Abstract
Chronic inflammation plays a significant role in tumor promotion, migration and invasion. Using microarray analysis, we observed a profound increase in genes involved in pro-inflammatory pathways in epidermal growth factor receptor inhibitor (EGFRI)-treated head and neck squamous cell carcinoma (HNSCC) cell lines compared to their respective vehicle-treated cell lines. We hypothesized that the efficacy of EGFRIs may be offset by the pro-inflammatory response that these drugs produce in HNSCC tumor cells. We found that clinical EGFRIs such as erlotinib, cetuximab, lapatinib and panitumumab induced the secretion of pro-inflammatory cytokines such as IL-2, IL-4, IL-6, IL-8, GM-CSF, TNFα and IFNγ. Focusing on IL-6, we found that erlotinib induced a time-dependent increase in IL-6 mRNA and protein expression and exogenous IL-6 was able to protect HNSCC cells from erlotinib-induced cytotoxicity. Conversely, an IL-6 receptor antagonist tocilizumab, sensitized HNSCC cells to erlotinib in vitro and in vivo. Inhibitors of NFκB, p38 and JNK suppressed erlotinib-induced IL-6 expression, suggesting an important role of NFκB and MAPK pathways in IL-6 expression. Furthermore, knockdown of NADPH oxidase 4 (NOX4) suppressed erlotinib-induced pro-inflammatory cytokines expression. Taken together, these results suggest that clinical EGFRIs induce the expression of pro-inflammatory cytokines via NOX4. Therefore, the anti-tumor activity of EGFRIs may be partially reduced by activation of NOX4-mediated pro-inflammatory pathways in HNSCC.
Keywords: EGFR, Erlotinib, IL-6, inflammation, cytokines, NOX4, HNSCC
Introduction
EGFR is expressed at high levels in HNSCC tumors (1); however EGFR-based chemotherapy has limited results in HNSCC patients (2–6). Numerous studies have proposed mechanisms (i.e. alternate signaling pathways, mutations, EMT) that may be responsible for poor responses and treatment failures due to EGFRI treatment (2). However, these discoveries have not yet led to significant improvements in response rates to EGFRIs in clinical trials. Therefore, the identification and understanding of molecular mechanisms associated with HNSCC tumor response to EGFR inhibition could lead to improvements in drug efficacy and HNSCC patient survival.
In an effort to further investigate the mechanism of action of clinical EGFRIs and possibly identify novel signaling pathways affected by EGFR inhibition, we used gene expression profiling to study the EGFR TKI erlotinib. Erlotinib (marketed as Tarceva) is used to treat non-small cell lung cancer and pancreatic cancer (7). It is also being studied in clinical trials in other cancer disease sites including HNSCC (6, 8, 9). Pathway analysis of the resultant gene expression data (using MetaCore™ Genego software) indicated a significant upregulation in pro-inflammatory immune response pathways in 3 HNSCC tumor cell lines treated with erlotinib (unpublished data). These results led to the reasoning that a pro-inflammatory response may be responsible in part for the reduced efficacy of EGFRIs because of the well-known role of inflammation in tumor progression (10–12). The current study determines if clinical EGFRIs induce pro-inflammatory cytokine expression and if IL-6 signaling in particular reduces the efficacy of erlotinib in HNSCC cells. Additionally, since we have previously shown that erlotinib increased oxidative stress in HNSCC cells via NOX4 (22), and pro-inflammatory pathways may be induced by redox activation, this study will also investigate the role of NOX4 in pro-inflammatory cytokine expression.
Materials and Methods
Cells and Culture Conditions
FaDu, Cal-27 and SCC-25 human head and neck squamous carcinoma (HNSCC) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). SQ20B HNSCC cells (13) were a gift from Dr. Anjali Gupta (Department of Radiation Oncology, The University of Iowa). FaDu, Cal-27 and SQ20B cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 4 mM L-glutamine, 1 mM sodium pyruvate, 1.5 g/L sodium bicarbonate and 4.5 g/L glucose with 10% fetal bovine serum (FBS; Hyclone, Logan, UT). All HNSCC cell lines are EGFR positive and are sensitive to EGFR inhibitors. All cell lines were authenticated by the ATCC for viability (before freezing and after thawing), growth, morphology and isoenzymology. Cells were stored according to the supplier’s instructions and used over a course of no more than 3 months after resuscitation of frozen aliquots. Cultures were maintained in 5% CO2 and air humidified in a 37°C incubator.
Drug Treatment
Erlotinib (ERL; Tarceva), cetuximab (CET; Erbitux), lapatinib (LAP; Tykerb), panitumumab (VEC; Vectibix) and tocilizumab (TOC; Actemra) were obtained from the inpatient pharmacy at the University of Iowa Hospitals and Clinics. Human IgG and dimethyl sulfoximine (DMSO) was obtained from Sigma Aldrich. Inhibitors of JNK (SP6; SP600125), p38 (SB2; SB203580) and STAT3 (AG4; AG490) were obtained from Calbiochem. The NFκB inhibitor BAY117092 (BAY) was obtained from Santa Cruz Biotechnology and recombinant human IL-6 was obtained from R&D Systems. Drugs were added to cells at final concentrations of 5 μM ERL, 17 nM CET, 1 μM LAP, 100 nM VEC, 1 μM TOC, 5 μM SP6, 10 μM SB2, 10 μM BAY, and 100 ng/mL IL-6. The required volume of each drug was added directly to complete cell culture media on cells to achieve the indicated final concentrations.
Microarray Analyses
FaDu, Cal-27 and SQ20B HNSCC cells were treated with DMSO or erlotinib (5 μM, 48 h). Total RNA was extracted from treated cells using the manufacturer’s protocol RNeasy mini kit (Qiagen): DNA microarray sample processing. RNA sample preparation for hybridization and the subsequent hybridization to the Illumina beadchips were performed at the University of Iowa DNA Facility using the manufacturer’s recommended protocol. Briefly, 100 nanograms total RNA was converted to amplified biotin-cRNA using the Ambion TotalPrep RNA Amplification Kit for Illumina Expression BeadChip (Ambion, Inc., Austin, TX, Cat. #AMIL1791) according to the manufacturer’s recommended protocol. 750 ng of this product were mixed with Illumina hybridization buffer, placed onto Illumina HumanHT-12v4 BeadChips (Part No. BD-103-0204), and incubated at 58° C for 17h, with rocking, in an Illumina Hybridization Oven. Following hybridization, the arrays were washed, blocked, then stained with streptavidin-Cy3 (Amersham/GE Healthcare, Piscataway, NJ) according to the Illumina Whole-Genome Gene Expression Direct Hybridization Assay protocol. Beadchips were scanned with the Illumina iScan System (ID #N054) and data collected using the GenomeStudio software v2011.1.
Human Cytokine 8-plex Assay
Tissue culture supernatants from drug-treated and/or adenoviral transfected cells were collected and assayed for secreted cytokines using the Bio-Plex Pro™ Human cytokine 8-plex assay (Bio Rad Laboratories, Hercules, CA) according to the manufacturer’s recommended protocol.
Enzyme-linked immunosorbent assay (ELISA)
Levels of IL-6 of treated cells were determined by ELISA. The culture media of drug-treated and/or adenoviral-transfected cells were harvested and IL-6 was detected according to the manufacturer’s protocol using the Human IL-6 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN).
Western blot analysis
Cell lysates were standardized for protein content, resolved on 4% to 12% SDS polyacrylamide gels, and blotted onto nitrocellulose membranes. Membranes were probed with rabbit anti-STAT3, anti-pSTAT3, anti-β-actin (Cell Signaling) and anti-NOX4 (Abcam) antibodies. Antibody binding was detected by using an ECL Chemiluminescence Kit (Amersham).
Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was extracted from treated cells after indicated time points using the RNeasy mini kit (Qiagen). The cDNA was amplified from 800 ng of total RNA using iScript cDNA synthesis kit (Bio-Rad). Thermocycler conditions included a 5 min incubation at 25°C, a 30 min incubation at 42°C, and a 5 min incubation at 85°C. The cDNAs were subjected to qPCR analysis with the following 5′→3′ primers (sense and antisense respectively): Human IL-6: AGGACTGGAGATGTCTGAGGCTC and GCGCTTGTGGAGAAGGAGTTC, NOX4: CTCAGCGAATCAATCAGCTGTG and AGAGGAACACGACAATCAGCCTTAG, and human 18S: CCTTGGATGTGGTAGCCGTTT and AACTTTCGATGGTAGTCGCCG. The assay was performed in a 96-well optical plate, with a final reaction volume of 20 μL, including synthesized cDNA (20 ng), oligonucleotide primers (100 μM each), and 2× SYBR Green/ROX PCR master mix (Bio-Rad Laboratories). Samples were run on an ABI PRISM Sequence Detection System (model 7000, Applied Biosystems). PCR conditions were 50°C for 2 min, 95°C for 2 min and 30 s, 95°C for 15 s, and 60°C for 1 min for 40 cycles. Results were analyzed using ABI PRISM 7000 SDS software. The denaturation and annealing steps were carried out for 40 cycles to determine the threshold cycle (CT) values for all of the genes analyzed. Samples were checked for non-specific products or primer/dimer amplification by melting curve analysis. The CT values for the target genes in all of the samples (analysed in duplicate or triplicate) were normalized on the basis of the abundance of the 18S transcript, and the fold difference (relative abundance) was calculated using the formula 2–ΔΔCT and was plotted as the mean.
Clonogenic survival assay
Clonogenic survival was determined as previously described (14). Individual assays were performed with multiple dilutions with at least four cloning dishes per data point, repeated in at least 3 separate experiments.
Measurement of intracellular prooxidant levels
Attached cells were labeled with 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate, (DCFH, 10 μg/mL) dissolved in 0.1% DMSO for 15 minutes at 37°C. Culture plates were placed on ice, trypsinized, resuspended in ice cold PBS, and analyzed by using a FACScan flow cytometer [excitation 488 nm, emission 530 nm band-pass filter. The mean fluorescence intensity (MFI) of 10,000 cells was analyzed in each sample and corrected for autofluorescence from unlabeled cells.
Adenoviral Vectors
Wild-type (WT) human NOX4 cDNA (BC040105.1) was obtained from Open Biosystems (MHS1010-9204787; Huntsville, AL) in the pCMV-SPORT6 vector. cDNA for the dominant-negative (DN) form of NOX4 lacking the C-terminal NADPH binding domain was generated by introduction of a stop codon at amino acid 384 as described by Mahadev et al., (15) using Stratagene Quickchange XL site-directed mutagenesis (La Jolla, Ca) with forward CTG GAC AGA ACG ATT TCG AGA TTA ACT ACT GCC TCC and reverse GGA GGC AGT AGT TAA TCT CGA AAT CGT TCT GTC CAG primers (nucleotide change underlined). The entire cDNA for the WT and DN clones were subsequently sequence verified and subcloned into the pacAd5.CMV.K-NpA shuttle vector (U Iowa Gene Transfer Vector Core (GTVC); Iowa City, IA) using Kpn1/Not1 restriction. Viruses (AdNOX4wt and AdNOX4dn) were purified by the U Iowa GTVC using CsCl gradient centrifugation and dialyzed against buffer containing 3% sucrose/PBS and stored at −80°C. Each virus was tested for WT revertants and for titer by PCR and A549 plaque assay as described in Anderson et al., (16). An empty vector lacking the NOX4 construct was used as a control. Construction and characterization of recombinant E1-deleted adenoviral vectors encoding siRNA targeted against eGFP (AdsiCON) or Nox4 (AdsiNox4) have each been described previously (17). All vectors were obtained from the University of Iowa Gene Vector Core. HNSCC cells in serum free media were infected with 150 MOI of the above described adenoviral vectors for 24 hours. Biochemical analyses were performed 72–96 h after transfection.
Tumor cell implantation
Female 4–5 week old athymic-nu/nu nude mice were purchased from Harlan Laboratories (Indianapolis, IN). Mice were housed in a pathogen-free barrier room in the Animal Care Facility at the University of Iowa and handled using aseptic procedures. All procedures were approved by the IACUC committee of the University of Iowa and conformed to the guidelines established by the NIH. Mice were allowed at least 3 days to acclimate prior to beginning experimentation, and food and water were made freely available. Tumor cells were inoculated into nude mice by subcutaneous injection of 0.1 mL aliquots of saline containing 4 × 106 FaDu or SQ20B cells into the right flank using 26-gauge needles.
Tumor measurements
Mice started drug treatment 1 week after tumor inoculation. Mice were evaluated daily and tumor measurements taken three times per week using Vernier calipers. Tumor volumes were calculated using the formula: tumor volume = (length × width2)/2 where the length was the longest dimension and width was the dimension perpendicular to length.
In vivo drugs administration
Mice were divided into 4 groups (n = 8–9 mice/group). ERL group: ERL was suspended in water and administered orally 12.5 mg/kg every day for 10 days. TOC group: TOC was administered i.p. 1 mg/kg every other day for 10 days. ERL+TOC group: mice were administered ERL orally 12.5 mg/kg every day and 1 mg/kg TOC i.p. every other day for 10 days. Control group: Mice were administered orally 100 uL water every day and 1 mg/kg IgG i.p every other day for 10 days. Mice were euthanized via CO2 gas asphyxiation when tumor diameter exceeded 1.5 cm in any dimension.
Statistical Analysis
Statistical analysis was done using GraphPad Prism version 5 for Windows (GraphPad Software, San Diego, CA). Differences between 3 or more means were determined by one-way ANOVA with Tukey post-tests. Linear mixed effects regression models were used to estimate and compare the group-specific change in tumor growth curves. All statistical analysis was performed at the p<0.05 level of significance.
