Abstract
Despite considerable advances in surgical repairing procedures, congenital heart diseases (CHDs) remain the leading noninfectious cause of infant morbidity and mortality. Understanding the molecular/genetic mechanisms underlying normal cardiogenesis will provide essential information for the development of novel diagnostic and therapeutic strategies against CHDs. BMP signaling plays complex roles in multiple cardiogenic processes in mammals. SMAD1 is a canonical nuclear mediator of BMP signaling, the activity of which is critically regulated through its interaction partners. We screened a mouse embryonic heart yeast two-hybrid library using Smad1 as bait and identified SERTAD1 as a novel interaction partner of SMAD1. SERTAD1 contains multiple potential functional domains, including two partially overlapping transactivation domains at the C terminus. The SERTAD1-SMAD1 interaction in vitro and in mammalian cells was further confirmed through biochemical assays. The expression of Sertad1 in developing hearts was demonstrated using RT-PCR, western blotting and in situ hybridization analyses. We also showed that SERTAD1 was localized in both the cytoplasm and nucleus of immortalized cardiomyocytes and primary embryonic cardiomyocyte cultures. The overexpression of SERTAD1 in cardiomyocytes not only enhanced the activity of two BMP reporters in a dose-dependent manner but also increased the expression of several known BMP/SMAD regulatory targets. Therefore, these data suggest that SERTAD1 acts as a SMAD1 transcriptional co-activator to promote the expression of BMP target genes during mouse cardiogenesis.
Keywords: BMP signaling, SMAD1, cardiogenesis, transcriptional co-activator, yeast two-hybrid
Introduction
Congenital heart diseases (CHDs) occur in as many as 1 to 5% of newborns and remain the leading noninfectious cause of infant morbidity and mortality [1,2]. Understanding the molecular and genetic mechanisms governing normal cardiogenesis will provide critical information for the development of novel diagnostic and therapeutic strategies against CHDs.
Bone Morphogenetic Proteins (BMPs) play complex roles during mammalian heart development; mutations that disturb BMP signaling lead to various forms of CHDs in both animal models and human patients [3,4,5,6]. BMP signaling is transduced through heterodimeric complexes of type I and type II serine/threonine kinase receptors on the cell surface. After formation of the receptor-ligand complex, the type II receptor phosphorylates the type I receptor, which in turn phosphorylates a group of cytoplasmic proteins called BMP receptor-activated-SMADs (R-SMADs), including SMAD1, SMAD5, and SMAD8. Phosphorylated SMAD1/5/8 associates with SMAD4, a transcriptional co-activator of R-SMADs, and the SMAD complex translocates to the nucleus to regulate the expression of target genes [7,8]. To activate the transcription of target genes, BMP R-Smads form a large nucleoprotein complex with SMAD-binding DNA elements, sequence specific transcription factors, and transcriptional co-activators. Many cardiogenic genes, such as Nkx2.5 [9,10,11,12], are directly regulated through BMP R-SMADs.
It has long been hypothesized that SMAD4 is essential for nearly all R-SMAD-mediated transcription [13,14]; however, this idea has been challenged by later studies. The inactivation of Smad4 in epiblasts causes embryonic lethality at E8.5, yet heart rudiments are still formed in mutants, indicating that Smad4 is not required for the Bmpr1a-mediated induction of cardiomyocytes from the lateral mesoderm [15,16]. We recently showed that Smad4 promotes cardiomyocyte proliferation and survival through the upregulation of Mycn expression; however, many BMP-mediated processes, including AV canal specification, extracellular matrix deposition, and outflow tract remodeling are not compromised by the myocardial inactivation of Smad4 [17]. These results collectively suggest the presence of other co-activators of BMP R-SMADs that are also crucial for mediating the complex activities of BMP signaling during cardiogenesis.
In this study, we report the identification of SERTAD1 as a novel interaction partner of SMAD1 in mouse embryonic hearts. Sertad1 encodes a 236-a.a. protein with five potential functional domains [18,19,20,21]. The N terminus of SERTAD1 is homologous to the Cyclin A binding sequence (a.a.1–30); however, whether Sertad1 interacts with Cyclin A has not been experimentally determined. The SERTA domain (a.a.43–82) refers to a novel heptad hydrophobic repeat, which has been identified in multiple proteins from insects to humans. The exact biological activities of the SERTA domain remain unclear. The CDK4 binding domain (a.a.30–160) is responsible for the direct interaction between CDK4 and SERTAD1. The C-terminal half of SERTAD1 contains two partially overlapping transactivation domains: the PHD-bromodomain- interacting domain (a.a.161–178), which interacts with the bromodomain and/or PHD zinc finger-containing general transcriptional co-activators (such as p300/CBP), and the acidic region (a.a.167–220), which has intrinsic transactivation activity [18,19,20]. SERTAD1 does not contain any known DNA binding motifs, nor has this protein been reported to interact with DNA directly. The current view is that SERTAD1 forms a complex with transcription factors (such as E2F1) to co-activate the transcription of target genes [18,19]. In addition to regulating gene expression, SERTAD1 directly interacts with CDK4 to antagonize the activity of the cdk inhibitor p16INK4, thereby promoting cell proliferation [21,22]. Our study provides the first evidence that SERTAD1 acts as a transcriptional co-activator of SMAD1 to promote the expression of BMP target genes in mouse embryonic hearts.
Materials and Methods
1. Mouse maintenance
This study conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 1996). All protocols were approved through the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham. The CD1 mice were purchased from Charles River.
2. Detection of the SERTAD1-SMAD1 interaction
Yeast two-hybrid screening using Smad1 as bait, in vitro GST-pull down, and mammalian cell GST-pull down analyses were performed as previously described [23,24].
3. Seartad1 expression analysis
Total RNA from pooled embryonic hearts or whole embryonic bodies was isolated using the RNeasy mini kit (Qiagen). Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis was performed using the OneStep RT-PCR kit (Qiagen). Western analysis was performed as previously described [24]. Non-isotope section in situ analysis was performed as previously described [25]. The ~300bp-cDNA fragment from the 3’ untranslated region of Sertad1 was used as the probe.
4. Cell culture, SERTAD1 subcellular localization analysis and reporter analysis
NkL-TAg cells were cultured as previously described [26]. Primary cardiomyocytes were isolated from E13.5 embryos as previously described [27]. BMP4 was purchased from R&D. For immunostaining assays, the cells were cultured on glass coverslips and incubated with a primary antibody at 4°C overnight. The next day, the cells were incubated with an Alexa488- (or Alexa594-) conjugated secondary antibody (Life Technologies). The cells were further stained with DAPI to visualize total nuclei. The samples were examined using a Zeiss LSM 710 confocal microscope. Nuclear/cytoplasmic fractionation was achieved using a kit purchased from ThermoSceintific following manufacturer’s instructions. Western analysis was performed using antibodies against SERTAD1, MEK1/2 (a cytoplasmic marker) and LSD1 (a nuclear marker). For the reporter analysis, NkL-TAg cells were co-transfected with a reporter construct and different doses of a construct expressing SERTAD1 using Lipofectamine 2000 (Life Technologies). The luciferase activity was determined using the Luciferase Assay kit from Promega. A lacZ reporter plasmid was co-transfected to normalize the transfection efficiency. LacZ activity was measured using the Beta-Glo Assay System (Promega).
5. Primary antibodies used in this study
The HA antibody was purchased from Babco. The GST antibody was purchased from Amersham. The SERTAD1 antibody was obtained from AVIVA. Antibodies against phosphorylated SMAD1 (p-SMAD1), MEK1/2, LSD and the myc tag were purchased from Cell Signaling. Antibodies against TBX20 and ID2 were purchased from Sigma. Antibodies against ACTIN, TUBULIN and cardiac troponin T (cTnt) were purchased from the Iowa Hybridoma Bank.
Results
1. Identification of SERTAD1 as a novel interaction partner of SMAD1 in mouse embryonic hearts
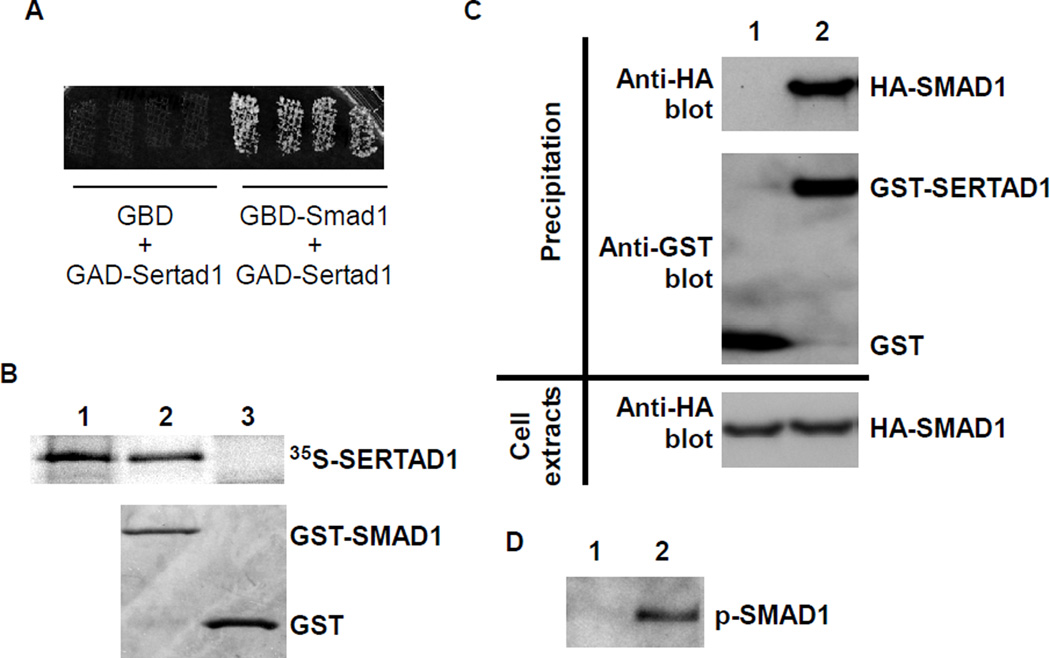
To identify novel interaction partners of BMP R-SMADs during cardiogenesis, we screened a mouse embryonic heart two-hybrid library [24] using Smad1 as bait. We acquired three identical clones that align to the region at −36 to +841 bp (relative to the start codon) of the Sertad1 cDNA. This fragment covers the entire coding region of Sertad1 (+1 to +708 bp). Fig. 1A shows that yeast cells harboring both the Sertad1 prey construct and the Smad1 bait construct grew on the selective medium, indicating a positive interaction between SERTAD1 and SMAD1. In the control experiment, the Sertad1 prey did not interact with the empty bait vector. To exclude the possibility that the SERTAD1-SMAD1 interaction was mediated through unknown yeast proteins, we performed an in vitro GST pull-down analysis (Fig. 1B). The 35S labeled SERTAD1 was pulled down with GST-SMAD1 but not with GST, demonstrating a direct interaction between the two proteins. We further performed a mammalian cell GST pull-down analysis to examine the interaction of these proteins in mammalian cells. GST-Sertad1 (or GST alone, negative control) and HA-Smad1 were co-expressed in HEK293 cells. A constitutively active form of ALK6 (Q203D) was co-transfected into HEK cells to activate the BMP intracellular signaling cascade. HA-Smad1 was co-purified with GST-Sertad1, but not with GST, from cell lysates (Fig. 1C), confirming that SERTAD1 interacts with SMAD1 in mammalian cells. In the absence of constitutively active ALK6, the association of SMAD1 with SERTAD1 was marginal (data not shown), indicating that the SMAD1-SERTAD1 interaction is enhanced through active BMP signaling. We further confirmed that active (phosphorylated) SMAD1 was co-purified with SERTAD1 through western analysis using a p-SMAD1 specific antibody (Fig. 1D). In both in vitro and in vivo pull down assays, the cDNA fragment encoding the full ORF of SERTAD1 (+1 to +708 bp) was used. Therefore, the interaction between the two proteins is not mediated through the extra 12 amino acids at the N-terminus of the protein encoded by the original clone identified from two-hybrid screening.
Figure 1. SERTAD1 interacts with SMAD1.
(A) AH109 yeast cells were transformed with various constructs as indicated and grown on selective medium. Growth indicates a positive interaction between the bait and prey. GBD: empty bait vector; GBD-Smad1: bait vector containing the full length of Smad1; GAD-Sertad1: the prey vector containing Sertad1. (B) SERTAD1 was labeled with [35S]methionine and incubated with bacterial-expressed GST (negative control) or GST-SMAD1. The signals were visualized through autoradiography (top panel). The bottom panel shows the Coomassie blue staining of a SDS-PAGE gel. Lane 1: free probe (10%); lane 2: 35S-SERTAD1 + GST-SMAD1; lane 3: 35S-SERTAD1 + GST. (C) A plasmid expressing HA-SMAD1 was co-transfected with a plasmid expressing GST alone (negative control, lane 1) or GST-SERTAD1 (lane 2) into HEK293 cells. The expression of HA-SMAD1 in HEK cells was confirmed through western analysis using an anti-HA antibody (bottom panel). The GST and GST-SERTAD fusion proteins were purified with GST-binding beads followed by western blot analysis. An HA antibody was applied to determine whether HA-SMAD1 was co-purified with GST-CHD7 (top panel). The successful purification of GST and GST-SERTAD1 was confirmed through western analysis using a GST antibody (middle panel). Lane 1: GST + HA-SMAD1; lane 2: GST-SERTAD1 + HA-SMAD1. (D) The leftover samples from the experiment shown in panel C were applied for an additional western analysis using a p-SMAD1-specific antibody. The active form of SMAD1 was co-purified with GST-SERTAD1. Lane 1: GST + HA-SMAD1; lane 2: GST-SERTAD1 + HA-SMAD1.
