Abstract
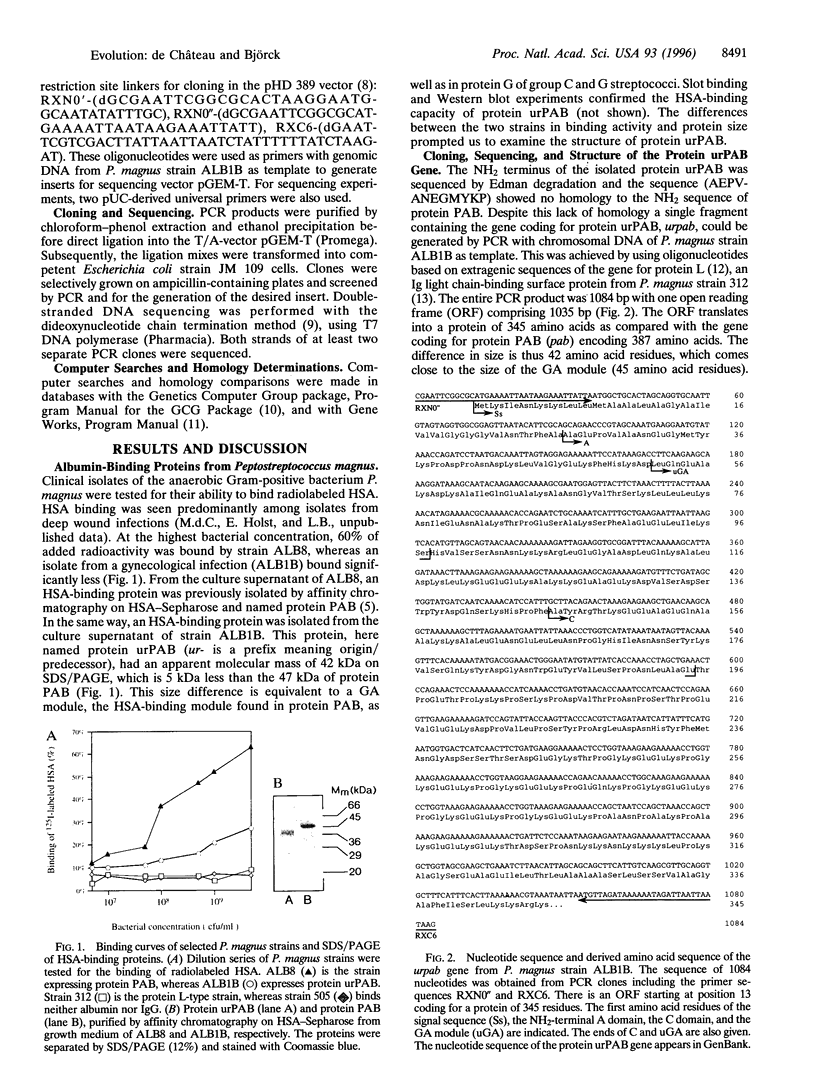
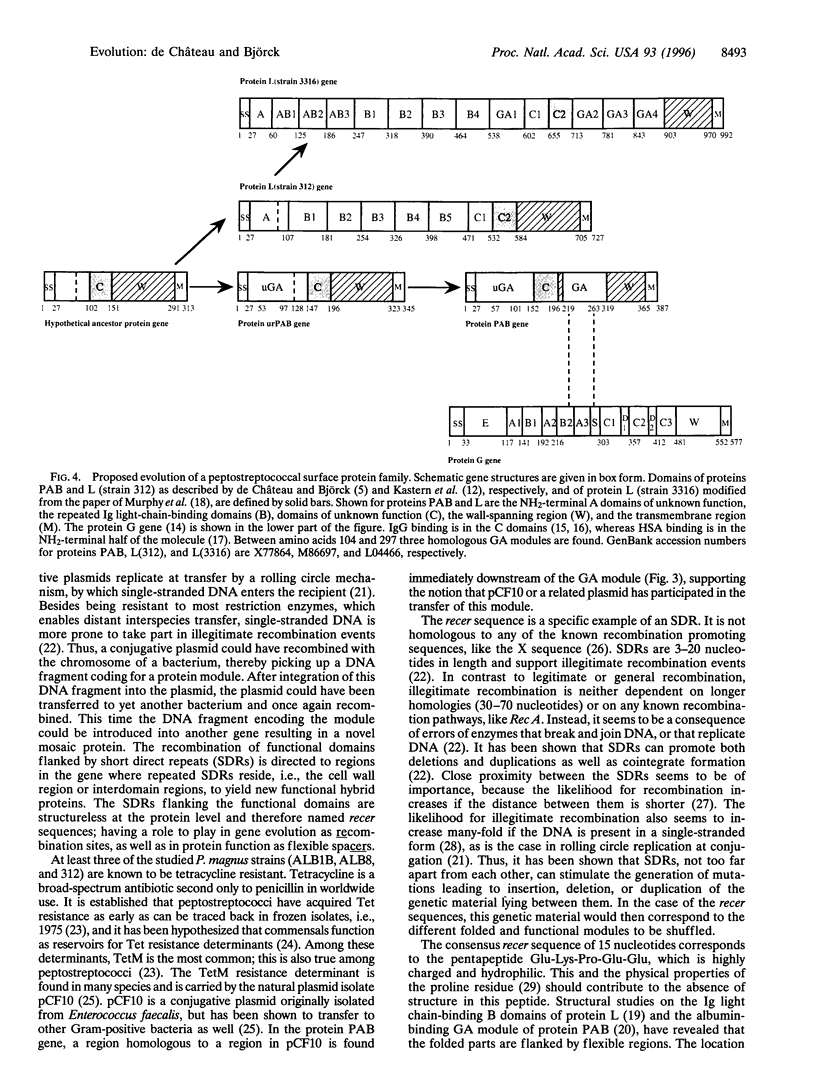
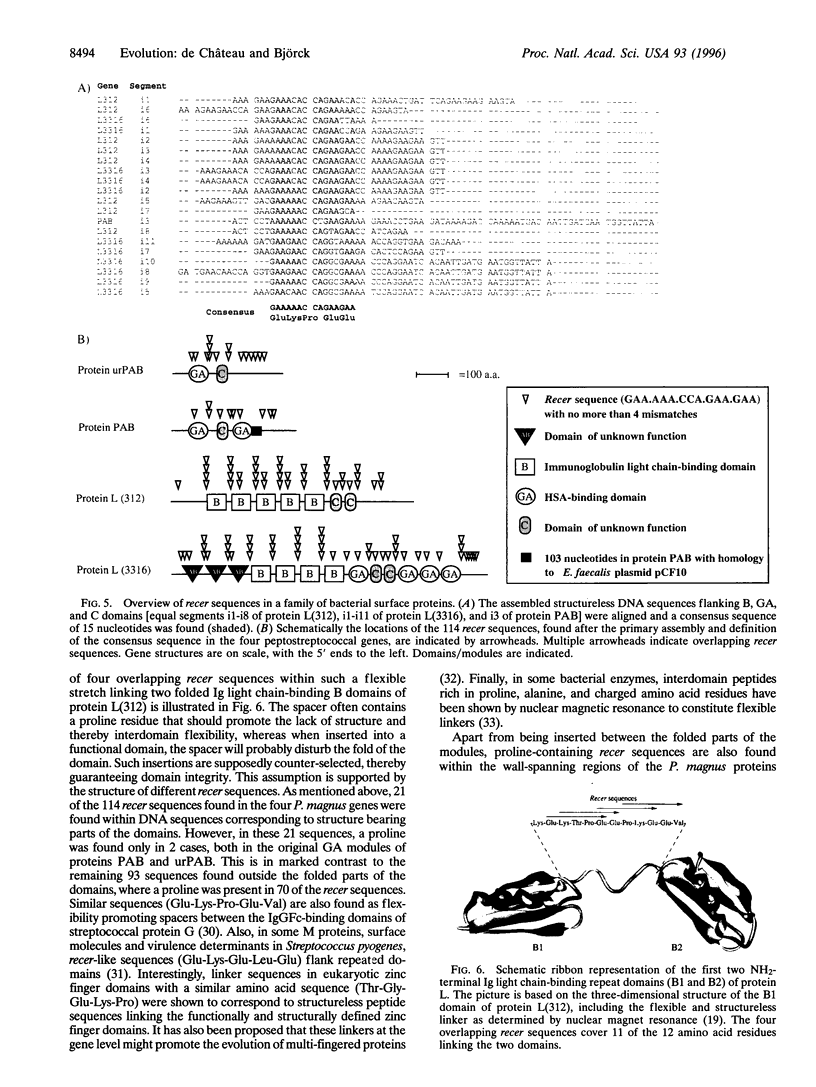
In the evolution of eukaryotic genes, introns are believed to have played a major role in increasing the probability of favorable duplication events, chance recombinations, and exon shuffling resulting in functional hybrid proteins. As a rule, prokaryotic genes lack introns, and the examples of prokaryotic introns described do not seem to have contributed to gene evolution by exon shuffling. Still, certain protein families in modern bacteria evolve rapidly by recombination of genes, duplication of functional domains, and as shown for protein PAB of the anaerobic bacterial species Peptostreptococcus magnus, by the shuffling of an albumin-binding protein module from group C and G streptococci. Characterization of a protein PAB-related gene in a P. magnus strain with less albumin-binding activity revealed that the shuffled module was missing. Based on this fact and observations made when comparing gene sequences of this family of bacterial surface proteins interacting with albumin and/or immunoglobulin, a model is presented that can explain how this rapid intronless evolution takes place. A new kind of genetic element is introduced: the recer sequence promoting interdomain, in frame recombination and acting as a structure-less flexibility-promoting spacer in the corresponding protein. The data presented also suggest that antibiotics could represent the selective pressure behind the shuffling of protein modules in P. magnus, a member of the indigenous bacterial flora.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achari A., Hale S. P., Howard A. J., Clore G. M., Gronenborn A. M., Hardman K. D., Whitlow M. 1.67-A X-ray structure of the B2 immunoglobulin-binding domain of streptococcal protein G and comparison to the NMR structure of the B1 domain. Biochemistry. 1992 Nov 3;31(43):10449–10457. doi: 10.1021/bi00158a006. [DOI] [PubMed] [Google Scholar]
- Akerström B., Nielsen E., Björck L. Definition of IgG- and albumin-binding regions of streptococcal protein G. J Biol Chem. 1987 Oct 5;262(28):13388–13391. [PubMed] [Google Scholar]
