Abstract
Four closely related facultative anaerobe, moderately thermophilic, Gram positive rods (JS1T, JS5, JS11, and JS15) were isolated from sediment samples from a hot spring at Suryakund, Jharkhand, India. Colonies were pale yellow, rough surface with uneven edges on TSA after 72 h incubation. Heterotrophic growth was observed at 40-60°C and pH 5.5-11.5; optimum growth occurred at 55°C and pH 7.5. 16S rRNA gene sequence analysis revealed the strains belong to genus Anoxybacillus. DNA-DNA homology values among strains were above 70% and showed distinct ERIC and REP PCR profile. On the basis of morphology and biochemical characteristics, strain JS1T was studied further. Strain JS1T showed 99.30% sequence similarity with A. flavithermus subsp. yunnanensis, 99.23% with A. mongoliensis, 99.16% with A. eryuanensis, 98.74% with A. flavithermus subsp. flavithermus, 98.54% with A. tengchongensis, 98.51% with A. pushchinoensis, 97.91% with A. thermarum, 97.82% with A. kaynarcensis, 97.77% with A. ayderensis and A. kamchatkensis, 97.63% with A. salavatliensis, 97.55% with A. kestanbolensis, 97.48% with A. contaminans, 97.27% with A. gonensis and 97.17% with A. voinovskiensis. In 16S rRNA secondary structure based phylogenetic comparison, strain JS1T was clustered with Anoxybacillus eryuanensis, A. mongoliensis, and A. flavithermus subsp. yunnanensis and showed 15 species specific base substitutions with maximum variability in helix 6. Moreover, DNA-DNA relatedness between JS1T and the closely related type strains were well below 70%. The DNA G+C content was 42.1 mol%. The major fatty acids were C15:0 iso, C16:0 iso and C17:0iso. The polar lipids were a phosphatidylgylycerol, a diphosphatidylglycerol, a phosphatidylethnolamine, a phosphatidylcholine, a phosphatidyl monomethylethanolamine and four unknown lipids. Based on polyphasic approach, strain JS1T represent a novel species of the genus Anoxybacillus for which Anoxybacillus suryakundensis sp. nov. is proposed. The type strain is JS1T (= DSM 27374T = LMG 27616T =JCM19211T).
Introduction
The genus Anoxybacillus was introduced by Pikuta et al. that comprised members showing anaerobic growth [1]. Later it was found that Anoxybacillus pushchinoensis strain K1T representing the genus Anoxybacillus can grow aerobically in ANX medium at pH 9-9.7 that led to amendment of genus description [2]. Members are Gram-positive, spore forming rods of size varying 0.4-0.85 x 2.5-5.0 µm, aerotolerant anaerobes or facultative anaerobes. Oxidase and catalase reactions are variable. DNA G+C content varies between 42-57 mol%. At the time of writing this manuscript, total 20 species was reported from different parts of the globe [1,3-14] being thermophilic and alkalitolerant.
Exploration of new microbes in hot springs is of importance owing to the intriguing biogeochemistry and extreme conditions of these environments. Extreme habitat inspired several studies on hot spring microbial diversity throughout the world [15,16]. Microorganisms that survive in hot springs have unique adaptations to high temperature and are an important bio-resource. Our recent studies revealed the presence of new species of bacterium from different tropical hot springs in India [17-20]. These observations inspired us to explore the microbial diversity in other hot springs.
In the present study we describe the phenotypic and genotypic characteristics of a novel strain, designated as JS1T, isolated from a hot spring at Suryakund, Jharkhand, India. Based on this polyphasic evidence, it is proposed that JS1T be assigned as the type strain of the novel species Anoxybacillus suryakundensis sp. nov.
Materials and Methods
Sample collection and isolation of bacterium
Sediment samples were collected twice from a hot spring at Suryakund ((24°08′58″N, 85°38′44″E), Jharkhand, India. The surface temperature of the sediment sample at the site of collection was 80°C and the pH was 7.4. No specific permissions were required for the collection of samples from this hot spring. Moreover, this location did not involve endangered and protected species and it is not under regulatory body concerned with protection of wildlife. Samples collected in a sterile container were transported to the laboratory without temperature control. 2.0 g (wet weight) sediment sample was transferred into a 250 ml conical flask containing 50 ml trypticase soy broth (Difco) pH 7.2 and incubated on a shaker (ISF-I-V; Adolf Kuhner AG) at 55°C and 200 rpm After overnight incubation, the suspension was serially diluted and plated onto trypticase soy agar (Difco) plates pH 7.2 and incubated at 55°C for three days. Morphologically similar pale yellow, rough surface circular colony that appeared on trypticase soy agar plate were picked and purified by repeated streaking on the same medium. For short term preservation, isolates were streaked on trypticase soy agar and stored at 4°C. For long-term preservation, the culture was stored at -80°C in 15% (v/v) glycerol.
For comparative study, closely related type strains were procured from Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Germany, National Collection of Industrial, Marine and Food Bacteria (NCIMB), UK, Korean Collection for Type Cultures (KCTC), Korea and from BCCM/LMG Bacteria collection, Universiteit Gent, Belgium. Unless indicated otherwise, all strains were grown and maintained as mentioned previously.
