Abstract
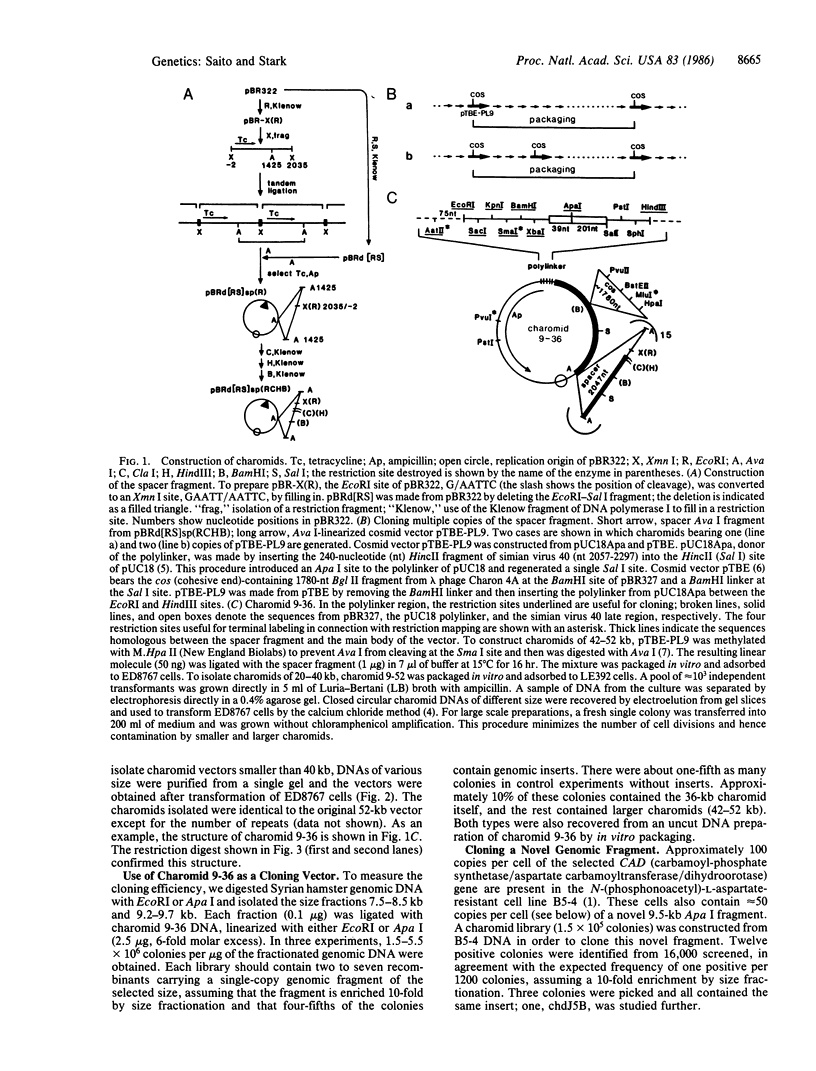
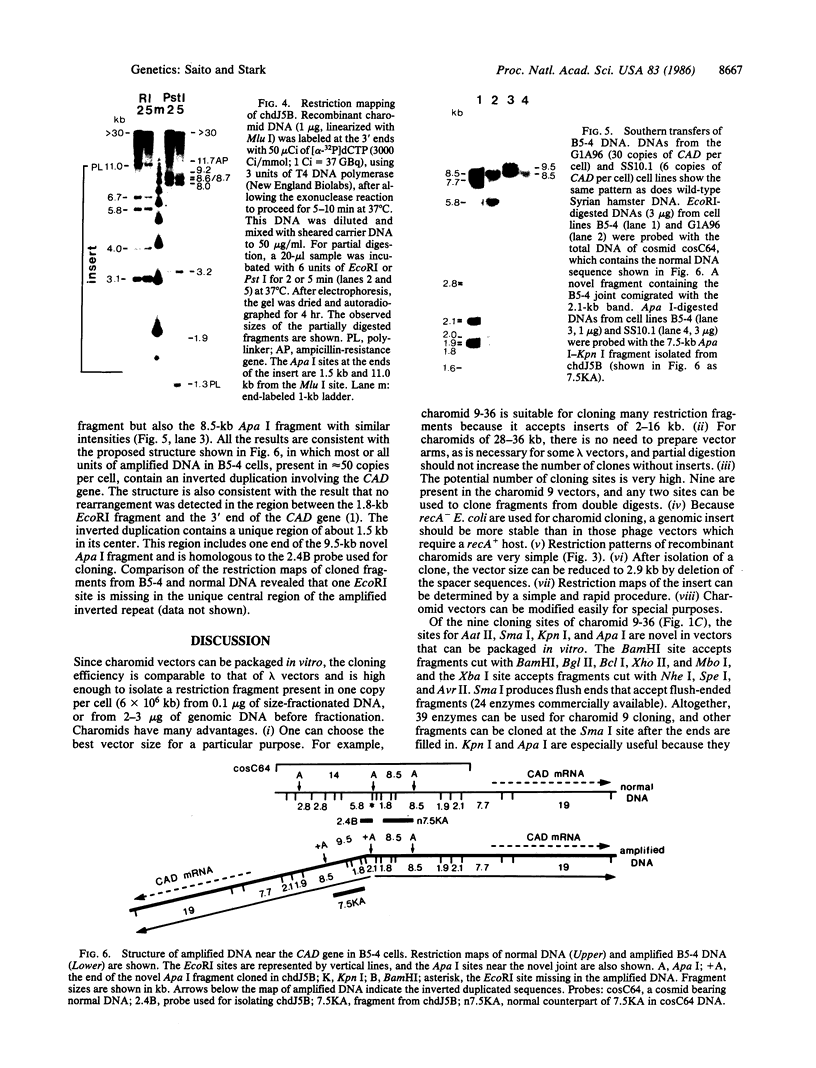
Charomids are cosmid vectors up to 52 kilobases (kb) long, bearing 1-23 copies of a 2-kb spacer fragment linked in head-to-tail tandem arrays. Like cosmids and lambda phage, charomids can be packaged in vitro for efficient introduction into bacteria. Charomids contain a polylinker with nine unique restriction sites for cloning and can be used without preparing vector arms. Using a charomid of appropriate size, one can clone inserts of any size up to 45 kb. For example, charomid 9-36 (9 cloning sites, 36 kb long) is too small to be packaged efficiently without an insert and can be used to clone fragments of 2-16 kb. The structure of charomids facilitates restriction mapping of the insert DNA and, after cloning, all the spacer fragments can be removed easily. After enrichment by size fractionation in an agarose gel, a specific single-copy genomic sequence can be cloned rapidly from approximately 3 micrograms of DNA. Using charomid 9-36, we have cloned and mapped an amplified novel DNA fragment from a cell line resistant to N-(phosphonoacetyl)-L-aspartate and carrying about 100 copies of the CAD (carbamoyl-phosphate synthetase/aspartate carbamoyltransferase/dihydroorotase) gene. The fragment lies at the center of an inverted duplication of this gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ardeshir F., Giulotto E., Zieg J., Brison O., Liao W. S., Stark G. R. Structure of amplified DNA in different Syrian hamster cell lines resistant to N-(phosphonacetyl)-L-aspartate. Mol Cell Biol. 1983 Nov;3(11):2076–2088. doi: 10.1128/mcb.3.11.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ford M., Davies B., Griffiths M., Wilson J., Fried M. Isolation of a gene enhancer within an amplified inverted duplication after "expression selection". Proc Natl Acad Sci U S A. 1985 May;82(10):3370–3374. doi: 10.1073/pnas.82.10.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Lund T., Murray E. J., Mellor A. L., Dahl H. H., Flavell R. A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982 Nov 11;10(21):6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Gregori T. J. Cloning multiple copies of a DNA segment. Gene. 1981 May;13(4):347–353. doi: 10.1016/0378-1119(81)90014-7. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz H. R., Zehetner G., Murialdo H., Delius H., Chai J. H., Poustka A., Frischauf A., Lehrach H. Analysis of cosmids using linearization by phage lambda terminase. Gene. 1985;40(2-3):259–266. doi: 10.1016/0378-1119(85)90048-4. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M. EcoK restriction during in vitro packaging of coliphage lambda DNA. Gene. 1985;39(2-3):313–315. doi: 10.1016/0378-1119(85)90330-0. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]