Results
Network analysis of Erlotinib-treated HNSCC cell lines
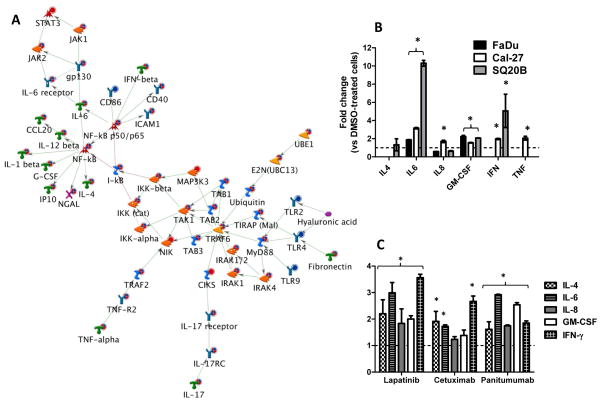
The gene expression profiles of FaDu, Cal-27 and SQ20B HNSCC cells exposed to erlotinib (5 μM, 48 hours) versus DMSO were analyzed by high-throughput microarray. Genetic network analysis of the resultant gene expression data for all 3 cell lines (n=3 experiments per cell line) was carried out using Metacore™ (GeneGo). Thirty networks were identified using the GeneGo tool (Supplementary Figure 1) that identified functional relationships between gene products based on known interactions in the scientific literature. Of these networks, we focused on the first scored (by the number of pathways) network with a p-value of 7.3×10−21 and z-score of 9.89 (Supplementary Table 1, Figure 1A). The genes in this network were related to positive regulation of immune response processes, response to stimulus and NFκB transcription factor activity. Additionally, signaling pathways including toll like receptor (TLR), IL-17 and TNFα pathways were implicated in the activation of NFκB (Figure 1A). According to the network shown in figure 1A, NFκB activation resulted in the expression of cytokines involved in pro-inflammatory pathways such as IL-1β, IL-4, IL-6, IL-12β, CCL20 (MIP3A), GM-CSF, IP10 and IFNγ. Of these cytokines, IL-6 appeared to be of importance since the IL-6/JAK/STAT3 pathway was also identified in this network (Figure 1A). Altogether, these results suggest that the induction of pro-inflammatory pathways may play a role in the mechanism of action of erlotinib.
Figure 1.
Pro-inflammatory cytokines are induced by EGFR inhibitors in HNSCC cells. A: Shown is the most significant (p = 7.27×10−21) network constructed from differentially regulated transcripts comparing microarray data from erlotinib (5 μM, 48 h) treated FaDu, Cal-27 and SQ20B head and neck squamous carcinoma (HNSCC) cells versus DMSO treated HNSCC cells. The microarray expression value changes were uploaded to and analyzed by MetaCore™ (GeneGo) software. Up regulated genes are marked with red circles; down regulated with blue circles. The ‘checkerboard’ color indicates mixed expression for the gene between cell lines. B: FaDu, Cal-27 and SQ20B cells were treated with 5 μM erlotinib (ERL) or DMSO for 48 h and then analyzed for proinflammatory cytokine production using an 8-plex human cytokine panel. SQ20B cells were treated with lapatinib (LAP, 5 μM), cetuximab (CET, 100 μg/ml) and panitumumab (VEC, 100 nM) for 48 h before analysis for proinflammatory cytokine production using an 8-plex cytokine panel (C). ERL and LAP-induced changes were compared to DMSO controls; CET and VEC-induced changes were compared to IgG controls. All controls were set at a value of 1 (hatched line). Error bars represent ± standard error of the mean (SEM) of N = 3 experiments. *:p<0.05 versus DMSO or IgG control.
Clinical EGFR inhibitors induce the expression of pro-inflammatory cytokines in HNSCC cells
In order to confirm that erlotinib may induce the expression of pro-inflammatory cytokines, levels of 8 cytokines (IL-2, IL-4, IL-6, IL-8, IL-10, IL-12, IFN-γ, and GM-CSF) were measured using a Human Cytokine 8-Plex Panel from the media of FaDu, Cal-27 and SQ20B cells treated 48 h with DMSO or 5 μM Erlotinib. Of these 8 cytokines, erlotinib increased levels of IL-6 and GM-CSF from FaDu cells; IL-6, GM-CSF and IFNγ from SQ20B cells; and IL-6, IL-8, GM-CSF, IFNγ and TNFα from Cal-27 cells compared to control treated cells (Figure 1B), which supports the network analysis shown in figure 1. IL-2 and IL-10 were below the limit of detection. Using SQ20B cells, we additionally observed that lapatinib and panitumumab increased levels of IL-4, IL-6, IL-8, GM-CSF and IFNγ; while cetuximab increased only IL-4, IL-6 and IFNγ (Figure 1C). IL-2, IL-10 and TNFα were below the limit of detection. These results suggest that all clinical EGFR inhibitors may induce the secretion of proinflammatory cytokines.
