Abstract
The last two decades have witnessed the development and application of nucleic acid aptamers in a variety of fields, including target analysis, disease therapy, and molecular and cellular engineering. The efficient and widely applicable aptamer selection, reproducible chemical synthesis and modification, generally impressive target binding selectivity and affinity, relatively rapid tissue penetration, low immunogenicity, and rapid systemic clearance make aptamers ideal recognition elements for use as therapeutics or for in vivo delivery of therapeutics. In this feature article, we discuss the development and biomedical application of nucleic acid aptamers, with emphasis on cancer cell aptamer isolation, targeted cancer therapy, oncology biomarker identification and drug discovery.
1. Introduction
Nucleic acid aptamers are single-stranded (ss) oligonucleotides with unique intramolecular conformations and specific recognition abilities to targets. They are isolated from large libraries containing 1013–1016 random nucleic acid sequences through Systematic Evolution of Ligands by Exponential Enrichment (SELEX).1–2 Since the pioneering isolations of aptamers against organic dyes1 and T4 DNA polymerase2, respectively, in 1990, a wide variety of aptamers have been identified using the SELEX technique. The targets of aptamers range from small molecules,1, 3–4 to proteins,2, 5–11 virus-infected cells,12 stem cells,13 and cancer cells14–21, and a wide variety of targets of high therapeutic interest were used as targets for aptamer identification (Table 1). The binding affinities of aptamers to their targets (Kds usually of picomolars-nanomolars) are comparable, or sometimes stronger than, those of other molecular recognition elements, such as antibodies.22
Table 1.
Examples of nucleic acid aptamers to targets of therapeutic interest
| Aptamers | Molecular targets | Associated diseases | Refs | |
|---|---|---|---|---|
| 1 | Macugen | Vascular endothelial growth factor | Age-related macular degeneration | 39 |
| 2 | AS1411 | Nucleolin | Cancer | 40 |
| 3 | sgc8 | Protein tyrosine Kinase 7 | Cancer | 41 |
| 4 | TD05 | Immunoglobulin μ Heavy Chains (IGHM) | Lymphoma | 42 |
| 5 | A20 | Prostate-specific membrane antigen | Cancer | 43 |
| 6 | TTA1 | Tenascin C | Cancer | 6 |
| 7 | S1.3/S2.2 | Mucin 1 | Cancer | 44 |
| 8 | IGEL1.2 | Immunoglobulin E | Allergy | 45 |
| 9 | Apt-αvβ3 | αvβ3 integrin | Cancer | 46 |
| 10 | TBA | α-thrombin | Thrombosis | 8 |
| 11 | B28 | HIV gp120 | