lop | Wild Type | D- | 1 | x | x | ||||
| Checkered | Wild Type | D- | 1 | x | x | ||||
| Giant | |||||||||
| Rex | Wild Type | D- | 1 | x | x | ||||
| Angora | Dilute | dd | 1 | x | x | ||||
| Lionhead | Dark-dilute | dd* | 1 | x | x | ||||
| Dwarf | |||||||||
| Loh | Dark-dilute | dd* | 1 | x | x | ||||
The dilution status and genotype with corresponding number of animals and SNP genotypes are given. Bold faced letters indicate the dilute individuals, in which SNP genotypes differ between c.111-5C>A and c.585delG. The two individuals with less obvious dilution status are given at the bottom of the table. Their dilution status is indicated as “dark-dilute” (dd*).
The alleles of the SNPs c.366CT and c.369AG were in high linkage disequilibrium with the alleles of c.111-5C>A, but not as strongly associated with the colour dilution as the c.111-5C>A mutation.
Therefore, genotyping the c.111-5C>A and c.585delG mutations in parallel allowed detection of the dilute phenotype in all rabbits analysed in this study. The c.111-5C>A dilution allele was homozygously or heterozygously detected in the breeds Netherland Dwarf, Lionhead Dwarf, Vienna Blue, Dwarf Lop, Checkered Giant, Rex, Angora, and Loh.
Discussion
In the present study we detected mutations within MLPH, co-segregating with the dilution phenotype in the nine rabbit breeds tested. MLPH was selected as candidate gene due to histological as well as clinical examination of the rabbits within a breeding group of Netherland Dwarf, Loh, and Lionhead Dwarf rabbits and their crosses. Only in blue rabbits, our histological analysis revealed clumps of melanin in hair matrix cells as well as a non-uniformly distribution pattern of the melanin granules throughout the hair bulb matrix. No abnormal clinical findings could be ascertained in the rabbits examined in the present study. Pigmentary dilution due to large agglutinations of pigment in the hair shafts and accumulation of melanosomes in melanocytes are characteristics of human Griscelli syndromes [11]. The dilution phenotype of the human Griscelli syndrome type 3 caused by MLPH mutations is usually not accompanied by severe clinical diseases [21]. This is in consistency with observations in the mouse [23]. Therefore, MLPH was sequenced as it was the most likely functional candidate gene.
The polymorphism c.111-5C>A is the most probable cause for dilute coat color in rabbits. Skipping of two exons was detected in 14/15 dilute rabbits, which is likely caused by the polymorphism c.111-5C>A. The exon skipping causes a premature stop codon at the fourtieth amino acid of the transcript (p.Q37QfsX4), while c.585delG is located further downstream. Both mutations are in most, but not all cases within the same linkage phase. In a single case of a dark-dilute rabbit showing a heterozygous genotype at c.111-5C>A and the homozygous deletion at c.585delG, the latter mutation might be of relevance, as it also leads to a frameshift and an altered amino acid sequence with a premature stop codon 123 or 166 amino acids downstream (p.L195LfsX123 or p.L195LfsX166), respectively (dependent on the polymorphism c.953TC). The c.585delG polymorphism therefore might have an effect on coat colour dilution when the individual has not the homozygous A/A genotype at c.111-5C>A.
Though c.366CT and c.369AG were in high linkage disequilibrium with c.111-5C>A, their wild type allele is not conserved among different species (Table S1). Therefore, they are not supposed to influence the exon skipping through, for example, the destruction of an exonic splicing enhancer. Further variants classified as potentially damaging and located within conserved regions of MLPH showed only low associations with the dilution phenotype. They may be assumed to have arisen subsequently to the causal mutations, due to missing selection pressure.
The MLPH two-exon skipping seen in dilute rabbits leads to a frameshift and results in a change in the amino acid sequence (p.QGL[37-39]QWA) followed by a premature stop codon (p.K40*). This mutation eliminates the myosin Va and the actin binding domain and leaves only 37/153 aa of the region containing the RAB27A binding domain [14,25,26]. As for the coordination of melanosome transport, and distribution, a tripartite protein complex of Rab27A, melanophilin, and Myosin Va is essential [14], and it can be assumed that the p.Q37QfsX4 protein is not functional. In case of a normal transcript without exon skipping, the frameshift mutation c.585delG (p.L195LfsX123 or p.L195LfsX166) also causes a change of the amino acid sequence and truncation of the protein, affecting the complete myosin Va and actin binding domain [25].
Mutations for the diluting genotype were determined in several other species. In cats, dilution of the coat color is caused by the deletion of one base-pair within exon 2, leading to a premature stop codon eleven amino acids downstream [26]. This mutation therefore shows a similarly truncated protein like the one in the present study. Most of the further mutations causing a dilution phenotype in different species are single base exchanges within the RAB27A binding domain. In lavender chicken and also in human Griscelli syndrome type 3, the causal mutation is a R35W substitution [3,22]. Leaden colored mice show a deletion of 6 amino acids on positions 31 to 37 [23]. In dogs and quails, however, other mutations have been described. In dogs, a polymorphism at the last nucleotide of the first, untranslated exon of MLPH was completely associated with coat color dilution [27] and in quails, the causal mutation is quite complex, as it includes different chromosomal rearrangements, which affect MLPH as well as three further genes [28].
The c.111-5C>A mutation appears as the most probable candidate to cause the two-exon skipping in dilute rabbits. It is located within the splice acceptor polypyrimidine tract of MLPH intron 2, five base pairs upstream of the start of exon 3, at a location which is supposed to contain a pyrimidine base. The percentage of occurrence of a pyrimidine in this position is at 87% in vertebrate genes [29]. While exon skipping due to mutations of the splice acceptor site usually affect the two invariant nucleotides A and G at positions -1 and -2, exon skipping defects resulting from mutations at positions -14 to -3 (Y(10)NC, [29]) are rare. However, mutations affecting different positions within the polypyrimidine tract have been previously described to cause exon skipping. A T>A mutation at position -6 within GHR was recently described to cause exon skipping by disruption of the polypyrimidine tract [30]. Further mutations causing partial or complete skipping of the subsequent exon were reported for the positions -11 (T>A [31]) within MLH1, and two mutations within intron 2 of ß globin at -8 (T>G [32-33] and -7 (C>G [34]), respectively. The polypyrimidine tract has been shown to be of importance during the splicing process and for branch site definition [35,36]. Efficient splicing depends on the AG dinucleotide at the 3' splice junction, but also on the length of the polypyrimidine stretch [37]. The total length of the polypyrimidine tract seems to play a major role for the impact of a pyrimidine to purine exchange within this tract [38]. Furthermore, the thymine content of the polypyrimidine tract may affect splicing efficiency [39]. The polypyrimidine tract of the second intron of MLPH counts 19 pyrimidines in rabbits, which is of medium size [37], including seven thymines and twelve cytosines. This number of thymines in the polypyrimidine tract seems low, as the probability of a thymine is higher than that of cytosine for the complete polypyrimidine tract except for position -6 [29]. Therefore, the polypyrimidine tract might already be weak. In dilute rabbits, the last cytosine at position -5 is changed to adenine, which might impede the splicing process. However, the effects of mutations affecting the polypyrimidine tract are not completely understood and also in silico predictions of mutation effects in this region have to be experimentally confirmed [30].
A further characteristic in the present study is the skipping of not only the third, but also of the fourth exon. Missing of more than one exon can be due to an inversion or a deletion of genomic DNA [40]. This was excluded in the present study by sequencing of genomic DNA and long range PCR. Two-exon skipping can also occur because of an alternatively spliced adjacent exon [41], which is also not known for exon 3 and 4 of MLPH. The remaining cases previously described are rare and due to splice mutations [40,42-46]. A two-exon skipping was explained by the order of intron removal [44]. A splice acceptor mutation caused skipping of exons 5 and 6 of COL5A1 due to rapid removal of intron 5 from the transcript relative to introns 4 and 6, which left a large composite exon 5/exon 6 construct that was skipped entirely. This might be also the case in the present study, as intron 3 between the two skipped exons only comprises 92 base pairs and therefore is the smallest within the complete gene, while intron 2 and 4 contained more than 6000 and 4500 base pairs, respectively. It may be assumed, that exons 3 and 4 fuse early during the splicing process and subsequently the c.111-5C>A mutation prevents splicing of the combined exon structure. Another explanation of a two-exon skipping was based on the detection of a splice donor (+5) mutation within intron 13 of OXCT1. This splice donor mutation led to retention of intron 13, thus causing the retention of intron 11 and 12. This so called splicing paralysis resulted in skipping the introns 11-13 and exons 12 and 13 [46]. In our data, we could not see retention of an intron in cDNA and thus, this latter explanation seems unlikely for the MLPH-associated dilute phenotype in rabbits.
In conclusion, the dilution phenotype in rabbits is likely caused by a two-exon skipping of exon 3 and 4 and a frameshift of the open reading frame causing a premature stop codon (p.Q37QfsX4). The c.111-5C>A mutation, located within intron 2 of MLPH, is the most probable cause for this exon skipping and therefore for the dilute phenotype in rabbits. The dilute phenotype seems to be further influenced by another frame-shift mutation (c.585delG), but only one individual was exclusively homozygous for this genotype. This deletion does not affect the RAB27A binding domain like the c.111-5C>A mutation. The mutations detected herein in rabbits expand the contingent of known dilution mutations in different species and provide an additional view on the splicing mechanism.
Materials and Methods
Ethics Statement
All animal work has been conducted according to the national and international guidelines for animal welfare. The Lower Saxony state veterinary office at the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany, was the responsible Institutional Animal Care and Use Committee (IACUC) for this specific study. The breeding experiment and EDTA-blood sampling of the rabbits for the present study had been approved by the IACUC of Lower Saxony, the state veterinary office Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany (registration number 33.9-42502-04-11/0563). Tissue samples were taken from rabbits, which had to be euthanized due to other medical reasons (after inducing a deep anesthesia by intramuscular injection of 5 mg/kg xylazine and 35 mg/kg ketamine, 2 ml of T61 was applied intrapulmonal for euthanasia). For blood or hair sampling of living animals, no anesthesia was applied as this would have meant at least equivalent stress than taking a sample. Each rabbit was only sampled once. Samples from rabbits of the further breeds were taken in the context of diagnostic analyses by a veterinarian at the Clinic for Pets, Reptiles and Pet and Feral Birds, University of Veterinary Medicine Hannover. We obtained informed consent from all owners of the rabbits.
Animals
We obtained blood or hair root samples of a total of 53 rabbits, whereof 23 animals were rabbits of the breeding trial (Figure S3). In this breeding trial, a Netherland Dwarf male showing fur dilution (blue coat color) and descending from a dilute x non-dilute mating was crossed with two black Loh females and one black Lionhead Dwarf female. None of the descendants showed diluted fur. Two Loh x Netherland Dwarf females were again crossed with the dilute Netherland Dwarf male. Of the resulting offspring, four individuals showed blue fur, five had black fur and one was completely white, which rendered a phenotypical determination of the dilution phenotype impossible. In total, of all these 23 rabbits, six showed fur dilution.
All rabbits were examined by veterinarians at the Clinic for Pets, Reptiles and Pet and Feral Birds and at the Institute for Animal Breeding and Genetics, University of Veterinary Medicine Hannover Foundation for defects accompanying the dilute coat color like in dogs [8,9,23] or in human Griscelli syndrome of types 1 [17] and 2 [18]. No difference in the health status was observed between rabbits with dilute and non-dilute color. The rabbits were between six months and four years of age when examined, the oldest dilute rabbit was three and a half years old.
The other 30 rabbits were from several breeds (Netherland Dwarf, Loh, Lionhead Dwarf, Vienna Blue, Rex, Angora, Dwarf Lop, Giant Lop, Checkered Giant). Of these rabbits, 21 were wild type colored, seven showed dilute fur color and two rabbits showed a darker blue or blue-brownish coat color, which was classified as dark-dilute and might be a variant of the dilution phenotype.
Genomic DNA was extracted from EDTA-blood or hair samples using routine procedures. Tissue samples for the RNA extraction were skin tissue of one black and two dilute rabbits. These samples were stored in RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) for stabilization and protection of cellular RNA in situ for storing at -20°C. The RNA was extracted from tissue samples using the RNeasy Lipid Tissue Mini Kit (Qiagen) according to manufacturer's protocol and transcribed into cDNA using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Fermentas Life Sciences, St. Leon-Rot, Germany).
Histological analysis
Skin samples of six black and four blue rabbits were fixed in Bouin’s solution and Ca-formol and were embedded in paraffin wax (Paraplast Standard, Leica Microsystems, Wetzlar, Germany) according to standard protocols [47]. The paraffin blocks were sectioned with a rotary microtome (Leica Microsystems, Wetzlar, Germany), and sections not thicker than 5 μm were used for staining.
Moreover, small Ca-formol fixed tissue blocks of about 0.5 mm3 were prepared, carefully dehydrated with graded ethanol (70, 80, 90, 2×96, 2×100%) and embedded in the water-soluble and rather shrinkage-free 2-hydroxy-methacrylate Technovit 7100 (Heraeus-Kulzer, Wehrheim, Germany [48,49]). Two µm plastic sections were cut with a motor-driven rotation microtome (Model 1140, Autocut, Reichert-Jung) and transferred to slides.
Paraffin sections of all collected samples were stained with haematoxylin and eosin (H&E, haematoxylin according to Delafield) and plastic sections were stained with Toluidine blue O [47].
All skin samples (six black rabbits, four blue rabbits) were investigated semi-quantitatively for their melanin granule distribution in hair matrix cells. A total of sixty longitudinal histological hair bulb sections per phenotype were separately evaluated by three scientists using a qualitative scoring system. The scoring system distinguished four categories including “-” = no melanin granules, “+” = few melanin granules, “++” = several melanin granules, “+++” = many melanin granules (several appear as melanin clumps). Within each category, the presence of the respective score was then subjected to a statistical analysis using a one-way ANOVA test. P-values<0.05 were defined as significant with * = P-value<0.05 and ** = P-value<0.01.
Sequence and mutation analysis
For mutation analysis of rabbit MLPH, we used cDNA of one black Loh and two dilute rabbits whereof one was a Netherland Dwarf and the other one was a cross of a Netherland Dwarf (75%) x Loh (25%). Four pairs of primers were designed based using the rabbit MLPH cDNA sequence obtained by Ensembl (www.ensembl.org) and the Primer3 software (http://frodo.wi.mit.edu/). These primers yielded products covering the complete MLPH open reading frame and all exon-exon boundaries, as well as 51 bp of the 5’ UTR and 60 bp of the 3’ UTR. The exons 3 and 4 of MLPH, which were skipped in dilute rabbit cDNA, were sequenced on genomic DNA of the same three animals used for cDNA sequence analysis and in addition, a black Netherland Dwarf x Loh crossbred with the genotype Dd was sequenced. For this purpose, two pairs of primers were designed as described above. To exclude an inversion including exon 3 and 4, we further performed a long range PCR on genomic DNA of two black (DD and Dd) and two dilute (dd) rabbits, with the forward primer located within exon 3 and the reverse primer located within exon 5. Products of the appropriate size in all individuals in combination with the genomic sequences containing exons 3 and 4 would exclude the presence of an inversion. All primer pairs are shown in Table S4.
PCRs were carried out according to the standard protocol advised by the manufacturer of the TaqDNA polymerase (Qbiogene Heidelberg, Germany). Sequencing of the PCR products was performed using the ABI BigDye Terminator v3.1 sequencing kit (Life Technologies, Darmstadt, Germany). The products were analysed on an automated ABI 3500 capillary sequencer (Life Technologies). Mutation analysis was performed using Sequencher 4.8 (GeneCodes, Ann Arbor, MI, USA). The open reading frame was determined using Open Reading Frame Finder (http://www.ncbi.nlm.nih.gov/gorf/). Translation into protein sequence was done using CLUSTALW (http://www.genome.jp/tools/clustalw/). In the next step, all detected polymorphisms were analysed for their potentially damaging effect using PolyPhen2 [50]. All polymorphisms, which were classified as probably damaging and all polymorphisms located outside of the reading frame but within highly conserved areas were genotyped in the 23 rabbits from the breeding trial using PCR-RFLPs. Then all polymorphisms associated with the dilute phenotype were genotyped in all 52 rabbit samples with a colored coat. All primer pairs for genotyping are given in Table S4. Mismatch PCR was partly used to create restriction sites for specific enzymes [51]. Digestion took place using specific restriction enzymes (Table S4) and the resulting products were separated on agarose gels. Association of SNPs with the dilute phenotype was tested using the CASECONTROL procedure of SAS/Genetics (Statistical Analysis System, version 9.3, Cary, NC, USA, 2013). Statistical calculation of pairwise linkage disequilibrium (LD) was performed and pictured using HAPLOVIEW 4.0 [52].
Accession Numbers
Sequence data has been deposited at the GenBank Data Libraries (KC791692, KC791692).
Supporting Information
Comparison of the different variants of truncated melanophilin proteins in dilute rabbits (variants 2-4) with the complete protein in wild type rabbits (1). Variant 2 is a result from skipping of exon 3 and 4 of MLPH (p.Q37QfsX4). Exon skipping is presumably caused by the c.111-5CγA transversion. Variants 3 and 4 (p.L195LfsX123 and p.L195LfsX166, respectively) are caused by the c.585delG mutation. Which one of these variants is generated depends on a further polymorphism affecting the premature stop codon of variant 3 (c.953T>C). The RAB27A domain is highlighted in green.
(DOC)
Linkage disequilibrium (LD) of the nine SNPs selected for genotyping in the 23 rabbits of the breeding trial (c.1-10AγG, c.214A>G, c.215A>G and c.262A>G) and in all 53 rabbits of different breeds (c.1-1GγA, c.111-5CγA, c.366C>T, c.369A>G and c.585delG). The latter five SNPs showed strong LD with the dilute phenotype. The pairwise r2-values are shown for each SNP pair. Red squares indicate complete linkage disequilibrium.
(DOC)
Pedigree of the rabbits from the breeding trial and the haplotypes of nine polymorphisms genotyped for these animals. Haplotypes include the polymorphisms c.1-10AγG, c.1-1GγA, c.111-5CγA, c.214A>G, c.215A>G, c.262A>G, c.366C>T, c.369A>G and c.585delG.