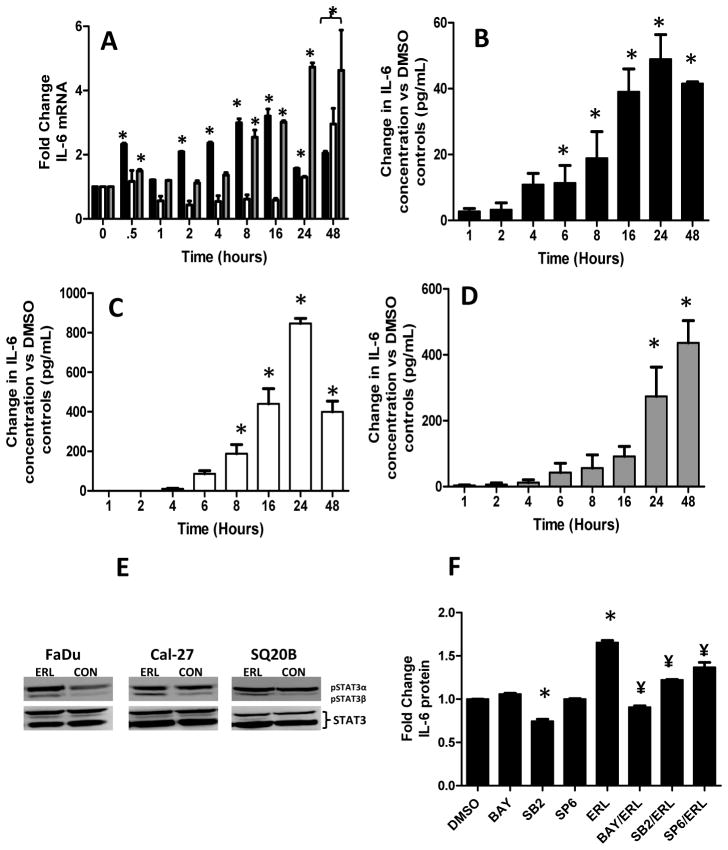
Erlotinib induces a time-dependent increase in IL-6 expression in HNSCC cells
Given that the IL-6 signaling pathway was identified in the microarray network analysis as important in the mechanism of action of erlotinib (Figure 1A), we analyzed the expression of IL-6 mRNA (using RT-PCR) and protein (using ELISA) over 48 hours in erlotinib-treated HNSCC cells. IL-6 mRNA expression increased over time in erlotinib-treated Cal-27 and SQ20B cells (Figure 2A). However, in FaDu cells, IL-6 mRNA levels initially peaked at 30 min then peaked again at 16 hours before declining at 24 and 48 h (Figure 2A). We observed that erlotinib significantly increased IL-6 protein secretion (compared to control-treated cells) beginning at 6 h in FaDu cells (Figure 2B); 8 h in Cal-27 cells (Figure 2C); and 24 h in SQ20B cells (Figure 2D) and increased gradually over a 24 h period. Erlotinib induced IL-6 secretion in other EGFR-positive cell lines such as A549 (lung cancer) and MiaPaca2 (pancreatic cancer) cells (Supplementary Figure 1) suggesting that erlotinib-induced IL-6 is not specific to HNSCC cell lines. Knockdown of EGFR using siRNA induced IL-6 in a similar manner to erlotinib and other EGFRIs in FaDu cells (Supplementary Figure 2) suggesting that the effects we see with erlotinib are likely due to EGFR inhibition. However in siEGFR-transfected FaDu cells which were treated with erlotinib, we observed an additional increase in IL-6 secretion compared to siEGFR-transfected FaDu cells treated with DMSO suggesting that some off-target effects of erlotinib may contribute to IL-6 production in this cell line (Supplementary Figure 2). Additionally, erlotinib increased STAT3 tyrosine phosphorylation in FaDu, Cal-27 and SQ20B cells after 48 hours as observed by increased pSTAT3β (79 Kd) isoform expression but not pSTAT3a isoform expression (86 Kd). In FaDu cells, erlotinib increased both pSTAT3 isoforms. Total STAT3 expression was not affected by erlotinib in all cell lines (Figure 2E). These results suggest that EGFR pathway inhibition with erlotinib induces IL-6 mRNA and protein expression and possibly downstream IL-6 signaling in HNSCC cells and this phenomenon may be observed in other EGFR-positive tumor cell models.
Figure 2.
Erlotinib induced IL-6 mRNA and protein expression in HNSCC cells. FaDu, Cal-27 and SQ20B cells were treated with 5 μM erlotinib (ERL) or DMSO for 48 h and then analyzed for IL-6 mRNA levels using qRT-PCR at 0.5, 1, 2, 4, 8, 16, 24 and 48 h after treatment (A). *:p<0.05 versus respective treatment at time 0. FaDu (B), Cal-27 (C) and SQ20B (D) cells were treated as described above and analyzed for IL-6 protein levels using ELISA at 1, 2, 4, 6, 8, 16, 24 and 48 h after treatment. *:p<0.05 versus time = 1 h. FaDu, Cal-27 and SQ20B cells were treated with 5 μM ERL for 48 h then analyzed for pSTAT3 and STAT3 expression by Western blot (E). FaDu cells were pretreated with inhibitors of NFκB (BAY 11-7082 [BAY]), p38 (SB230580 [SB2]) and JNK (SP600125 [SP6]) signaling for 2 h before treatment with or without 5 μM erlotinib (ERL) for 48 h (F). IL-6 protein secretion was measured by ELISA. Error bars represent ± standard error of the mean (SEM) of N = 3 experiments. *:p<0.05 versus DMSO, ¥:p<0.05 versus ERL.
Inhibitors of NFκB, p38 and JNK suppress erlotinib-induced IL-6 in FaDu cells
To confirm that erlotinib-induced IL-6 was a result of NFκB activation we pretreated FaDu cells with the NFκB inhibitor BAY117082 before treatment with erlotinib then analyzed IL-6 expression by ELISA. We observed that NFκB inhibition completely reversed erlotinib-induced IL-6 expression confirming that NFκB activation was involved in erlotinib-induced IL-6 activation (Figure 2F). Given that both NFκB and AP-1 binding sites are present on the promoter region of IL-6 (18) and MAPK signaling pathways have been shown to affect AP-1 activity (19), we determined if MAPK activation was also involved in erlotinib-induced IL-6 expression. We observed that pretreatment of cells with inhibitors of p38 (SB203580) and JNK (SP600125) partially suppressed erlotinib-induced IL-6 expression suggesting that MAPK activation and thus AP-1 activation was also involved in IL-6 expression (Figure 2F).
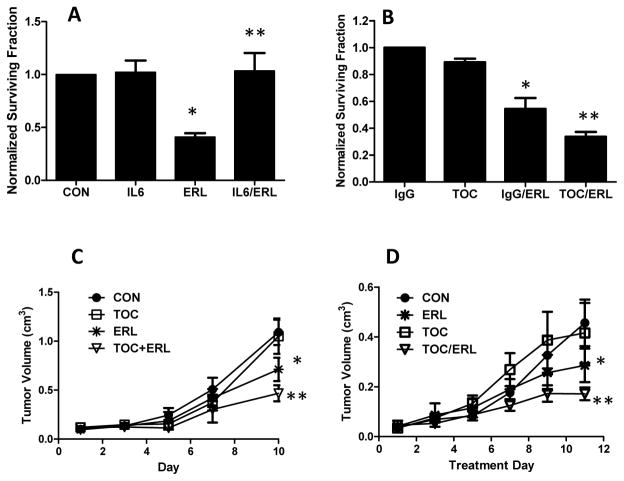
IL-6 expression affects HNSCC response to erlotinib
In order to determine the significance of erlotinib-induced IL-6 expression, we pretreated FaDu cells with 100 ng/ml of human recombinant IL-6 for 2 h before treatment with erlotinib in serum free media then analyzed for cell survival (via clonogenic assay). We found that exogenous IL-6 was able to completely protect against erlotinib-induced cytotoxicity (Figure 3A). Conversely, to determine if inhibition of the IL-6 pathway enhances HNSCC tumor cell response to erlotinib, we treated FaDu cells with the IL-6 receptor antagonist tocilizumab with or without erlotinib in vitro. We observed that tocilizumab was able to sensitize FaDu cells to erlotinib in vitro (Figure 3B). The combination of erlotinib and tocilizumab was tested in FaDu and SQ20B xenografts (Figure 3C, D). The results indicated that tocilizumab was able to increase the efficacy of erlotinib after 10 days of treatment in both xenograft models (Figure 3C, D). These results suggest that IL-6 pathway inhibition may be a viable strategy to enhance HNSCC tumor response to EGFRIs.