- Björck L., Kastern W., Lindahl G., Widebäck K. Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Mol Immunol. 1987 Oct;24(10):1113–1122. doi: 10.1016/0161-5890(87)90080-0. [DOI] [PubMed] [Google Scholar]
- Björck L., Kronvall G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J Immunol. 1984 Aug;133(2):969–974. [PubMed] [Google Scholar]
- Björck L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988 Feb 15;140(4):1194–1197. [PubMed] [Google Scholar]
- Bron S., Holsappel S., Venema G., Peeters B. P. Plasmid deletion formation between short direct repeats in Bacillus subtilis is stimulated by single-stranded rolling-circle replication intermediates. Mol Gen Genet. 1991 Apr;226(1-2):88–96. doi: 10.1007/BF00273591. [DOI] [PubMed] [Google Scholar]
- Choo Y., Klug A. A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 1993 Jul 25;21(15):3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Korman R. Z., Zahler S. A., Adsit J. C., Dunny G. M. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J Bacteriol. 1987 Jun;169(6):2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin F., Dervyn E., Dervyn R., Ehrlich S. D., Noirot P. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol Microbiol. 1994 May;12(4):561–569. doi: 10.1111/j.1365-2958.1994.tb01042.x. [DOI] [PubMed] [Google Scholar]
- Dalbøge H., Jensen E. B., Tøttrup H., Grubb A., Abrahamson M., Olafsson I., Carlsen S. High-level expression of active human cystatin C in Escherichia coli. Gene. 1989 Jul 15;79(2):325–332. doi: 10.1016/0378-1119(89)90214-x. [DOI] [PubMed] [Google Scholar]
- De Château M., Nilson B. H., Erntell M., Myhre E., Magnusson C. G., Akerström B., Björck L. On the interaction between protein L and immunoglobulins of various mammalian species. Scand J Immunol. 1993 Apr;37(4):399–405. doi: 10.1111/j.1365-3083.1993.tb03310.x. [DOI] [PubMed] [Google Scholar]
- Diamond J. M. Extinctions, catastrophic and gradual. Nature. 1983 Aug 4;304(5925):396–397. doi: 10.1038/304396a0. [DOI] [PubMed] [Google Scholar]
- Fahnestock S. R., Alexander P., Nagle J., Filpula D. Gene for an immunoglobulin-binding protein from a group G streptococcus. J Bacteriol. 1986 Sep;167(3):870–880. doi: 10.1128/jb.167.3.870-880.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Guss B., Eliasson M., Olsson A., Uhlén M., Frej A. K., Jörnvall H., Flock J. I., Lindberg M. Structure of the IgG-binding regions of streptococcal protein G. EMBO J. 1986 Jul;5(7):1567–1575. doi: 10.1002/j.1460-2075.1986.tb04398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. U., de Château M., Björck L., Forsén S., Drakenberg T., Wikström M. The GA module, a mobile albumin-binding bacterial domain, adopts a three-helix-bundle structure. FEBS Lett. 1995 Oct 30;374(2):257–261. doi: 10.1016/0014-5793(95)01121-t. [DOI] [PubMed] [Google Scholar]
- Kastern W., Sjöbring U., Björck L. Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J Biol Chem. 1992 Jun 25;267(18):12820–12825. [PubMed] [Google Scholar]
- Koch A. L. Genetic response of microbes to extreme challenges. J Theor Biol. 1993 Jan 7;160(1):1–21. doi: 10.1006/jtbi.1993.1001. [DOI] [PubMed] [Google Scholar]
- Levinton J. S. The big bang of animal evolution. Sci Am. 1992 Nov;267(5):84–91. doi: 10.1038/scientificamerican1192-84. [DOI] [PubMed] [Google Scholar]
- Little E., Bork P., Doolittle R. F. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J Mol Evol. 1994 Dec;39(6):631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994 Sep;58(3):563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw A. R., Beachey E. H., Burdett V. Molecular evolution of streptococcal M protein: cloning and nucleotide sequence of the type 24 M protein gene and relation to other genes of Streptococcus pyogenes. J Bacteriol. 1988 Feb;170(2):676–684. doi: 10.1128/jb.170.2.676-684.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. P., Duggleby C. J., Atkinson M. A., Trowern A. R., Atkinson T., Goward C. R. The functional units of a peptostreptococcal protein L. Mol Microbiol. 1994 Jun;12(6):911–920. doi: 10.1111/j.1365-2958.1994.tb01079.x. [DOI] [PubMed] [Google Scholar]
- Olsson A., Eliasson M., Guss B., Nilsson B., Hellman U., Lindberg M., Uhlén M. Structure and evolution of the repetitive gene encoding streptococcal protein G. Eur J Biochem. 1987 Oct 15;168(2):319–324. doi: 10.1111/j.1432-1033.1987.tb13423.x. [DOI] [PubMed] [Google Scholar]
- Pancholi V., Fischetti V. A. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J Bacteriol. 1988 Jun;170(6):2618–2624. doi: 10.1128/jb.170.6.2618-2624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao G. M., Saier M. H., Jr Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J Mol Evol. 1995 Feb;40(2):136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Martin S. R., Appella E. Conformational flexibility and folding of synthetic peptides representing an interdomain segment of polypeptide chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Biol Chem. 1989 Jan 15;264(2):767–775. [PubMed] [Google Scholar]
- Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993 Jan;9(1):17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- Reis K. J., Ayoub E. M., Boyle M. D. Streptococcal Fc receptors. I. Isolation and partial characterization of the receptor from a group C streptococcus. J Immunol. 1984 Jun;132(6):3091–3097. [PubMed] [Google Scholar]
- Roberts M. C., Hillier S. L. Genetic basis of tetracycline resistance in urogenital bacteria. Antimicrob Agents Chemother. 1990 Feb;34(2):261–264. doi: 10.1128/aac.34.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C. Tetracycline resistance in Peptostreptococcus species. Antimicrob Agents Chemother. 1991 Aug;35(8):1682–1684. doi: 10.1128/aac.35.8.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol Microbiol. 1994 May;12(3):343–350. doi: 10.1111/j.1365-2958.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O., Mihaylova-Petkov D., Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993 Dec;12(12):4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., Dowson C. G., Spratt B. G. Localized sex in bacteria. Nature. 1991 Jan 3;349(6304):29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- Wikström M., Drakenberg T., Forsén S., Sjöbring U., Björck L. Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G. Biochemistry. 1994 Nov 29;33(47):14011–14017. doi: 10.1021/bi00251a008. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Château M., Björck L. Protein PAB, a mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J Biol Chem. 1994 Apr 22;269(16):12147–12151. [PubMed] [Google Scholar]