(DOC)
Conservation of nucleotides in different species at the melanophilin (MLPH) SNP positions detected in rabbits. Nucleotides in brackets were located at position c.-9, but in that species, this position matched with c.1-10 in other species due to the surrounding sequence.
(DOC)
Case-control analysis for five melanophilin-associated polymorphisms with the dilution phenotype in 15 dilute and 37 colored non-dilute rabbits. The χ2- and P-values are shown for genotype, allele and trend tests.
(DOC)
Distribution of the melanophilin (MLPH) c.111-5CγA, c.585delG and c.953T>C mutations in rabbits of different breeds. The dilution status and genotype with corresponding number of animals and SNP genotypes are given. Bold faced letters indicate the dilute individuals, in which SNP genotypes differ between c.111-5C>A and c.585delG. The two individuals with less obvious dilution status are given at the bottom of the table. Their dilution status is indicated as “dark-dilute” (dd*).
(DOC)
Primer sequences with their product sizes (P) for amplification of sequences within melanophilin (MLPH) to be used for sequencing, long range PCR or genotyping as well as the targeted region and the appropriate template. The primers were designed to work at an annealing temperature of 60°C. In the case of performing a mismatch PCR for digestion, the base pairs, which were changed within a primer relative to the reference sequence, are highlighted in gray. If the product of a pair of primers was enzymatically digested for genotyping, the enzyme used is given.
(DOC)
Acknowledgments
We thank Heike Klippert-Hasberg and Mogens Kilian Drabert for their expert technical support in sequence analyses. We also thank Franziska von Graevemeyer and Birte Erhardt for their support in histological evaluations. Holger Lönneker, Kerstin Liebig, Ann-Kathrin Kachler and Mogens Kilian Drabert are thanked for rabbit care.
Funding Statement
The project has been supported by internal funds of the University of Veterinary Medicine Hannover. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pap E (1921) Über Vererbung von Farbe und Zeichnung bei dem Kaninchen. Z Indukt Abstamm Vererbungsl 26: 185-270. [Google Scholar]
- 2. Prieur DJ, Collier LL (1981) Morphologic basis of inherited coat-color dilutions of cats. J Hered 72: 178-182. PubMed: 7276525. [DOI] [PubMed] [Google Scholar]
- 3. Vaez M, Follett SA, Bed'hom B, Gourichon D, Tixier-Boichard M et al. (2008) A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet 9: 7. doi: 10.1186/1471-2156-9-7. PubMed: 18197963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Minvielle F, Cecchi T, Passamonti P, Gourichon D, Renieri C (2009) Plumage colour mutations and melanins in the feathers of the Japanese quail: a first comparison. Anim Genet 40: 971-974. doi: 10.1111/j.1365-2052.2009.01929.x. PubMed: 19496774. [DOI] [PubMed] [Google Scholar]
- 5. Russel ES (1948) A quantitative histological study of the pigment found in the coat color mutants of the house mouse. II. Estimates of the total volume of pigment. Genetics 33: 228-236. [DOI] [PubMed] [Google Scholar]
- 6. Bradbury MW, Fabricant JD (1988) Changes in melanin granules in the fox due to coat color mutations. J Hered 79: 133-136. [DOI] [PubMed] [Google Scholar]
- 7. Anistoroaei R, Christensen K (2007) Mapping of the silver gene in mink and its association with the dilution gene in dog. Cytogenet Genome Res 116: 316-318. doi: 10.1159/000100417. PubMed: 17431331. [DOI] [PubMed] [Google Scholar]
- 8. Philipp U, Hamann H, Mecklenburg L, Nishino S, Mignot E et al. (2005) Polymorphisms within the canine MLPH gene are associated with dilute coat color in dogs. BMC Genet 6: 34. doi: 10.1186/1471-2156-6-S1-S34. PubMed: 15960853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JH, Kang KI, Sohn HJ, Woo GH, Jean YH et al. (2005) Color-dilution alopecia in dogs. J Vet Sci 6: 259-261. PubMed: 16131833. [PubMed] [Google Scholar]
- 10. Perego R, Proverbio D, Roccabianca P, Spada E (2009) Color dilution alopecia in a blue Doberman pinscher crossbreed. Can Vet J 50: 511–514. PubMed: 19436637. [PMC free article] [PubMed] [Google Scholar]
- 11. Griscelli C, Durandy A, Guy-Grand D, Daguillard F, Herzog C et al. (1978) A syndrome associating partial albinism and immunodeficiency. Am J Med 65: 691-702. doi: 10.1016/0002-9343(78)90858-6. PubMed: 707528. [DOI] [PubMed] [Google Scholar]
- 12. Markert CL, Silvers WK (1956) The effects of genotype and cell environment on melanoblast differentiation in the house mouse. Genetics 41: 429-450. PubMed: 17247639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. von Bomhard W, Mauldin EA, Schmutz SM, Leeb T, Casal ML (2006) Black hair follicular dysplasia in Large Münsterländer dogs: clinical, histological and ultrastructural features. Vet Dermatol 17: 182-188. doi: 10.1111/j.1365-3164.2006.00517.x. PubMed: 16674733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fukuda M, Kuroda TS, Mikoshiba K (2002) Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem 277: 12432-12436. doi: 10.1074/jbc.C200005200. PubMed: 11856727. [DOI] [PubMed] [Google Scholar]
- 15. Wu X, Bowers B, Rao K, Wei Q, Hammer JA III (1998) Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J Cell Biol 143: 1899-1918. doi: 10.1083/jcb.143.7.1899. PubMed: 9864363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu XS, Rao K, Zhang H, Wang F, Sellers JR et al. (2002) Identification of an organelle receptor for myosin-Va. Nat Cell Biol 4: 271-278. doi: 10.1038/ncb760. PubMed: 11887186. [DOI] [PubMed] [Google Scholar]
- 17. Nagashima K, Torii S, Yi Z, Igarashi M, Okamoto K et al. (2002) Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett 517: 233-238. doi: 10.1016/S0014-5793(02)02634-0. PubMed: 12062444. [DOI] [PubMed] [Google Scholar]
- 18. Van Gele M, Dynoodt P, Lambert J (2009) Griscelli syndrome: a model system to study vesicular trafficking. Pigment. Cell - Melanoma Res 22: 268-282. doi: 10.1111/j.1755-148X.2009.00558.x. [DOI] [PubMed] [Google Scholar]
- 19. Pastural E, Barrat FJ, Dufourcq-Lagelouse R, Certain S, Sanal O, et al. (1997) Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat Genet 16: 289-292. Erratum: Nat Genet 23: 373 [DOI] [PubMed] [Google Scholar]
- 20. Ménasché G, Pastural E, Feldmann J, Certain S, Ersoy F et al. (2000) Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet 25: 173-176. doi: 10.1038/76024. PubMed: 10835631. [DOI] [PubMed] [Google Scholar]
- 21. Ménasché G, Ho CH, Sanal O, Feldmann J, Tezcan I et al. (2003) Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). J Clin Invest 112: 450-456. doi: 10.1172/JCI200318264. PubMed: 12897212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanal O, Ersoy F, Tezcan I, Metin A, Yel L et al. (2002) Griscelli disease: genotype-phenotype correlation in an array of clinical heterogeneity. J Clin Immunol 22: 237-243. doi: 10.1023/A:1016045026204. PubMed: 12148598. [DOI] [PubMed] [Google Scholar]
- 23. Matesic LE, Yip R, Reuss AE, Swing DA, O'Sullivan TN et al. (2001) Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci U S A 98: 10238-10243. doi: 10.1073/pnas.181336698. PubMed: 11504925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooks SA, Gabreski N, Miller D, Brisbin A, Brown HE et al. (2010) Whole-genome SNP association in the horse: identification of a deletion in myosin Va responsible for lavender foal syndrome. PLoS Genet 4: e1000909 PubMed: 20419149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hume AN, Tarafder AK, Ramalho JS, Sviderskaya EV, Seabra MC (2006) A coiled-coil domain of melanophilin is essential for Myosin Va recruitment and melanosome transport in melanocytes. Mol Biol Cell 17: 4720-4735. doi: 10.1091/mbc.E06-05-0457. PubMed: 16914517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishida Y, David VA, Eizirik E, Schäffer AA, Neelam BA et al. (2006) A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 88: 698-705. doi: 10.1016/j.ygeno.2006.06.006. PubMed: 16860533. [DOI] [PubMed] [Google Scholar]
- 27. Drögemüller C, Philipp U, Haase B, Günzel-Apel A-R, Leeb T (2007) A noncoding melanophilin gene (MLPH) SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J Hered 98: 468-473. doi: 10.1093/jhered/esm021. PubMed: 17519392. [DOI] [PubMed] [Google Scholar]
- 28. Bed'hom B, Vaez M, Coville JL, Gourichon D, Chastel O et al. (2012) The lavender plumage colour in Japanese quail is associated with a complex mutation in the region of MLPH that is related to differences in growth, feed consumption and body temperature. BMC Genomics 13: 442. doi: 10.1186/1471-2164-13-442. PubMed: 22937744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA (1986) Splicing of messenger RNA precursors. Annu Rev Biochem 55: 1119-1150. doi: 10.1146/annurev.bi.55.070186.005351. PubMed: 2943217. [DOI] [PubMed] [Google Scholar]
- 30. David A, Miraki-Moud F, Shaw NJ, Savage MO, Clark AJ et al. (2010) Identification and characterisation of a novel GHR defect disrupting the polypyrimidine tract and resulting in GH insensitivity. Eur J Endocrinol 162: 37-42. doi: 10.1530/EJE-09-0583. PubMed: 19812236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clarke LA, Veiga I, Isidro G, Jordan P, Ramos JS et al. (2000) Pathological exon skipping in an HNPCC proband with MLH1 splice acceptor site mutation. Genes Chromosomes Cancer 29: 367-370. doi: 10.1002/1098-2264(2000)9999:9999. PubMed: 11066084. [DOI] [PubMed] [Google Scholar]
- 32. Beldjord C, Lapoumeroulie C, Pagnier J, Benabadji M, Krishnamoorthy R et al. (1988) A novel beta thalassemia gene with a single base mutation in the conserved polypyrimidine sequence at the 3' end of IVS 2. Nucleic Acids Res 16: 4927-4935. doi: 10.1093/nar/16.11.4927. PubMed: 3387213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sébillon P, Beldjord C, Kaplan JC, Brody E, Marie J (1995) A T to G mutation in the polypyrimidine tract of the second intron of the human beta-globin gene reduces in vitro splicing efficiency: evidence for an increased hnRNP C interaction. Nucleic Acids Res 23: 3419-3425. doi: 10.1093/nar/23.17.3419. PubMed: 7567451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murru S, Loudianos G, Deiana M, Camaschella C, Sciarratta GV et al. (1991) Molecular characterization of beta-thalassemia intermedia in patients of Italian descent and identification of three novel beta-thalassemia mutations. Blood 77: 1342-1347. PubMed: 2001456. [PubMed] [Google Scholar]
- 35. Reed R (1996) Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev 6: 215-220. doi: 10.1016/S0959-437X(96)80053-0. PubMed: 8722179. [DOI] [PubMed] [Google Scholar]
- 36. Smith CWJ, Valcárcel J (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25: 381-388. doi: 10.1016/S0968-0004(00)01604-2. PubMed: 10916158. [DOI] [PubMed] [Google Scholar]
- 37. Reed R (1989) The organization of 3' splice-site sequences in mammalian introns. Genes Dev 3: 2113-2123. doi: 10.1101/gad.3.12b.2113. PubMed: 2628164. [DOI] [PubMed] [Google Scholar]
- 38. Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90: 41-54. PubMed: 1427786. [DOI] [PubMed] [Google Scholar]
- 39. Roscigno RF, Weiner M, Garcia-Blanco MA (1993) A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem 268: 11222-11229. PubMed: 8496178. [PubMed] [Google Scholar]
- 40. Haire RN, Ohta Y, Strong SJ, Litman RT, Liu Y et al. (1997) Unusual patterns of exon skipping in Bruton tyrosine kinase are associated with mutations involving the intron splice site. Am J Hum Genet 60: 798-807. p. 17 3 [PMC free article] [PubMed] [Google Scholar]
- 41. Schneider S, Wildhardt G, Ludwig R, Royer-Pokora B (1993) Exon skipping due to a mutation in a donor splice site in the WT-1 gene is associated with Wilms' tumor and severe genital malformations. Hum Genet 91: 599-604. PubMed: 8393425. [DOI] [PubMed] [Google Scholar]
- 42. Aoshima M, Nunoi H, Shimazu M, Shimizu S, Tatsuzawa O et al. (1996) Two-exon skipping due to a point mutation in p67-phox--deficient chronic granulomatous disease. Blood 88: 1841-1845. PubMed: 8781442. [PubMed] [Google Scholar]
- 43. Fang LJ, Simard MJ, Vidaud D, Assouline B, Lemieux B et al. (2001) A novel mutation in the neurofibromatosis type 1 (NF1) gene promotes skipping of two exons by preventing exon definition. J Mol Biol 307: 1261-1270. doi: 10.1006/jmbi.2001.4561. PubMed: 11292340. [DOI] [PubMed] [Google Scholar]
- 44. Takahara K, Schwarze U, Imamura Y, Hoffman GG, Toriello H et al. (2002) Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal pro-alpha1(V) N-propeptides and Ehlers-Danlos syndrome type I. Am J Hum Genet 71: 451-465. doi: 10.1086/342099. PubMed: 12145749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamada K, Fukao T, Zhang G, Sakurai S, Ruiter JP et al. (2007) Single-base substitution at the last nucleotide of exon 6 (c.671GA), resulting in the skipping of exon 6, and exons 6 and 7 in human succinyl-CoA:3-ketoacid CoA transferase (SCOT). Gene - Mol Genet Metab 90: 291-297. doi: 10.1016/j.ymgme.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 46. Hori T, Fukao T, Murase K, Sakaguchi N, Harding CO et al. (2013) Molecular basis of two-exon skipping (exons 12 and 13) by c.1248+5ga in OXCT1 gene: Study on intermediates of OXCT1 transcripts in fibroblasts. Hum Mutat 34: 473-480. doi: 10.1002/humu.22258. PubMed: 23281106. [DOI] [PubMed] [Google Scholar]
- 47. Romeis B, Boeck P (1989) Romeis Mikroskopische Technik. 17th edn.. Munich: Urban & Fischer; . p. 657 [Google Scholar]
- 48. Gerrits PO, Smid L (1983) A new, less toxic polymerisation system for the embedding of soft tissues in glycol methacrylate and subsequent preparing of serial sections. J Microsc 132: 81-85. doi: 10.1111/j.1365-2818.1983.tb04711.x. PubMed: 6361264. [DOI] [PubMed] [Google Scholar]
- 49. Hanstede JG, Gerrits PO (1983) The effects of embedding in water-soluble plastics on the final dimensions of liver sections. J Microsc 131: 79–86. doi: 10.1111/j.1365-2818.1983.tb04233.x. PubMed: 6350599. [DOI] [PubMed] [Google Scholar]
- 50. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248-249. doi: 10.1038/nmeth0410-248. PubMed: 20354512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quadros L, Ghosh K, Shetty S (2008) Establishment of a new mismatch PCR-RFLP technique for detection of G10430A common mutation present in moderate to mild haemophilia B patients belonging to Gujarati community from the western part of India. Haemophilia 14: 628-629. doi: 10.1111/j.1365-2516.2008.01704.x. PubMed: 18393981. [DOI] [PubMed] [Google Scholar]
- 52. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263-265. doi: 10.1093/bioinformatics/bth457. PubMed: 15297300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the different variants of truncated melanophilin proteins in dilute rabbits (variants 2-4) with the complete protein in wild type rabbits (1). Variant 2 is a result from skipping of exon 3 and 4 of MLPH (p.Q37QfsX4). Exon skipping is presumably caused by the c.111-5CγA transversion. Variants 3 and 4 (p.L195LfsX123 and p.L195LfsX166, respectively) are caused by the c.585delG mutation. Which one of these variants is generated depends on a further polymorphism affecting the premature stop codon of variant 3 (c.953T>C). The RAB27A domain is highlighted in green.
(DOC)
Linkage disequilibrium (LD) of the nine SNPs selected for genotyping in the 23 rabbits of the breeding trial (c.1-10AγG, c.214A>G, c.215A>G and c.262A>G) and in all 53 rabbits of different breeds (c.1-1GγA, c.111-5CγA, c.366C>T, c.369A>G and c.585delG). The latter five SNPs showed strong LD with the dilute phenotype. The pairwise r2-values are shown for each SNP pair. Red squares indicate complete linkage disequilibrium.
(DOC)
Pedigree of the rabbits from the breeding trial and the haplotypes of nine polymorphisms genotyped for these animals. Haplotypes include the polymorphisms c.1-10AγG, c.1-1GγA, c.111-5CγA, c.214A>G, c.215A>G, c.262A>G, c.366C>T, c.369A>G and c.585delG.
(DOC)
Conservation of nucleotides in different species at the melanophilin (MLPH) SNP positions detected in rabbits. Nucleotides in brackets were located at position c.-9, but in that species, this position matched with c.1-10 in other species due to the surrounding sequence.
(DOC)
Case-control analysis for five melanophilin-associated polymorphisms with the dilution phenotype in 15 dilute and 37 colored non-dilute rabbits. The χ2- and P-values are shown for genotype, allele and trend tests.
(DOC)
Distribution of the melanophilin (MLPH) c.111-5CγA, c.585delG and c.953T>C mutations in rabbits of different breeds. The dilution status and genotype with corresponding number of animals and SNP genotypes are given. Bold faced letters indicate the dilute individuals, in which SNP genotypes differ between c.111-5C>A and c.585delG. The two individuals with less obvious dilution status are given at the bottom of the table. Their dilution status is indicated as “dark-dilute” (dd*).
(DOC)
Primer sequences with their product sizes (P) for amplification of sequences within melanophilin (MLPH) to be used for sequencing, long range PCR or genotyping as well as the targeted region and the appropriate template. The primers were designed to work at an annealing temperature of 60°C. In the case of performing a mismatch PCR for digestion, the base pairs, which were changed within a primer relative to the reference sequence, are highlighted in gray. If the product of a pair of primers was enzymatically digested for genotyping, the enzyme used is given.
(DOC)