Figure 3.
IL6 signaling affects response to erlotinib in head and neck (HNSCC) cells. FaDu cells were pre-treated with 100 ng/mL of human recombinant IL-6 for 2 h before treatment with or without 5 μM erlotinib (ERL) for 48 h. Treated cells were analyzed for cytotoxicity via clonogenic analysis (A). DMSO was used as a control (CON). FaDu cells were treated with 1 μM of the IL-6 receptor antagonist tocilizumab (TOC) or IgG with or without 5 μM erlotinib (ERL) for 48 hours then analyzed by clonogenic assay (B). Athymic (nu/nu) mice bearing FaDu (C) and SQ20B (D) xenograft tumors were treated with 1 mg/mouse tocilizumab (TOC) and/or 12 mg/kg erlotinib (ERL) for 10 days. Tocilizumab was given i.p. every other day and erlotinib was given by oral gavage every day. Control mice (CON) received IgG. Data points represent the average values for 9 mice. Error bars represent the standard error of the mean (SEM). *:p<0.05 versus DMSO or IgG control. **:p<0.05 versus ERL.
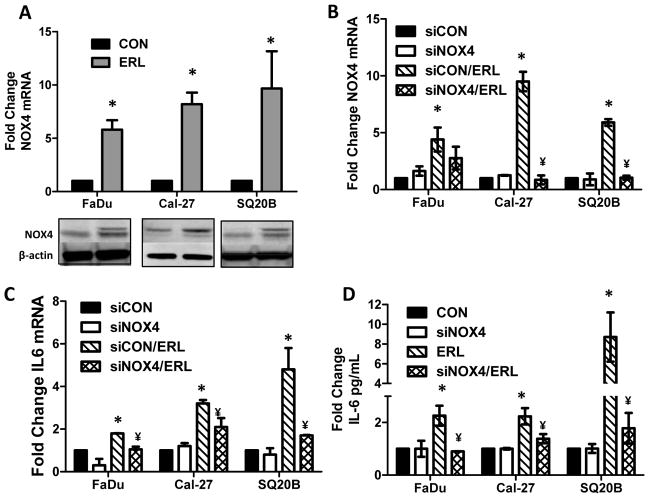
Knockdown of NOX4 suppresses erlotinib-induced pro-inflammatory cytokine expression
Oxidative stress has been implicated in inflammatory responses through NFKB activation in various chronic diseases (20, 21). Our previous work has shown that erlotinib induced metabolic oxidative stress in HNSCC cells via NOX4 (22). In order to determine if NOX4 contributed to the increase in pro-inflammatory cytokines induced by erlotinib, we knocked down NOX4 using adenoviral siRNA targeted to NOX4 before treatment with erlotinib. Erlotinib increased NOX4 mRNA (Figure 4A, B) and protein expression (Figure 4A) in all 3 HNSCC cell lines, and transfection with AdsiNOX4 was effective at suppressing the NOX4 mRNA (Figure 4B), IL-6 mRNA (Figure 4C) and IL-6 protein (Figure 4D). Additionally, knockdown of NOX4 expression was able to suppress erlotinib-induced expression of IL-2, IL-4, IFNγ and TNFα in Cal-27 cells and IFNγ in SQ20B cells (Table 1). Even though AdsiNOX4 was able to consistently decrease erlotinib-induced NOX4 mRNA expression, we were unable to detect similar changes in NOX4 protein expression. We did observe a partial decrease in erlotinib-induced NOX4 protein expression with AdsiNOX4 in FaDu cells, however, we were unable to detect decreased NOX4 protein expression in Cal-27 and SQ20B cells (data not shown) even though the effect of AdsiNOX4 was clearly observed by mRNA levels (Figure 4A) in these cell lines. This suggests that NOX4 may have a long half-life in our HNSCC cells and may be resistant to proteolytic degradation as observed in THP-1 monocytes (23) and human monocyte-derived macrophages (24). Altogether, these results suggest that NOX4-mediated oxidative stress may be involved in the increase in pro-inflammatory cytokines induced by EGFR inhibitors.
Figure 4.
Erlotinib induces IL-6 production via NOX4. FaDu, Cal27 and SQ20B cells were treated with 5 μM erlotinib (ERL) or DMSO (CON) for 48 h and then analyzed for NOX4 mRNA and protein levels using qRT-PCR and Western blot (A). β-actin was used as a loading control. FaDu, Cal27 and SQ20B cells were transfected with a control adenoviral vector (siCON) or adenoviral siNOX4 before treatment with DMSO or 5 μM ERL for 48 hours. NOX4 mRNA expression (B), IL-6 mRNA expression (C) and IL-6 protein secretion (D) were analyzed by Western blot, qRT-PCR and ELISA respectively. Error bars represent the standard error of the mean (SEM). *:p<0.05 versus DMSO control. ¥:P<0.05 versus ERL.
Table 1.
Fold Change in Cytokine Levels versus EMPTY
| Cytokine | Cal-27 | SQ20B | ||||
|---|---|---|---|---|---|---|
| siNOX4 | EMPTY+ERL | siNOX4+ERL | siNOX4 | EMPTY+ERL | siNOX4+ERL | |
| IL-2 | 1.0±0.1 | 2.4±0.1* | 1.3±0.5** | ND | ND | ND |
| IL-4 | 0.9±0.2 | 1.8±0.2* | 1.0±0.2** | 0.6±0.5* | 1.7±0.5* | 2.2±2.4 |
| IL-8 | 0.6±0.1* | 1.0±0.6 | 0.3±0.2** | 1.0±0.1 | 0.5±0.12* | 0.2±0.1 |
| IL-10 | ND | ND | ND | ND | ND | ND |
| GM-CSF | 0.5±0.1* | 0.9±0.2 | 0.4±0.1** | 1.0±0.1 | 1.1±1.0 | 0.6±0.3** |
| IFN-γ | 1.0±0.1 | 1.6±0.4* | 0.9±0.1** | 1.0±0.1 | 2.6±0.9* | 0.2±0.3** |
| TNF-α | 0.8±0.1 | 1.4±0.8* | 0.7±0.6** | ND | ND | ND |
p<0.05 vs EMPTY,
:p<0.05 vs EMPTY+ERL, ND:non-detectable
NOX4 overexpression increases ROS and pro-inflammatory cytokine production in HNSCC cells
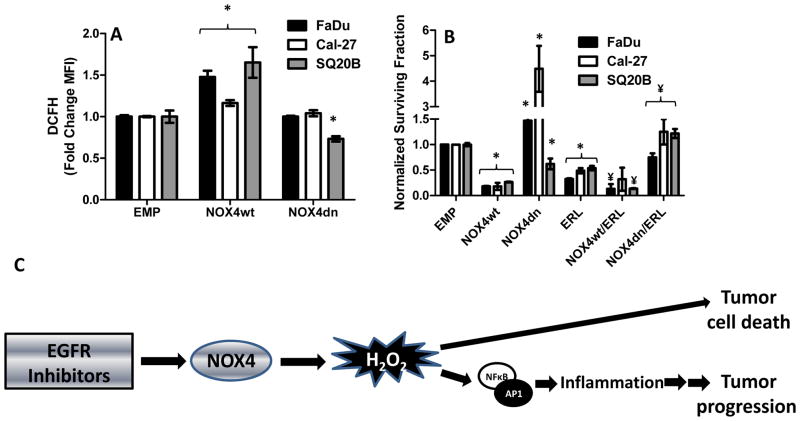
To confirm that NOX4 was involved in ROS and cytokine production we transfected HNSCC cells with adenoviral vectors containing wildtype NOX4 (AdNOX4wt), dominant negative NOX4 (AdNOX4dn) and empty (AdEMPTY) before measurement of DCFH oxidation and cytokine production. Overexpression of NOX4 resulted in an increase in DCFH oxidation in all 3 cell lines (Figure 5A) and also increased IL-6, IL-8 and GM-CSF in FaDu cells; IL-2, IL-6, GM-CSF and TNFα in Cal-27 cells; and IL-6 and IFNγ in SQ20B cells (Table 2) suggesting that NOX4-induced ROS was capable of inducing pro-inflammatory cytokine expression. On the other hand, overexpression of NOX4dn either did not affect or suppressed ROS (Figure 5A) and pro-inflammatory cytokine levels (Table 2).