2. Sertad1 is expressed in embryonic hearts
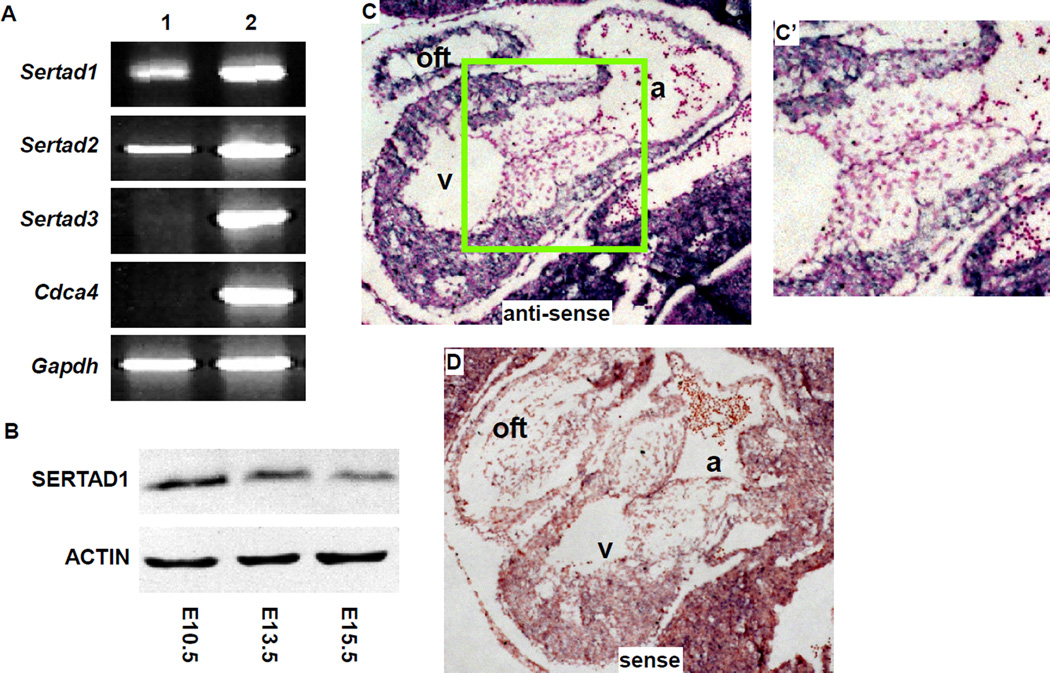
To directly examine whether Sertad1 is expressed during mouse cardiogenesis, we first performed RT-PCR analysis using RNA samples extracted from pooled embryonic hearts (E9.5–E11.5) or embryonic bodies. We detected the expression of Sertad1 and its homolog Sertad2 in embryonic hearts, whereas the cardiac expression of the other two Sertad1 homologs, Sertad3 and Cdca4, was not detected (Fig. 2A). To examine SERTAD1 expression at the protein level, we performed western analysis. The expression of SERTAD1 was detected in embryonic hearts at all stages tested (E10.5 to E15.5) (Fig. 2B). The expression level was reduced at E15.5 compared with earlier stages. The expression of Sertad1 in embryonic hearts was further verified through in situ hybridization analysis (Fig. 2C, C’). In embryonic hearts at E10.5, Sertad1 transcripts were observed only in the myocardium, and not in the endocardial or cushion mesenchymal cells, suggesting that at this stage, Sertad1 primarily regulates the cardiogenic processes in myocardial cells, but not in endocardial/mesenchymal cells.
Figure 2. Sertad1 is expressed in developing mouse hearts.
(A) RT-PCR analysis was performed using RNA samples extracted from pooled E9.5–11.5 embryonic hearts (lane 1) or whole embryos (lane 2). No signal was detected from the “no RT” control reactions (data not shown). Gapdh was used as a loading control. (B) Western analysis was performed on proteins extracted from embryonic hearts using anti-SERTAD1 or anti-ACTIN antibodies (loading control). (C, C’) In situ hybridization analysis was performed on sagittal sections of E10.5 embryos using a Sertad1 anti-sense probe (purple). Total nuclei were visualized with nuclear-fast-red staining (red). (C’) corresponds to the boxed area of panel C. (D) The sense probe was used as a negative control. a: atrium; oft: outflow tract; v: ventricle.