Phenotypic, biochemical and physiological characterization
Cell morphology was examined by transmission electron microscopy (model FEI Morgagni 268D) as described by Jyoti et al. [18]. Gram character was determined using Gram staining kit (Becton-Dickinson, USA) following manufacturer’s instruction. The formation of spores was observed by growing culture for 48 h in half strength TSB supplemented with 5 mg/l MnSO4, 4 H2O. Spores were detected by using Schaeffer and Fulton Spore Stain Kit (Hi-Media) following manufacturer’s instruction. For phenotypic characterization, all isolates and reference type strains were tested under same laboratory conditions. Tests like oxidase, catalase indole, methyl red, Voges Proskauer, lysine decarboxylase, ornithine decarboxylase, citrate and malonate utilization, hydrolysis of starch, urea, ONPG and nitrate reduction was performed by previously described methods [20]. Utilization of different carbohydrates as sole source of carbon and energy with concomitant production of acid was tested in peptone water basal media (Hi-Media) supplemented with substrates at concentration of 5.0 g/l and phenol red of 0.18 g/l. Tests were considered positive, weakly positive and negative based on the media colour (yellow, light yellow or pink and red respectively). In addition to classical tests, oxidation of organic compounds as sole source of carbon was determined using API50CH, API20E (bioMérieux) and Hi25 identification kit (Hi-Media) following manufacturer’s instruction except incubation was performed at 55°C for 18 h. Yeast extract-peptone broth containing 0-5.0% (w/v) NaCl was inoculated and incubated at 55°C for 4 days to test for salt tolerance. Ethanol tolerance was determined by growing the cells in TSB medium containing ethanol at a final concentration of 0-15% (v/v). Anaerobic growth was tested using the BD GasPak EZ system (Becton-Dickinson). The temperature for growth in the range of 30-60°C was examined in TSB medium at pH 7.5 for 2 days. Similarly, the pH range for growth was examined in TSB medium prepared in buffered solutions [21] in the pH range 3.5-11.5 in steps of 0.5 pH unit at 55°C for 48 h.
DNA isolation, PCR amplification and sequencing of 16S rRNA
Isolation of chromosomal DNA, amplification and sequencing of 16S rRNA gene of the isolates were performed by the methods described earlier [22]. Primers used for the amplification of 16S rRNA were 5’-GAG TTT GAT CCT GGC TCA-3’ (forward primer) and 5’-AGA AAG GAG GTG ATC CAG CC-3’ (reverse primer) [23]. The nucleotide sequences thus obtained were assembled using the sequence alignment editor program Bioedit (http://www.mbio.ncsu.edu/Bio-Edit/bioedit.html) and was compared with those in GenBank after BLAST searches [24] and using the EzTaxon-e server [25]. 16S rRNA sequences of valid strains obtained from EzTaxon-e server were considered for secondary structure based phylogenetic analysis where E.coli 16S rRNA was used as reference template. All sequences were aligned using template based alignment algorithm CRWalign [26], edited and refined manually. The base positions were assigned according to E.coli numbering and helix numbering was adopted from ARB database [27]. The 16S rRNA based phylogenetic tree was constructed according to the Kimura two-parameter model using the MEGA 5 [28] software package (The Biodesign Institute, Arizona, USA). A phylogenetic tree was constructed using the neighbor joining method of Saitou & Nei [29] and with the maximum likelihood algorithms. The statistical significance of branch points was calculated by 1,000 bootstrap re-samplings of the data [30].
Determination of DNA base ratio and DNA-DNA homology study
For determination of DNA G+C content, DNA was degraded enzymatically into nucleosides as described by Mesbah et al. [31]. The obtained nucleoside mixture was then separated by HPLC (Shimadzu) using an analytical column (Vydac 201 SP54, C18, 5μm; 250 x 4.6 mm) equipped with a guard column (201 GD54H; Vydac). Non methylated lambda phage DNA (Sigma) was used as the calibration reference.
DNA-DNA re-association study is considered as standard method for bacterial species delineation [32]. Hence, DNA-DNA re-association study was performed among the isolates and between strain JS1T and other closely related type species showing 16S rRNA sequence similarity more than 97% following the method described by Ezaki et al. [33]. DNA from E. coli strain HB101 was taken as an unrelated negative control. Hybridization was performed following the methods described earlier [20].
ERIC and REP-PCR
ERIC-PCR (enterobacterial repetitive intergenic consensus sequence PCR) and REP-PCR (repetitive extragenic palindromic PCR) was performed with the isolated strains by method as described previously [34]. For ERIC PCR, two oligonucleotide primers were 5ʹ-ATG TAA GCT CCT GGG GAT TCA C-3ʹ and 5ʹ-AAG TAA GTG ACT GGG GTG AGC G-3ʹ. For Rep PCR two (18-mer) oligonucleotide primers were REP 1R-1 (5ʹ-III ICG ICG ICA TCI GGC-3’) and REP-21 (5ʹ-ICG ICT TAT CIG GCC TAC-3ʹ) [35]. The fingerprints were analyzed by using Bionumerics software (Applied Maths, Bio-Rad).
Chemotaxonomic analysis
For the analysis of polar lipids, and nature of diaminoacids in the peptidoglycan layer, cells were grown in TSB medium to mid log phase in a rotary shaker at 55°C. Polar lipids were determined by 2D-TLC following the method of Bligh & Dyer [36]. Cellular fatty acids were extracted from stationary phase cells grown on TSA at 55°C for 3 days and analyzed by previously described method [19]. Diaminopimelic acid (DAP) and cell wall sugar was detected following the method of Staneck & Roberts [37] by using the TLC on cellulose plates (Merck, cat no. 1.05577.0001, Germany).
Nucleotide sequence accession number
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene of strain JS1T, JS5, JS11 and JS15 are KC958552, KF772607, KF772610 and KF772608 respectively.
Results and Discussion
Abundance of bacterial population in the hot spring sample was very low for which enrichment procedure [5,12] was followed. Four pale yellow, rough surface circular colonies with uneven edges were picked for 16S rRNA sequence analysis in order to determine their phylogenetic position. 16S rRNA sequence of JS1T (1435 nt), JS5 (1427 nt), JS11 (1396 nt), and JS15 (1458 nt) were obtained in this study. The levels of 16S rRNA gene sequence similarity among the isolates were between 97.13 and 99.43% (Table S1). Biochemical tests with all isolates exhibited similar phenotype except hydrolysis of starch and utilization of sugars (Table S2). DNA-DNA homology values were well above 70% (Table S1). Looking into the phenotypic characteristics, 16S rRNA sequence similarity and DNA-DNA homology values, all isolates can be considered to represent same species of the genus Anoxybaciilus. Although, heterogeneity among the isolates were confirmed by ERIC and REP-PCR that produced distinct banding pattern (Figure S1A & B) indicating all four isolates were of non clonal origin and strain JS1T was considered for further characterization.
Strain JS1T was non motile, straight rod of length 1-4 µm and diameter 0.3-0.8 µm (Figure S2). It formed spherical spores located centrally or terminally (Figure S3). Phenotypically all strains were negative for indole, Voges Proskauer, H2S production, arginine dihydrolase, ornithine decarboxylase, lysine decarboxylase, phenylalanine deaminase and urease activity. All strains were positive for utilization of fructose, sucrose, trehalose; negative for utilization of citrate and malonate. However, considering the phenotypic and chemotaxonomic properties, the strain JS1T can be differentiated from other type strains of the genus Anoxybacillus. Phenotypic properties that differentiate strain JS1T from other closely related strains are listed in Table 1.
Table 1. Features that differentiate strain JS1T with its closely related species of Anoxybacillus.
| Features | 1 | 2 | 3 | 4 | 5 | 6 | 7*e | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Motility | - | + | + | + | + | + | - | + | + | + | + | + | + | + | - | + |
| Endospore | ||||||||||||||||
| Shape | S | E*a | O*b | E*j | E/Cy*d | E/Cy*d | S | E/C*f | S*g | O*h | E*i | S*g | E*j | S*k | ND | E*m |
| Position | T/C | T*a | T*b | T*j | T*d | T*d | T | T*f | T*g | T*h | T*i | T*g | T/ST*j | T*k | ND | C/ST*m |
| Swelling of sporangia | Slightly | ND | Slightly | +*j | ND | ND | - | ND | ND | ND | - | ND | Slightly*j | ND | ND | +*m |
| Biochemical tests | ||||||||||||||||
| Catalase | + | + | + | + | + | + | - | + | + | - | + | + | + | + | + | - |
| Oxidase | + | + | + | + | + | + | ND | - | + | - | + | + | - | + | + | - |
| Nitrate reduction | - | - | - | + | - | + | + | - | + | - | + | + | + | - | + | + |
| Methyl red | + | - | + | - | + | + | ND | - | - | - | - | - | + | - | - | + |
| Growth characteristics | ||||||||||||||||
| Temperature range (°C) | 40-60 | 30-66 | 30-75 | 30-72 | 35-70 | 30-70 | 37-65 | 55–67 | 30-70 | 37-66 | 37-69 | 40-70 | 37- 60 | 40-70 | 30-64 | 35-70 |
| Temperature optima (°C) | 55 | 60 | 60 | 60-65 | 55 | 50 | 62 | 65 | 50 | 57-62 | 60 | 50-55 | 50 | 55-60 | 54 | 60 |
| pH range | 5.5-11.5 | 5.5-10*a | 5.0-10.8*b | 5.5-9*c | 7-11*d | 7-11*d | 8-10.5 | 6-7.5*f | 6-11*g | 5.7-9.9*h | 5.5-9.5*i | 6-10.5*g | 4-9.5*j | 6-10*k | 7-8*l | 6-10*m |
| pH optima | 7.5 | 7-7.5*a | 8*b | 7*c | 8*d | 8.5*d | 9.5-9.7 | 7.2*f | 7.5-8.5*g | 6.8-8.5*h | 8-9*i | 7.5-8.5*g | 7*j | 7.5-8*k | ND | 7*m |
| NaCl tolerance (%) | 3.5 | 3.5 | 5 | 2.5 | 3 | 4 | 3 | 2.5 | 2.5 | 3 | 4.5 | 4 | 5 | 4 | 3 | 4 |
| Ethanol tolerance (%) | 2 | 12 | 2 | 2 | 2 | 4 | ND | 2 | 2 | - | 2 | 4 | - | 2 | - | 3 |
| Hydrolysis of | ||||||||||||||||
| Gelatin | + | - | + | - | + | + | - | - | + | - | + | - | + | + | - | + |
| Starch | + | - | + | + | + | + | + | - | + | + | + | + | + | + | - | - |
| Esculin | + | + | - | + | + | + | ND | + | + | + | + | + | - | - | + | + |
| DNA | + | + | - | - | - | - | ND | - | + | - | + | + | - | + | + | + |
| Acid production from D-glucose | + | - | + | + | + | + | ND | - | + | + | + | + | + | + | + | + |
| Utilization of | ||||||||||||||||
| Glucose | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + |
| Raffinose | - | w | w | - | - | - | - | - | - | - | - | + | + | + | + | |
| Mannose | + | w | w | + | + | + | - | + | + | + | + | + | + | w | + | |
| Mannitol | + | + | + | + | + | + | - | - | + | + | + | + | + | + | + | + |
| Lactose | + | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Galactose | - | + | + | + | - | + | - | - | + | + | + | + | + | + | + | |
| Xylose | - | + | + | - | - | - | - | - | + | - | - | - | + | + | + | + |
| Rhamnose | - | - | - | - | - | w | - | w | - | w | + | - | - | - | - | + |
| Maltose | + | + | + | + | + | + | ND | + | + | - | + | + | + | + | + | |
| Arabinose | - | + | - | w | - | - | ND | - | + | w | - | - | + | w | + | |
| G+C content (mol%) | 42.1 | 42.3*a | 44.2*b | 41.6*c | 42.6*d | 41.4*d | 42.2+/- 0.2 | 53.5*f | 54*g | 42.3*h | 45.1*i | 50*g | 44.4*j | 57*k | 43.9*l | 42.9*m |
All data are from present study unless indicated.
Strains: 1, A. suryakundensis strain JS1T; 2, A. flavithermus subsp. yunnanensis DSM 23293T; 3, A. mongoliensis DSM 19169T; 4, A. flavithermus subsp. flavithermus DSM 2614T; 5, A. eryuanensis KCTC 13720T; 6, A. tengchongensis KCTC 13721T; 7, A. pushchinoensis DSM 12423T; 8, A. thermarum DSM 17141T; 9, A. ayderensis NCIMB 13972T; 10, A. kamchatkensis DSM 14988T; 11, A. salavatliensis DSM 22626T; 12, A. kestanbolensis NCIMB 13971T; 13, A. contaminans DSM 15866T; 14, A. gonensis NCIMB 13933T; 15, A. voinovskiensis DSM 17075T; 16, A. kaynarcensis LMG 25303T.
Data from *a, Dai et al. [10]; *b, Namsaraev et al. [13]; *c, Heinen et al. [3]; *d, Zhang et al. [12]; *e, Pikuta et al. [1]; *f, Poli et al. [8]; *g, Dulger et al. [5]; *h, Kevbrin et al. [9]; *i, Cihan et al. [11]; *j, De Clerck et al. [6]; *k, Belduz et al. [4]; *l, Yumoto et al. [7]; *m, Inan et al. [14]
S, spherical; E, ellipsoidal; O, oval; Cy, cylindrical; ST, subterminal; C, central; +, positive reaction; w, weakly positive reaction; -, negative reaction; ND, not determined.
16S rRNA sequence analysis revealed strain JS1T showed 99.30% similarity with A. flavithermus subsp. yunnanensis DSM 23293T, 99.23% with A. mongoliensis DSM 19169T, 99.16% with A. eryuanensis KCTC 13720T, 98.74% with A. flavithermus subsp. flavithermus DSM 2614T, 98.54% with A. tengchongensis KCTC 13721T, 98.51% with A. pushchinoensis DSM 12423T, 97.91% with A. thermarum DSM 17141T, 97.82% with A. kaynarcensis LMG 25303T, 97.77% with A. ayderensis NCIMB 13972T, and A. kamchatkensis DSM 14988T, 97.63% with A. salavatliensis DSM 22626T, 97.55% with A. kestanbolensis NCIMB 13971T, 97.48% with A. contaminans DSM 15866T, 97.27% with A. gonensis NCIMB 13933T and 97.17% with A. voinovskiensis DSM 17075T. In the phylogenetic tree, strain JS1T clustered with A. eryuanensis KCTC 13720T with 49% bootstrap confidence (Figure 1). The phylogenetic tree presented in this paper was similar in its topology to the tree generated by the maximum likelihood algorithm. Further, phylogenetic diversity was confirmed by comparing 16S rRNA secondary structures to find genus specific and species specific base substitutions or deletions (signature positions). By comparing 16S rRNA sequence of A. suryakundnensis strain JS1T, 15 species specific base substitutions (signature positions) were determined with those of the closely related strains. Maximum variability (8 out of 15 signatures) was confined within region 70-100 i.e. helix 6 (Table 2).
Figure 1. Neighbour-joining phylogenetic tree showing the position of Anoxybacillus suryakundensis strain JS1T among the related taxa based on 16S rRNA secondary structure information.
E. coli 16S rRNA sequence has been selected as reference template. Bootstrap values expressed as percentages of 1000 replications are given at branch points. Accession numbers are given in parentheses. Bar 2 substitutions per 100 nucleotide position.
Table 2. Species specific base substitutions in 16S rRNA secondary structure between A. suryakundensis strain JS1T and phylogenetically closest members.
| Signature | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| helix | 6 | |||||||
| Position | 71 | 72 | 77 | 78 | 90 | 92 | 93 | 96 |
| A. suryakundensis | U | C | A | G | C | A | U | G |
| A. eryuanensis | C | G | A | A | U | G | A | C |
| A. mongoliensis | C | G | A | A | U | G | A | C |
| A. flavithermus subsp. yunnanensis | C | G | G | A | U | G | A | C |
| Signature | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| helix | 18 | 37 | 44 | 49 | ||||
| Position | 478 | 479 | 1011 | 1018 | 1134 | 1135 | 1453 | |
| A. suryakundensis | U | G | A | U | A | C | C | |
| A. eryuanensis | U | A | A | U | A | C | U | |
| A. mongoliensis | C | U | G | U | A | C | C | |
| A. flavithermus subsp. yunnanensis | U | G | G | C | G | G | C | |
The G+C content of strain JS1T was 42.1 mol%, a value consistent with those of the genus Anoxybacillus (Table 1).
In DNA-DNA hybridization study, using DNA of strain JS1T as a labeled probe, the mean DNA-DNA re-association values were well below 70% for all type strains tested (Table S3). Higher re-association values were obtained with A. flavithermus subsp. yunnanensis DSM 23293T (48.9±0.9%), A. mongoliensis DSM 19169T (39±3.6%) and A. eryuanensis KCTC 13720T (41.2±6.6%). Reciprocal probe were used in such cases that also indicated reliability of homology values. Therefore, given the recommended DNA–DNA relatedness cut-off point for species delineation of 70% [32], strain JS1T should be regarded as a novel species of the genus Anoxybacillus.
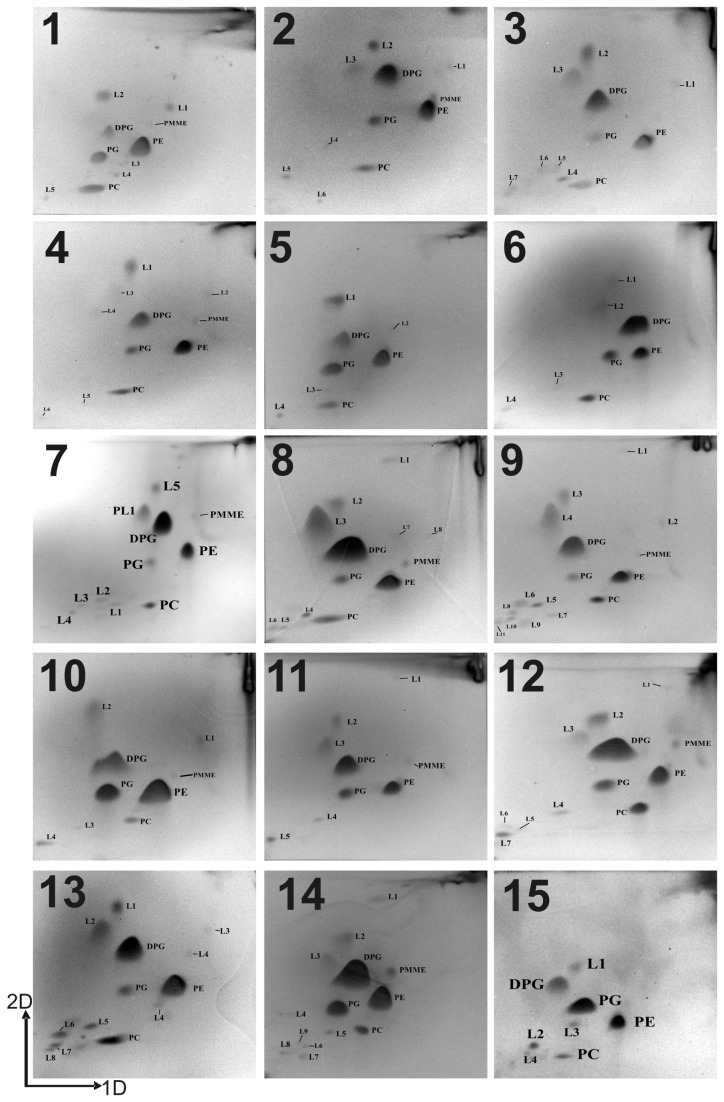
The major fatty acids of strain JS1T were C16:0iso, C15:0iso, C17:1anteiso w9c and C17:0iso. Fatty acid compositions were almost identical to those of the closely related reference strains used in this study. However, strain JS1T can be differentiated from other species by the absence of C17:0anteiso, C17:1anteisoA and C18:1w9c (Table 3). Polar lipids of strain JS1T and the closely related reference strains comprised a phosphatidylgylycerol (PG), a diphosphatidylglycerol (DPG), a phosphatidylethnolamine (PE) and a phosphatidylcholine (PC) (Figure 2). Besides, strain JS1T contained a phosphatidyl monomethylethanolamine (PMME) which was absent in A. contaminans, A. gonensis, A. voinovskiensis, and A. tengchongensis. In addition, several unidentified lipids were detected in the strain JS1T. Diaminopimelic acid was of meso type and glucose was sole sugar in cell wall in all strains that indicate genus uniformity.
Table 3. Fatty acid profiles of Anoxybacillus suryakundensis strain JS1T and the type strains of related Anoxybacillus species.
| Peak Name | 1 | 2 | 3 | 4 | 5 | 6 | 7* | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12:0 | tr | tr | tr | tr | tr | tr | 6.9 | tr | tr | tr | Tr | Tr | tr | tr | tr | 1.75 |
| 12:0 aldehyde? | 1.79 | 1.46 | 2.37 | tr | tr | 2.56 | - | tr | 4.16 | - | Tr | 2.65 | tr | - | 9.95 | tr |
| 14:0 | tr | 2.78 | 0.66 | - | 1.87 | 1.02 | 7.3 | 3.05 | tr | 1.02 | 1.10 | 2.02 | 1.13 | 1.09 | 1.24 | 1.09 |
| 14:0 3OH/16:1 iso I | 1.79 | 1.46 | 2.37 | tr | tr | 2.56 | - | tr | 4.16 | tr | tr | 2.65 | tr | 1.26 | 9.95 | tr |
| 14:0 iso | tr | 1.29 | tr | tr | tr | 1.87 | - | tr | 2.64 | tr | tr | 1.35 | tr | 1.95 | 5.25 | tr |
| 15:0 iso | 17.96 | 34.59 | 42.16 | 54.80 | 56.64 | 59.36 | 38.70 | 48.44 | 55.44 | 57.06 | 60.92 | 57.07 | 56.48 | 53.79 | 37.94 | 52.34 |
| 15:0 anteiso | tr | 4.16 | tr | 2.93 | 2.15 | 2.17 | 2.0 | 2.20 | 1.18 | 3.15 | 1.76 | 3.08 | 3.23 | 2.05 | 5.98 | 2.55 |
| 15:1 w5c | - | tr | - | tr | - | tr | - | 0.58 | tr | tr | tr | - | tr | tr | 2.25 | tr |
| 16:0 | tr | 9.42 | - | 6.01 | 6.16 | 3.69 | 14.5 | 9.17 | 2.13 | 2.70 | 2.99 | 2.77 | 5.15 | 3.53 | 3.33 | 3.52 |
| 16:0 iso | 56.47 | 9.47 | 24.09 | tr | 1.93 | 4.73 | tr | 1.40 | 10.84 | 1.71 | 1.29 | 2.33 | 3.29 | 8.29 | 13.22 | 1.92 |
| 16:1 w6c/16:1 w7c | 1.20 | 9.44 | 2.00 | 5.96 | 2.04 | 1.77 | - | 5.73 | 3.14 | 3.41 | 3.10 | 6.45 | 1.88 | 2.13 | 2.40 | 2.32 |
| 16:1 | - | - | - | - | - | - | 2.6 | - | - | - | - | - | - | - | - | - |
| 16:1 2OH | 1.03 | - | tr | - | - | - | - | tr | - | - | - | - | - | - | - | - |
| 17:0 iso | 3.61 | 12.95 | 10.80 | 7.33 | 9.46 | 11.99 | tr | 9.67 | 8.33 | 11.32 | 11.05 | 5.01 | 14.05 | 13.84 | 6.39 | 11.62 |
| 17:0 anteiso | - | 4.81 | 2.34 | 2.37 | 1.96 | 2.62 | tr | 2.98 | 1.02 | 3.30 | 1.43 | 1.62 | 3.54 | 2.91 | 3.30 | 3.32 |
| 17:1 iso w5c | 1.61 | 3.47 | 5.52 | 5.56 | 2.91 | 3.33 | - | 4.89 | 5.50 | 6.59 | 8.72 | 8.67 | 4.40 | 3.35 | 3.50 | 5.59 |
| 17:1 anteiso A | - | 1.95 | - | 1.58 | tr | tr | - | 1.87 | tr | 2.53 | 1.07 | 2.15 | tr | tr | 2.54 | 1.71 |
| 17:1 anteiso w9c | 6.14 | - | 2.46 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 18:0 | tr | tr | tr | 1.13 | - | tr | 10.4 | 1.12 | tr | tr | tr | - | tr | tr | tr | tr |
| 18:1 2OH | tr | - | - | tr | tr | - | - | 0.76 | 2.29 | - | tr | - | tr | - | - | 3.84 |
| 18:1 iso H | 2.11 | - | - | tr | - | - | - | - | - | - | - | - | - | - | - | - |
| 18:1 w9c | - | tr | tr | 4.04 | 3.76 | tr | 4.3 | 0.73 | tr | 1.15 | tr | tr | tr | tr | tr | tr |
| 18:2 | - | - | - | - | - | - | 2.2 | - | - | - | - | - | - | - | - | - |
Fatty acids were extracted from cells grown on tryptic soy agar plates at 55°C for 2 days. All data in the table are from the present study unless mentioned. Fatty acids amounting to less than 1.0% of the total fatty acids not mentioned in this table. Strains: 1, A. suryakundensis; 2, A. flavithermus subsp. yunnanensis DSM 23293T; 3, A. mongoliensis DSM 19169T; 4, A. flavithermus subsp. flavithermus DSM 2614T; 5, A. eryuanensis KCTC 13720T; 6, A. tengchongensis KCTC 13721T; 7, A. pushchinoensis DSM 12423T; 8, A. thermarum DSM 17141T; 9. A. ayderensis NCIMB 13972T; 10, A. kamchatkensis DSM 14988T; 11, A. salavatliensis DSM 22626T; 12, A. kestanbolensis NCIMB 13971T; 13, A .contaminans DSM 15866T; 14, A. gonensis NCIMB 13933T; 15, A. voinovskiensis DSM 17075T; 16, A. kaynarcensis LMG 25303T.
* Data from Pikuta et al. [1].
tr, trace amount i.e. <1%; -, not detected.
Figure 2. Polar lipid profile of strain JS1T and its closely related species.
Strains: 1, A. suryakundensis JS1T; 2, A. flavithermus subsp. yunnanensis DSM 23293T; 3, A. mongoliensis DSM 19169T; 4, A. flavithermus subsp. flavithermus DSM 2614T; 5, A. eryuanensis KCTC 13720T; 6, A. tengchongensis KCTC 13721T; 7, A. thermarum DSM 17141T; 8. A. ayderensis NCIMB 13972T; 9, A. kamchatkensis DSM 14988T; 10, A. salavatliensis DSM 22626T; 11, A. kestanbolensis NCIMB 13971T; 12, A. contaminans DSM 15866T; 13, A. gonensis NCIMB 13933T; 14, A. voinovskiensis DSM 17075T; 15, A. kaynarcensis LMG 25303T.
Based on phylogentic, phenotypic and genotypic differences presented in this study by polyphasic approach, it can be concluded that strain JS1T represents a novel species of genus Anoxybacillus for which the name Anoxybacillus suryakundensis is proposed.
Description of Anoxybacillus suryakundensis sp. nov
Anoxybacillus suryakundensis (sur.ya.kund.en’sis. N.L. masc. adj. suryakundensis pertaining to Suryakund in Jharkhand, India, the geographical origin of isolation of the type strain).
Cells are non motile, straight rod of length 1-4 µm and diameter 0.3-0.8 µm. Occurs single or in pair. Facultative anaerobe. Gram-positive. Endospores are spherical, located centrally or terminally and slightly swell sporangia. Aerobically grown colonies on TSA are pale yellow, rough surface, round with an uneven edge of diameter 1-2 mm. Moderately thermophilic and alkalitolerant. Grows at 40-60°C and pH 5.5-11.5 and can tolerate 3.5% NaCl. Optimum growth was observed at 55°C and pH 7.5. Catalase and oxidase positive. Positive for methyl red test and hydrolysis of gelatin, starch, esculin and DNA. Negative for reduction of nitrate, lysine decarboxylase, arginine dihydrolase, ornithine decarboxylase, phenylalanie deaminase, H2S production, indole, Voges Proskauer and hydrolysis of ONPG and urea. Acid is produced from oxidation of glucose, sucrose, fructose, trehalose, mannose, mannitol, lactose, maltose and starch. Negative for utilization of raffinose, galactose, xylose, rhamnose, malonate, citrate and arabinose. Cell wall contains glucose as sole sugar and DAP is of meso type. The major fatty acids are C16:0iso, C15:0iso, C17:1anteisow9c and C17:0iso. Polar lipids include a phosphatidylgylycerol (PG), a diphosphatidylglycerol (DPG), a phosphatidylethnolamine (PE), a phosphatidylcholine (PC), a phosphatidyl monomethylethanolamine (PMME) and four unidentified lipids (L1-4). The G+C content of DNA is 42.1 mol%. Anoxybacillus suryakundensis strain JS1T, was isolated from a hot spring at Suryakund, Jharkhand, India.
Supporting Information
Dendrogram based on (A) ERIC-PCR and (B) REP-PCR. The similarity between the species was calculated using the Pearson correlation coefficient for the range from 0.25 to 10 kb (optimization, 1%; position tolerance, 1%), and the species were grouped according to their similarities using UPGMA algorithm.
(TIF)
Electron micrograph of strain JS1T.
(TIF)
Bright field image of strain JS1T showing formation of endospore. TS, terminal spore; CS, central spore; ES, exospores.
(TIF)
16S rRNA similarity and DNA-DNA homology values among strain JS1T with other isolated strains. DNA-DNA homology values are mean of two replicates. Standard deviation values are given in parentheses.
(DOCX)
Physiological and biochemical properties of isolated strains.
(DOCX)
DNA-DNA homology values among strain JS1T and closely related species of Anoxybacillus. Reverse probe were used in cases when re-association values were intermediate i.e. near or above 40% Values are mean of two replicates. Standard deviation values are given in parentheses.
(DOCX)
Acknowledgments
We are thankful to Prof. J. P. Euzéby (Ecole Nationale Vétérinaire, France) for etymological advice. We are grateful to identification services at DSMZ, Germany for determination of G+C content of the strain JS1T. We are grateful to All India Institute of Medical Sciences, New Delhi, for electron microscopic facility. This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India. The authors, KD and AP acknowledge the University Grant Commission and Council of Scientific and Industrial Research, Government of India, New Delhi respectively for providing the fellowship.
Funding Statement
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India. The authors, KD and AP acknowledge the University Grant Commission and Council of Scientific and Industrial Research, Government of India, New Delhi respectively for providing the fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pikuta E, Lysenko A, Chuvilskaya N, Mendrock U, Hippe H et al. (2000) Anoxybacillus pushchinensis gen. nov., sp. nov., a novel anaerobic, alkaliphilic, moderately thermophilic bacterium from manure, and description of Anoxybacillus flavithermus comb. nov. Int J Syst Evol Microbiol 50: 2109–2117. doi: 10.1099/00207713-50-6-2109. PubMed: 11155986. [DOI] [PubMed] [Google Scholar]
- 2. Pikuta E, Cleland D, Tang J (2003) Aerobic growth of Anoxybacillus pushchinoensis K1T: emended descriptions of A. pushchinoensis and the genus Anoxybacillus . Int J Syst Evol Microbiol 53: 1561–1562. [DOI] [PubMed] [Google Scholar]
- 3. Heinen W, Lauwers AM, Mulders JWM (1982) Bacillus flavothermus, a newly isolated facultative thermophile. Antonie Van Leeuwenhoek 48: 265–272. doi: 10.1007/BF00400386. PubMed: 7125637. [DOI] [PubMed] [Google Scholar]
- 4. Belduz AO, Dulger S, Demirbag Z (2003) Anoxybacillus gonensis sp. nov., a moderately thermophilic, xylose-utilizing, endospore-forming bacterium. Int J Syst Evol Microbiol 53: 1315–1320. doi: 10.1099/ijs.0.02473-0. PubMed: 13130012. [DOI] [PubMed] [Google Scholar]
- 5. Dulger S, Demirbag Z, Belduz AO (2004) Anoxybacillus ayderensis sp. nov. and Anoxybacillus kestanbolensis sp. nov. Int J Syst Evol Microbiol 54: 1499–1503. doi: 10.1099/ijs.0.02863-0. PubMed: 15388701. [DOI] [PubMed] [Google Scholar]
- 6. De Clerck E, Rodríguez-Díaz M, Vanhoutte T, Heyrman J, Logan NA et al. (2004) Anoxybacillus contaminans sp. nov. and Bacillus gelatini sp. nov., isolated from contaminated gelatin batches. Int J Syst Evol Microbiol 54: 941-946. doi: 10.1099/ijs.0.02960-0. PubMed: 15143046. [DOI] [PubMed] [Google Scholar]
- 7. Yumoto I, Hirota K, Kawahara T, Nodasaka Y, Okuyama H et al. (2004) Anoxybacillus voinovskiensis sp. nov., a moderately thermophilic bacterium from a hot spring in Kamchatka. Int J Syst Evol Microbiol 54: 1239-1242. doi: 10.1099/ijs.0.02889-0. PubMed: 15280298. [DOI] [PubMed] [Google Scholar]
- 8. Poli A, Romano I, Cordella P, Orlando P, Nicolaus B et al. (2009) Anoxybacillus thermarum sp. nov., a novel thermophilic bacterium isolated from thermal mud in Euganean hot springs, Abano Terme, Italy. Extremophiles 13: 867-874. doi: 10.1007/s00792-009-0274-y. PubMed: 19710998. [DOI] [PubMed] [Google Scholar]
- 9. Kevbrin VV, Zengler K, Lysenko AM, Wiegel J (2005) Anoxybacillus kamchatkensis sp. nov., a novel thermophilic facultative aerobic bacterium with a broad pH optimum from the Geyser valley, Kamchatka. Extremophiles 9: 391-398. doi: 10.1007/s00792-005-0479-7. PubMed: 16142505. [DOI] [PubMed] [Google Scholar]
- 10. Dai J, Liu Y, Lei Y, Gao Y, Han F et al. (2011) A new subspecies of Anoxybacillus flavithermus ssp. yunnanensis ssp. nov. with very high ethanol tolerance. FEMS Microbiol Lett 320: 72-78. PubMed: 21521361. [DOI] [PubMed] [Google Scholar]
- 11. Cihan AC, Ozcan B, Cokmus C (2011) Anoxybacillus salavatliensis sp. nov., an α-glucosidase producing, thermophilic bacterium isolated from Salavatli, Turkey. J Basic Microbiol 50: 1-11. [DOI] [PubMed] [Google Scholar]
- 12. Zhang CM, Huang XW, Pan WZ, Zhang J, Wei KB et al. (2011) Anoxybacillus tengchongensis sp. nov. and Anoxybacillus eryuanensis sp. nov., facultatively anaerobic, alkalitolerant bacteria from hot springs. Int J Syst Evol Microbiol 61: 118-122. doi: 10.1099/ijs.0.020834-0. PubMed: 20173008. [DOI] [PubMed] [Google Scholar]
- 13. Namsaraev ZB, Babasanova OB, Dunaevsky YE, Akimov VN, Barkhutova DD et al. (2011) Anoxybacillus mongoliensis sp. nov., a novel thermophilic proteinase producing bacterium isolated from alkaline hot spring, central Mongolia. Microbiology 79: 491-499. [PubMed] [Google Scholar]
- 14. Inan K, Belduz AO, Canakci S (2013) Anoxybacillus kaynarcensis sp. nov., a moderately thermophilic, xylanase producing bacterium. J Basic Microbiol 53: 410-419. doi: 10.1002/jobm.201100638. PubMed: 22736500. [DOI] [PubMed] [Google Scholar]
- 15. Hou W, Wang S, Dong H, Jiang H, Briggs BR et al. (2013) A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLOS ONE 8: e53350. doi: 10.1371/journal.pone.0053350. PubMed: 23326417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coman C, Drugă B, Hegedus A, Sicora C, Dragoş N (2013) Archaeal and bacterial diversity in two hot spring microbial mats from a geothermal region in Romania. Extremophiles 17: 523-534. doi: 10.1007/s00792-013-0537-5. PubMed: 23568449. [DOI] [PubMed] [Google Scholar]