Figure 5.
NOX4 manipulation affects ROS production and erlotinib-induced cytotoxicity in HNSCC cells. FaDu, Cal27 and SQ20B cells were transfected with empty (EMP), wildtype NOX4 (NOX4wt) or dominant negative NOX4 (NOX4dn) adenoviral vectors before analysis for DCFH oxidation (A). FaDu, Cal27 and SQ20B cells were transfected with EMP, NOX4wt or NOX4dn adenoviral vectors before treatment with DMSO or 5 μM erlotinib (ERL) for 48 h, then analysis by clonogenic assay (B). All treatment groups were normalized to their respective EMP control. Error bars represent the standard error of the mean (SEM). *:p<0.05 versus DMSO control. ¥:p<0.05 versus ERL. C: NOX4 may play a dual role in EGFR inhibitor-induced cytotoxicity and inflammation in HNSCC cells. Exposure of HNSCC cells to EGFR inhibitors stimulates the expression and activation of NOX4 resulting in hydrogen peroxide-induced oxidative stress. This oxidative stress can lead to both cell death and the redox activation of NFKB and AP-1 leading to the induction of pro-inflammatory mediators which may mediate tumor progression.
Table 2.
Fold Change in Cytokine Levels versus EMPTY
| Cytokine | FaDu | Cal-27 | SQ20B | |||
|---|---|---|---|---|---|---|
| NOX4wt | NOX4dn | NOX4wt | NOX4dn | NOX4wt | NOX4dn | |
| IL-2 | ND | ND | 2.0±0.5* | ND | ND | ND |
| IL-4 | 0.7±0.6 | ND | 1.3±0.4 | 0.9±0.3 | 0.9±0.2 | ND |
| IL-6 | 1.9±1.1* | 1.0±0.2 | 1.6±0.3* | 0.6±0.1* | 2.0±0.2* | 0.8±0.2 |
| IL-8 | 2.2±0.2* | 0.8±0.1 | 1.3±0.2 | 0.5±0.1* | 1.1±0.2 | ND |
| IL-10 | ND | ND | ND | ND | ND | ND |
| GM-CSF | 2.0±0.5* | 3.5±0.8* | 2.0±0.1* | 0.3±0.1* | 0.7±0.2 | 0.3±0.1* |
| IFN-γ | 1.3±0.9 | 0.8±1.2 | 1.3±0.2 | 0.9±0.3 | 1.5±0.9* | ND |
| TNF-α | ND | ND | 2.3±0.2* | 0.3±0.1* | ND | ND |
p<0.05 vs EMPTY, ND:non-detectable
NOX4 expression affects HNSCC cell response to Erlotinib
To investigate how NOX4 expression affects cell survival and HNSCC cell sensitivity to erlotinib, FaDu, Cal-27 and SQ20B cells were transfected with AdEMPTY, AdNOX4wt and AdNOX4dn as mentioned above before treatment with DMSO and erlotinib then analyzed by clonogenic assay. We observed that overexpression of NOX4wt significantly increased cytotoxicity of all 3 cell lines and increased the sensitivity of FaDu and SQ20B cells to erlotinib (Figure 5B). Overexpression of NOX4dn increased cell survival in FaDu and Cal-27 cells and significantly protected all 3 cell lines from erlotinib-induced cytotoxicity (Figure 5B). These results suggest that NOX4 expression may affect HNSCC response to erlotinib.
Discussion
These studies presented here indicate that EGFRIs increase the expression of pro-inflammatory mediators through a mechanism involving NOX4-mediated oxidative stress. Chronic inflammation may play a significant role in tumor promotion, migration and invasion in some cancers (25), and many of the cytokines found to be increased by EGFRIs (e.g. IL-2, IL-4, IL-6, IL-8, GM-CSF, IFN and TNF [Figures 1,2]) are involved in immune/inflammatory responses, angiogenesis, metastasis and tumor progression (25). This implies that the pressure of EGFRI treatment itself may drive the reduction in drug efficacy by inducing oxidative stress and inflammation in the tumor microenvironment.
The observation that EGFRIs induce immune and inflammatory responses is not unheard of since EGFRIs are known to promote skin inflammation in a majority of patients (26). What is less known however, is how these inflammatory pathways affect the anti-tumor activity of EGFRIs. For example, we show that exogenous IL-6 was able to completely protect FaDu cells from the anti-tumor effect of erlotinib (Figure 3A), which supports prior work demonstrating the ability of IL-6 to protect lung cancer cells against erlotinib in lung cancer cells (27). Interleukin-6 (IL-6) is a pleotropic cytokine with a wide range of biological activities and is well known for its role in inflammation, tumor progression and chemoresistance in HNSCC (28–33). Increases in IL-6 expression correlate with poor prognosis in HNSCC patients and patients resistant to chemotherapy have shown significantly higher serum IL-6 levels than those who did respond (28, 29, 31).
Crosstalk exists between the EGFR and IL-6 signaling pathways since both pathways are able to phosphorylate STAT3 (33). STAT3 activation has been shown to mediate cancer cell proliferation and carcinogenesis (34). This crosstalk is initiated by the secretion of IL-6, resulting from NFκB activation, and the subsequent phosphorylation of STAT3 leading to STAT3-dependent gene expression (33). EGFR activation also leads to phosphorylation of STAT3 (35). Therefore in the presence of high levels of IL-6, STAT3 expression is sustained in the presence of EGFRIs and the antitumor effect of EGFRIs may be reduced. In our studies, the ability of erlotinib to not change or surprisingly increase STAT3 tyrosine phosphorylation (Figure 2E) support the idea of crosstalk between the IL-6 and EGFR signaling systems. Given the crosstalk between the EGFR and the IL-6 pathways, IL-6 pathway inhibitors should be considered a promising strategy to enhance tumor response to EGFRIs. The IL-6 pathway inhibitor tocilizumab is already approved for use in rheumatoid arthritis and systemic juvenile idiopathic arthritis as an anti-inflammatory agent in combination with methotrexate (36). Tocilizumab is a humanized monoclonal antibody against the interleukin-6 receptor (IL-6R) that blocks IL-6 from binding to its receptor (36–38). Tocilizumab has shown antitumor and anti-angiogenic activity as a single agent in glioma cells and oral squamous carcinoma cells in vitro and in vivo (37, 38). We have now shown that tocilizumab was able to effectively sensitize FaDu and SQ20B HNSCC xenografts to erlotinib in vivo (Figure 3C, D), suggesting that the combination of EGFRIs and IL-6 pathway inhibitors may be a viable strategy for the treatment of EGFR-positive tumors.
The second major finding of these studies is the role that NOX4 plays in inflammation in HNSCC cells. NOX enzymes are a family of transmembrane enzymes (NOX1-5, DUOX1 and 2) that produce reactive oxygen species (ROS) in response to stimuli including growth factors (39). We have previously shown that erlotinib, induces oxidative stress via NOX4 in FaDu HNSCC cells and that NOX4 expression was necessary for the anti-tumor activity of erlotinib (22). In support of this, we have now duplicated these results in other HNSCC cell lines (Figure 4A) and additionally have shown that overexpression of NOX4 increased the anti-tumor effect of erlotinib in all 3 HNSCC cell lines (Figure 5B). Furthermore, overexpression of a dominant negative form of NOX4 (NOX4dn) lacking the NADPH binding domain and thus ROS producing capacity (Figure 5A), significantly protected cells from erlotinib-induced toxicity (Figure 5B) suggesting that NOX4 plays a vital role in erlotinib-induced cytotoxicity.
Given that NFκB is redox regulated (40), we proposed that NOX4-mediated oxidative stress may activate NFκB leading to cytokine expression. Suppression of NOX4 expression was indeed effective at suppressing erlotinib-induced IL-6 expression in all 3 cell lines (Figure 4C, D) suggesting that suppression of NOX4 would also suppress NFkB activation. These studies are currently underway. Interestingly decreased NOX4 expression also suppressed many of the other pro-inflammatory cytokines induced by erlotinib in Cal-27 and SQ20B cells, but this effect was cell line specific (Table 1). Additionally, in the absence of erlotinib, NOX4 overexpression in itself was able to increase the expression of pro-inflammatory cytokines (Table 2) further supporting the role of NOX4 in inflammation. Overall, these results suggest a dual role of NOX4 in the mechanism of action of erlotinib and possibly other EGFRIs. NOX4-induced oxidative stress has already been established by our laboratory as being important for the anti-tumor activity of erlotinib (Figure 5B, (22)). However, NOX4 appears to also induce a cytoprotective mechanism via the induction of pro-inflammatory cytokines such as IL-6, which may reduce the anti-tumor effect of erlotinib (Figure 5C).
Overall these studies will shed some light on the mechanism of action of EGFRIs and how we can improve targeted therapy for HNSCC patients. Cisplatin and radiation have long been the backbone and standard of care of HNSCC patients presenting with locally advanced disease. Cisplatin-based chemotherapy and radiation have already been shown to induce IL-6 secretion (41, 42) implying that EGFRI-induced IL-6 may not be unique to EGFR inhibitors. It is possible that the cell death caused by standard chemo-/radiotherapy in HNSCC may be activating pathways leading to IL-6 and other pro-inflammatory cytokines. Nevertheless, the use of IL-6 inhibitors may be an important adjuvant for not only EGFRIs but for other chemo-/radiotherapy regimens used for HNSCC treatment.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Allan Shepard for providing the NOX4wt and NOX4dn adenoviral vectors and Dr. Thomas Bair in the Bioinformatics Division at The University of Iowa for his assistance in running the microarray studies. The authors also acknowledge the Flow Cytometry Facility, which is a Carver College of Medicine Core Research Facilities/ Holden Comprehensive Cancer Center Core Laboratory at the University of Iowa. The Facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center
Grant Support
This work was supported by the Department of Pathology and grants NIH K01CA134941 (ALS), R01CA127958 (AG) and IRG-77-004-34 from the American Cancer Society, administered through the Holden Comprehensive Cancer Center at the University of Iowa.
Footnotes
The authors have no conflict of interest.
References
- 1.Bei R, Budillon A, Masuelli L, Cereda V, Vitolo D, Di Gennaro E, et al. Frequent overexpression of multiple ErbB receptors by head and neck squamous cell carcinoma contrasts with rare antibody immunity in patients. J Pathol. 2004;204:317–25. doi: 10.1002/path.1642. [DOI] [PubMed] [Google Scholar]
- 2.Rexer BN, Engelman JA, Arteaga CL. Overcoming resistance to tyrosine kinase inhibitors: lessons learned from cancer cells treated with EGFR antagonists. Cell Cycle. 2009;8:18–22. doi: 10.4161/cc.8.1.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007;25:2171–77. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- 4.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 5.Cohen EE, Kane MA, List MA, Brockstein BE, Mehrotra B, Huo D, et al. Phase II trial of gefitinib 250 mg daily in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:8418–24. doi: 10.1158/1078-0432.CCR-05-1247. [DOI] [PubMed] [Google Scholar]
- 6.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 7.Pollack VA, Savage DM, Baker DA, Tsaparikos KE, Sloan DE, Moyer JD, et al. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J Pharmacol Exp Ther. 1999;291:739–48. [PubMed] [Google Scholar]
- 8.Siu LL, Soulieres D, Chen EX, Pond GR, Chin SF, Francis P, et al. Phase I/II trial of erlotinib and cisplatin in patients with recurrent or metastatic squamous cell carcinoma of the head and neck: a Princess Margaret Hospital phase II consortium and National Cancer Institute of Canada Clinical Trials Group Study. J Clin Oncol. 2007;25:2178–83. doi: 10.1200/JCO.2006.07.6547. [DOI] [PubMed] [Google Scholar]