3. SERTAD1 is localized in both the cytoplasm and nucleus of cardiomyocytes
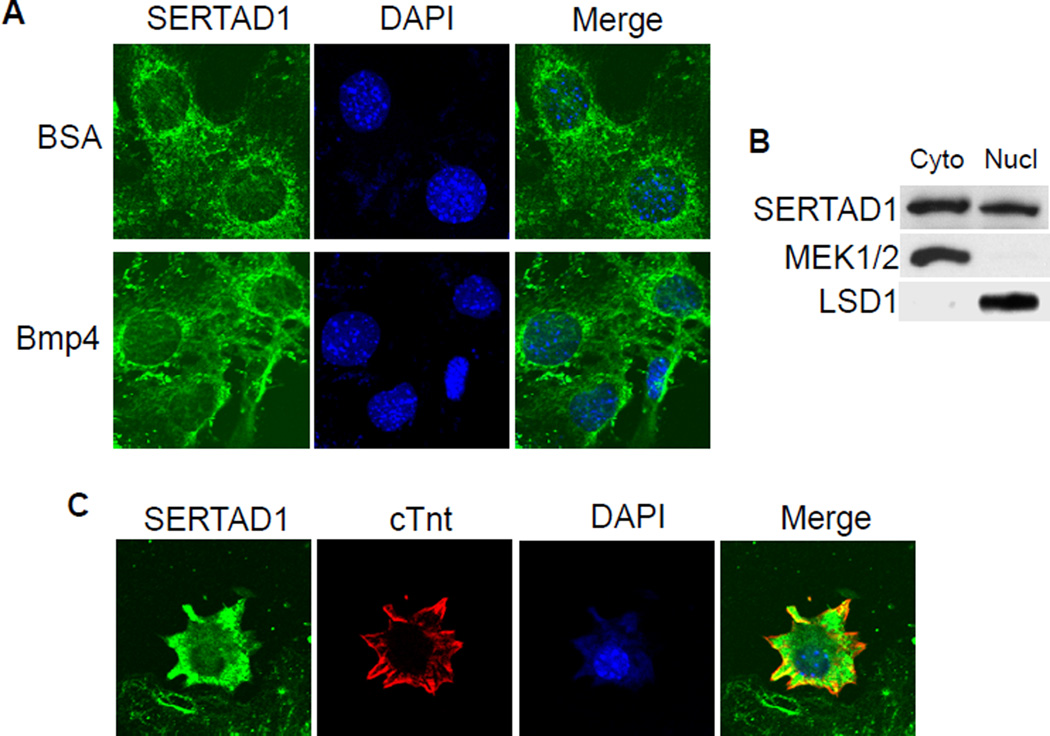
Depending on the cell types examined, SERTAD1 has been reported as localized to the nucleus [20,22,28] or cytoplasm [29]. We therefore determined the subcellular localization of SERTAD1 in cardiomyocytes. NkL-TAg cells are immortalized cardiomyocytes widely used to study various biological processes in heart muscle cells [26,30]. We immunostained NkL-TAg cells using a SERTAD1-specific antibody and detected positive signals in both the nucleus and cytoplasm (Fig. 3A). The subcellular localization of SERTAD1 was not altered by BMP stimulation. The western analysis confirmed that SERTAD1 is detected in both the cytoplasmic and nuclear fractions of NkL-TAg cells (Fig. 3B). The subcellular localization of SERTAD1 was further confirmed through immunostaining primary embryonic cardiomyocyte cultures (E13.5) (Fig. 3C). Although the signal detected in the cytoplasm was stronger, positive signals were clearly detected in nuclei.
Figure 3. SERTAD1 is localized in both the cytoplasm and nucleus of cardiomyocytes.
(A) NkL-TAg cells were treated with BSA (negative control) or 100ng/ml BMP4 for 2 hours, followed by immunostaining using an anti-SERTAD1 antibody (green). The nuclei were visualized with DAPI staining (blue). The samples were examined under a Zeiss confocal microscope. Green signals were detected in both the cytoplasm and nucleus. No difference was observed between the BSA- and BMP4-treated samples. (B) The cytoplasmic and nuclear fractions of NkL-TAg cells were subjected to western analysis using a SERTAD1 antibody. MEK1/2 and LSD1 were used as cytoplasmic and nuclear controls, respectively. (C) Embryonic cardiomyocytes (E13.5) were isolated and cultured for 24 hours. Immunostaining was performed using antibodies against SERTAD1 (green) and cTnt (red). Total nuclei were visualized with DAPI staining (blue). Green signals were detected in both the cytoplasm and nucleus.
4. SERTAD1 enhances BMP/SMAD-mediated transcription
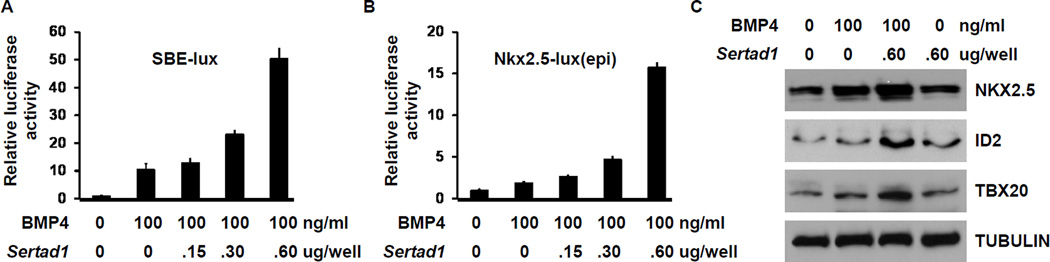
Considering the function of SERTAD1 as a transcriptional co-activator of E2F1 [18,19], we examined whether Sertad1 enhances BMP/SMAD-mediated transcription. We first performed reporter analyses using two BMP-responsive reporters in NkL-TAg cells. The SBE-lux construct contains 4 copies of SMAD binding elements from the JunB promoter and has been widely used to study the transcriptional regulation of TGFβ/BMP signaling [31,32]. In the Nkx2.5-lux(epi) construct, the genomic DNA containing the G/S and AR2 enhancers of Nkx2.5 was fused with the SV40 minimum promoter and cloned into the pREP4-luc reporter vector. The G/S and AR2 enhancers of Nkx2.5 harbor functional BMP R-SMAD binding elements demonstrated through in vitro and transgenic studies [9,10]. pREP4-luc is an Epstein-Barr virus-based episomal vector that undergoes chromatinization after transfected into mammalian cells [33]. Therefore, the Nkx2.5-lux(epi) construct is expected to more closely resemble activities of enhancers in the context of chromosomes. Overexpression of SERTAD1 enhanced the activities of both reporters in a dose-dependent manner (Fig. 4A,B). We further showed that the overexpression of SERTAD1 upregulated the expression of endogenous NKX2.5, ID2 and TBX20 upon BMP4 stimulation in NkL-TAg cells (Fig. 4C). Nkx2.5, Id2, and Tbx20 are direct regulatory targets of BMP R-SMADs in embryonic hearts.
Figure 4. Overexpression of SERTAD1 enhanced BMP R-SMAD mediated transcription in cardiomyocytes.
(A) NkL-TAg cells were cultured in 24-well plates and were co-transfected with SBE-lux and different doses of the construct expressing SERTAD1. The cells were treated with or without BMP4 (100ng/ml) for 48 hours, followed by luciferase analysis. The luciferase activity from cells that were not transfected with the Sertad1 construct and were not treated with BMP4 was set at 1.0. The data were averaged from three independent experiments. The error bars represent standard deviation. (B) Same as panel A except that the Nkx2.5-lux(epi) reporter construct was used. (C) NkL-TAg cells were transfected with an empty vector or a plasmid expressing SERTAD1 and treated with or without BMP4 (100ng/ml) for 48 hours. Total protein extracts were subjected to western analysis using antibodies as indicated. The overexpression of SERTAD1 upregulated the expression of the three BMP targeted genes. TUBULIN was used as a loading control.
Discussion
The activities of R-SMADs are critically regulated through their interaction partners [7,8]. The present study provides convincing evidence supporting the idea that SERTAD1 acts as a co-activator of SMAD1 to promote BMP-mediated gene expression in mouse embryonic hearts. The SERTAD1-SMAD1 interaction is demonstrated with multiple complementary approaches, including yeast two-hybrid, in vitro GST pull-down and mammalian cell co-purification analyses. Our sub-cellular localization analysis showed that SERTAD1 was localized in both the cytoplasm and nucleus in cardiomyocytes. Consistent with the presence of SERTAD1 in the nucleus, we showed that activated (phosphorylated) SMAD1, localized in the nucleus, was co-purified with SERTAD1 from mammalian cells. The nuclear localization of SERTAD1 and its interaction with p-SMAD supports SERTAD1 as a transcriptional co-activator. Furthermore, our functional analysis showed that the overexpression of SERTAD1 in immortalized cardiomyocytes not only enhanced the activities of two BMP-responsive reporters but also upregulated the expression of endogenous NKX2.5, ID2 and TBX20 upon BMP stimulation.
In a previous study, we showed that the inactivation of Smad4 in the myocardium of embryonic hearts led to severe myocardial wall defects and embryonic lethality [17], supporting a critical role for SMAD-mediated transcription during cardiomyogenesis. Notably, many cardiogenic activities of BMP signaling were not impaired by the deletion of Smad4. For example, Nkx2.5 and some other known BMP/SMAD target cardiogenic genes remain normally expressed in Smad4-deleted hearts [17]. As a novel SMAD1 co-transcriptional activator, we speculate that SERTAD1 mediates certain SMAD4-independent cardiogenic activities of BMP signaling. Consistently, we observed that the overexpression of SERTAD1 upregulated NKX2.5 expression. We are currently examining the interrelationship between SERTAD1 and SMAD4.
The subcellular localization of SERTAD1 is currently under debate. The nuclear localization of SERTAD1 has been reported in multiple studies [20,22,28]. However, a recent study using an anti-SERTAD1 antibody showed that endogenous SERTAD1 is exclusively localized in the cytoplasm of primary fibroblasts and some cancer cell lines [29]. These authors argued that the differential results reflect the use of tagged versions (HA or Flag tag) of SERTAD1 to examine its cellular localization. They speculated that tagging SERTAD1 somehow altered the distribution of the protein in cells. To avoid any potential complication from tagging SERTAD1, we used a SERTAD1-specific antibody to examine the subcellular distribution of endogenous SERTAD1. In a cardiomyocyte cell line and primarily cultured embryonic cardiomyocytes, SERTAD1 is localized in both the cytoplasm and nucleus. This result is supported through western analysis of nuclear/cytoplasmic fractions of NkL-TAg cells. Thus, we speculate that the different cellular localization of SERTAD1 reported in different studies reflect the use of different cell types, suggesting that the cellular localization of SERTAD1 is cell-type dependent. Consistent with our speculation, myc-tagged SERTAD1 was also localized in both the cytoplasm and nucleus of NkL-TAg cardiomyocytes (Sup. Fig. 1). The differential results between the present study and the previous study [29] might also reflect the different types of antibodies used. In the previous study, a mouse monoclonal SERTAD1 antibody was used, whereas in the present study we used a rabbit polyclonal antibody. It is possible that the monoclonal antibody detects only cytoplasmic SERTAD1, but not nuclear SERTAD1, due to the conformational differences between the two forms of the protein. Further studies are warranted to study the subcellular localization of SERTAD1 in different cell types under different pathophysiological conditions.
Studies using Sertad1-inactivated mice showed that this gene is important for maintaining the proper number of pancreatic beta cells [34]. However, no embryonic phenotype was reported. The lack of embryonic heart defects in Sertad1 knockout mice likely reflects the redundant functions of Sertad2, another SERTA gene expressed in embryonic hearts (Fig. 2). Sertad2 regulates fat storage but is not an essential gene for embryonic development [35]. We speculate that Sertad1 and Sertad2 share redundant functions in embryos and further double gene inactivation experiments are required to reveal the embryonic activities of the two genes.
In summary, we identified SERTAD1 as a novel nuclear interaction partner of SMAD1 in mouse embryonic hearts. SERTAD1 acts as a co-activator of SMAD1 to enhance BMP/SMAD-mediated transcription. These results provide crucial information concerning the BMP/SMAD pathway-mediated regulation of cardiogenic gene expression in embryonic hearts.
Supplementary Material
SERTAD1 interacts with SMAD1.
Sertad1 is expressed in mouse embryonic hearts.
SERTAD1 is localized in both cytoplasm and nucleus of cardiomyocytes.
SERTAD1 enhances expression of BMP target cardiogenic genes as a SMAD1 co-activator.
Acknowledgments
The authors would like to thank Drs. E. Olson (UT Southwestern) and K. Zhao (NIH) for providing reagents. The authors would also like to thank past and present members of the Jiao lab for assistance with this project. This project was supported through a Grant-in-Aid (09GRNT2060268) from the American Heart Association and an R01 grant (R01HL095783-01A1) from NIH awarded to KJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. doi: 10.1146/annurev.physiol.68.040104.113828. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Greene SB, Martin JF. BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res A Clin Mol Teratol. 2011;91:441–448. doi: 10.1002/bdra.20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Euler-Taimor G, Heger J. The complex pattern of SMAD signaling in the cardiovascular system. Cardiovasc Res. 2006;69:15–25. doi: 10.1016/j.cardiores.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Waite KA, Eng C. From developmental disorder to heritable cancer: it's all in the BMP/TGF-beta family. Nat Rev Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 6.Harradine KA, Akhurst RJ. Mutations of TGFbeta signaling molecules in human disease. Ann Med. 2006;38:403–414. doi: 10.1080/07853890600919911. [DOI] [PubMed] [Google Scholar]
- 7.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 9.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 10.Brown CO, 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 11.Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 12.Lee KH, Evans S, Ruan TY, Lassar AB. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- 13.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 14.Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Miura S, Davis S, Klingensmith J, Mishina Y. BMP signaling in the epiblast is required for proper recruitment of the prospective paraxial mesoderm and development of the somites. Development. 2006 doi: 10.1242/dev.02552. [DOI] [PubMed] [Google Scholar]
- 16.Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- 17.Song L, Yan W, Chen X, Deng CX, Wang Q, Jiao K. Myocardial Smad4 Is Essential for Cardiogenesis in Mouse Embryos. Circ Res. 2007;101:277–285. doi: 10.1161/CIRCRESAHA.107.155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. Embo J. 2001;20:2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe-Fukunaga R, Iida S, Shimizu Y, Nagata S, Fukunaga R. SEI family of nuclear factors regulates p53-dependent transcriptional activation. Genes Cells. 2005;10:851–860. doi: 10.1111/j.1365-2443.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- 20.Lai IL, Wang SY, Yao YL, Yang WM. Transcriptional and subcellular regulation of the TRIP-Br family. Gene. 2007;388:102–109. doi: 10.1016/j.gene.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Melvin WS, Tsai MD, Muscarella P. The nuclear protein p34SEI-1 regulates the kinase activity of cyclin-dependent kinase 4 in a concentration-dependent manner. Biochemistry. 2004;43:4394–4399. doi: 10.1021/bi035601s. [DOI] [PubMed] [Google Scholar]
- 22.Sugimoto M, Nakamura T, Ohtani N, Hampson L, Hampson IN, Shimamoto A, Furuichi Y, Okumura K, Niwa S, Taya Y, Hara E. Regulation of CDK4 activity by a novel CDK4-binding protein, p34(SEI-1) Genes Dev. 1999;13:3027–3033. doi: 10.1101/gad.13.22.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao K, Zhou Y, Hogan BL. Identification of mZnf8, a mouse Kruppel-like transcriptional repressor, as a novel nuclear interaction partner of Smad1. Mol Cell Biol. 2002;22:7633–7644. doi: 10.1128/MCB.22.21.7633-7644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debenedittis P, Harmelink C, Chen Y, Wang Q, Jiao K. Characterization of the novel interaction between muskelin and TBX20, a critical cardiogenic transcription factor. Biochem Biophys Res Commun. 2011;409:338–343. doi: 10.1016/j.bbrc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L, Fassler R, Mishina Y, Jiao K, Baldwin HS. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Rybkin, Markham DW, Yan Z, Bassel-Duby R, Williams RS, Olson EN. Conditional expression of SV40 T-antigen in mouse cardiomyocytes facilitates an inducible switch from proliferation to differentiation. J Biol Chem. 2003;278:15927–15934. doi: 10.1074/jbc.M213102200. [DOI] [PubMed] [Google Scholar]
- 27.Gruber PJ, Kubalak SW, Chien KR. Downregulation of atrial markers during cardiac chamber morphogenesis is irreversible in murine embryos. Development. 1998;125:4427–4438. doi: 10.1242/dev.125.22.4427. [DOI] [PubMed] [Google Scholar]
- 28.Hirose T, Fujii R, Nakamura H, Aratani S, Fujita H, Nakazawa M, Nakamura K, Nishioka K, Nakajima T. Regulation of CREB-mediated transcription by association of CDK4 binding protein p34SEI-1 with CBP. Int J Mol Med. 2003;11:705–712. [PubMed] [Google Scholar]
- 29.Zang ZJ, Gunaratnam L, Cheong JK, Lai LY, Hsiao LL, O'Leary E, Sun X, Salto-Tellez M, Bonventre JV, Hsu SI. Identification of PP2A as a novel interactor and regulator of TRIP-Br1. Cell Signal. 2009;21:34–42. doi: 10.1016/j.cellsig.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 30.el Azzouzi H, Leptidis S, Bourajjaj M, Armand AS, van der Nagel R, van Bilsen M, Da Costa Martins PA, De Windt LJ. Peroxisome proliferator-activated receptor (PPAR) gene profiling uncovers insulin-like growth factor-1 as a PPARalpha target gene in cardioprotection. J Biol Chem. 2011;286:14598–14607. doi: 10.1074/jbc.M111.220525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci. 1999;112(Pt 24):4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 32.Grandclement C, Pallandre JR, Valmary Degano S, Viel E, Bouard A, Balland J, Remy-Martin JP, Simon B, Rouleau A, Boireau W, Klagsbrun M, Ferrand C, Borg C. Neuropilin-2 expression promotes TGF-beta1-mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS ONE. 2011;6:e20444. doi: 10.1371/journal.pone.0020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Marcos PJ, Pantoja C, Gonzalez-Rodriguez A, Martin N, Flores JM, Valverde AM, Hara E, Serrano M. Normal proliferation and tumorigenesis but impaired pancreatic function in mice lacking the cell cycle regulator sei1. PLoS ONE. 2010;5:e8744. doi: 10.1371/journal.pone.0008744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liew CW, Boucher J, Cheong JK, Vernochet C, Koh HJ, Mallol C, Townsend K, Langin D, Kawamori D, Hu J, Tseng YH, Hellerstein MK, Farmer SR, Goodyear L, Doria A, Bluher M, Hsu SI, Kulkarni RN. Ablation of TRIP-Br2, a regulator of fat lipolysis, thermogenesis and oxidative metabolism, prevents diet-induced obesity and insulin resistance. Nat Med. 2013;19:217–226. doi: 10.1038/nm.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.