- 17. Panda SK, Jyoti V, Bhadra B, Nayak KC, Shivaji S et al. (2009) Thiomonas bhubaneswarensis sp. nov., an obligately mixotrophic, moderately thermophilic, thiosulfate-oxidizing bacterium. Int J Syst Evol Microbiol 59: 2171-2175. doi: 10.1099/ijs.0.007120-0. PubMed: 19605731. [DOI] [PubMed] [Google Scholar]
- 18. Jyoti V, Narayan KD, Das SK (2010) Gulbenkiania indica sp. nov., isolated from a sulfur spring. Int J Syst Evol Microbiol 60: 1052-1055. doi: 10.1099/ijs.0.014035-0. PubMed: 19666808. [DOI] [PubMed] [Google Scholar]
- 19. Panday D, Das SK (2010) Chelatococcus sambhunathii sp. nov., a moderately thermophilic alphaproteobacterium isolated from hot spring sediment. Int J Syst Evol Microbiol 60: 861–865. doi: 10.1099/ijs.0.013466-0. PubMed: 19661510. [DOI] [PubMed] [Google Scholar]
- 20. Bandyopadhyay S, Schumann P, Das SK (2013) Pannonibacter indica sp. nov., a highly arsenate-tolerant bacterium isolated from hot spring in India. Arch Microbiol 195: 1-8. doi: 10.1007/s00203-012-0840-z. PubMed: 22940883. [DOI] [PubMed] [Google Scholar]
- 21. Jung YT, Oh TK, Yoon JH (2012) Marinomonas hwangdonensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol 62: 2062–2067. doi: 10.1099/ijs.0.036582-0. PubMed: 22021582. [DOI] [PubMed] [Google Scholar]
- 22. Panday D, Schumann P, Das SK (2011) Rhizobium pusense sp. nov., isolated from the rhizosphere of chickpea (Cicer arietinum L.). Int J Syst Evol Microbiol 61: 2632-2639. [DOI] [PubMed] [Google Scholar]
- 23. Das SK, Mishra AK, Tindall BJ, Rainey FA, Stackebrandt E (1996) Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans (strain BI-42) gen. nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int J Syst Bacteriol 46: 981–987. doi: 10.1099/00207713-46-4-981. PubMed: 8863427. [DOI] [PubMed] [Google Scholar]
- 24. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402. doi: 10.1093/nar/25.17.3389. PubMed: 9254694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M et al. (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62: 716–721. doi: 10.1099/ijs.0.038075-0. PubMed: 22140171. [DOI] [PubMed] [Google Scholar]
- 26. Gardner DP, Xu W, Miranker DP, Ozer S, Cannone JJ et al. (2012) An Accurate Scalable Template-Based Alignment Algorithm. Proceedings of 2012 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2012), Philadelphia, PA. October 4-7, IEEE Computer Society, Washington, DC, USA: pp. 237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ludwig W, Strunk O, Westram R, Richter L, Meier H et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363-1371. doi: 10.1093/nar/gkh293. PubMed: 14985472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al. (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 10: 2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425. PubMed: 3447015. [DOI] [PubMed] [Google Scholar]
- 30. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 31. Mesbah M, Premachandran U, Whitman W (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39: 159 - 167. doi: 10.1099/00207713-39-2-159. [DOI] [Google Scholar]
- 32. Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O et al. (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37: 463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 33. Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39: 224–229. doi: 10.1099/00207713-39-3-224. [DOI] [Google Scholar]
- 34. Narayan KD, Pandey SK, Das SK (2010) Characterization of Comamonas thiooxidans sp. nov., and comparison of thiosulfate oxidation with Comamonas testosteroni and Comamonas composti . Curr Microbiol 61: 248-253. doi: 10.1007/s00284-010-9602-9. PubMed: 20148250. [DOI] [PubMed] [Google Scholar]
- 35. Rivera IG, Chowdhury MAR, Huq A, Jacobs D, Martins MT et al. (1995) Enterobacterial repetitive intergenic consensus sequences and PCR to generate fingerprints of genomic DNAs from Vibrio cholera O1, O139 and non-1 strains. Appl Environ Microbiol 61: 2898–2904. PubMed: 7487023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911-917. doi: 10.1139/o59-099. PubMed: 13671378. [DOI] [PubMed] [Google Scholar]
- 37. Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28: 226-231. PubMed: 4605116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendrogram based on (A) ERIC-PCR and (B) REP-PCR. The similarity between the species was calculated using the Pearson correlation coefficient for the range from 0.25 to 10 kb (optimization, 1%; position tolerance, 1%), and the species were grouped according to their similarities using UPGMA algorithm.
(TIF)
Electron micrograph of strain JS1T.
(TIF)
Bright field image of strain JS1T showing formation of endospore. TS, terminal spore; CS, central spore; ES, exospores.
(TIF)
16S rRNA similarity and DNA-DNA homology values among strain JS1T with other isolated strains. DNA-DNA homology values are mean of two replicates. Standard deviation values are given in parentheses.
(DOCX)
Physiological and biochemical properties of isolated strains.
(DOCX)
DNA-DNA homology values among strain JS1T and closely related species of Anoxybacillus. Reverse probe were used in cases when re-association values were intermediate i.e. near or above 40% Values are mean of two replicates. Standard deviation values are given in parentheses.
(DOCX)