- 9.Thomas F, Rochaix P, Benlyazid A, Sarini J, Rives M, Lefebvre JL, et al. Pilot study of neoadjuvant treatment with erlotinib in nonmetastatic head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13:7086–92. doi: 10.1158/1078-0432.CCR-07-1370. [DOI] [PubMed] [Google Scholar]
- 10.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–90. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Weichselbaum RR, Dahlberg W, Beckett M, Karrison T, Miller D, Clark J, et al. Radiation-resistant and repair-proficient human tumor cells may be associated with radiotherapy failure in head- and neck-cancer patients. Proc Natl Acad Sci U S A. 1986;83:2684–88. doi: 10.1073/pnas.83.8.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitz DR, Malcolm RR, Roberts RJ. Cytotoxicity and metabolism of 4-hydroxy-2-nonenal and 2-nonenal in H2O2-resistant cell lines. Do aldehydic by-products of lipid peroxidation contribute to oxidative stress? Biochem J. 1990;267:453–59. doi: 10.1042/bj2670453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson JR, Burmeister MA, Tian X, Zhou Y, Guruju MR, Stupinski JA, et al. Genetic silencing of Nox2 and Nox4 reveals differential roles of these NADPH oxidase homologues in the vasopressor and dipsogenic effects of brain angiotensin II. Hypertension. 2009;54:1106–14. doi: 10.1161/HYPERTENSIONAHA.109.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahadev K, Motoshima H, Wu X, Ruddy JM, Arnold RS, Cheng G, et al. The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol Cell Biol. 2004;24:1844–54. doi: 10.1128/MCB.24.5.1844-1854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson RD, Haskell RE, Xia H, Roessler BJ, Davidson BL. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–38. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- 18.Squarize CH, Castilho RM, Sriuranpong V, Pinto DS, Jr, Gutkind JS. Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. 2006;8:733–46. doi: 10.1593/neo.06274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvers AL, Bachelor MA, Bowden GT. The role of JNK and p38 MAPK activities in UVA-induced signaling pathways leading to AP-1 activation and c-Fos expression. Neoplasia. 2003;5:319–29. doi: 10.1016/S1476-5586(03)80025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cachofeiro V, Goicochea M, de Vinuesa SG, Oubina P, Lahera V, Luno J. Oxidative stress and inflammation, a link between chronic kidney disease and cardiovascular disease. Kidney Int Suppl. 2008;111:S4–9. doi: 10.1038/ki.2008.516. [DOI] [PubMed] [Google Scholar]
- 21.Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381–86. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- 22.Orcutt KP, Parsons AD, Sibenaller ZA, Scarbrough PM, Zhu Y, Sobhakumari A, et al. Erlotinib-mediated inhibition of EGFR signaling induces metabolic oxidative stress through NOX4. Cancer Res. 2011;71:3932–40. doi: 10.1158/0008-5472.CAN-10-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullevig S, Zhao Q, Lee CF, Seok Kim H, Zamora D, Asmis R. NADPH oxidase 4 mediates monocyte priming and accelerated chemotaxis induced by metabolic stress. Arterioscler Thromb Vasc Biol. 2012;32:415–26. doi: 10.1161/ATVBAHA.111.238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CF, Qiao M, Schroder K, Zhao Q, Asmis R. Nox4 is a novel inducible source of reactive oxygen species in monocytes and macrophages and mediates oxidized low density lipoprotein-induced macrophage death. Circ Res. 2010;106:1489–97. doi: 10.1161/CIRCRESAHA.109.215392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keibel A, Singh V, Sharma MC. Inflammation, microenvironment, and the immune system in cancer progression. Curr Pharm Des. 2009;15:1949–55. doi: 10.2174/138161209788453167. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Perez-Soler R. Skin toxicities associated with epidermal growth factor receptor inhibitors. Target Oncol. 2009;4:107–119. doi: 10.1007/s11523-009-0114-0. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 2010;107:15535–40. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffy SA, Taylor JM, Terrell JE, Islam M, Li Y, Fowler KE, et al. Interleukin-6 predicts recurrence and survival among head and neck cancer patients. Cancer. 2008;113:750–57. doi: 10.1002/cncr.23615. [DOI] [PubMed] [Google Scholar]
- 29.Heimdal JH, Kross K, Klementsen B, Olofsson J, Aarstad HJ. Stimulated monocyte IL-6 secretion predicts survival of patients with head and neck squamous cell carcinoma. BMC Cancer. 2008;8:34. doi: 10.1186/1471-2407-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamimura D, Ishihara K, Hirano T. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev Physiol Biochem Pharmacol. 2003;149:1–38. doi: 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- 31.Riedel F, Zaiss I, Herzog D, Gotte K, Naim R, Hormann K. Serum levels of interleukin-6 in patients with primary head and neck squamous cell carcinoma. Anticancer Res. 2005;25:2761–65. [PubMed] [Google Scholar]
- 32.Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond) 2012;122:143–159. doi: 10.1042/CS20110340. [DOI] [PubMed] [Google Scholar]
- 33.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–56. [PubMed] [Google Scholar]
- 34.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 35.Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, Endo S, et al. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A. 2000;97:4227–32. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro-Millan I, Singh JA, Curtis JR. Systematic review of tocilizumab for rheumatoid arthritis: a new biologic agent targeting the interleukin-6 receptor. Clin Ther. 2012;34:788–802. doi: 10.1016/j.clinthera.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinriki S, Jono H, Ota K, Ueda M, Kudo M, Ota T, Oike Y, Endo M, et al. Humanized anti-interleukin-6 receptor antibody suppresses tumor angiogenesis and in vivo growth of human oral squamous cell carcinoma. Clin Cancer Res. 2009;15:5426–34. doi: 10.1158/1078-0432.CCR-09-0287. [DOI] [PubMed] [Google Scholar]
- 38.Shinriki S, Jono H, Ueda M, Ota K, Ota T, Sueyoshi T, et al. Interleukin-6 signalling regulates vascular endothelial growth factor-C synthesis and lymphangiogenesis in human oral squamous cell carcinoma. J Pathol. 2011;225:142–150. doi: 10.1002/path.2935. [DOI] [PubMed] [Google Scholar]
- 39.Kamata T. Roles of Nox1 and other Nox isoforms in cancer development. Cancer Sci. 2009;100:1382–88. doi: 10.1111/j.1349-7006.2009.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowie A, O’Neill LA. Oxidative stress and nuclear factor-kappaB activation: a reassessment of the evidence in the light of recent discoveries. Biochem Pharmacol. 2000;59:13–23. doi: 10.1016/s0006-2952(99)00296-8. [DOI] [PubMed] [Google Scholar]
- 41.Reers S, Pfannerstill AC, Rades D, Maushagen R, Andratschke M, Pries R, et al. Cytokine changes in response to radio-/chemotherapeutic treatment in head and neck cancer. Anticancer Res. 2013;33:2481–89. [PubMed] [Google Scholar]
- 42.Poth KJ, Guminski AD, Thomas GP, Leo PJ, Jabbar IA, Saunders NA. Cisplatin treatment induces a transient increase in tumorigenic potential associated with high interleukin-6 expression in head and neck squamous cell carcinoma. Mol Cancer Ther. 2010;9:2430–39. doi: 10.1158/1535-7163.MCT-10-0258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.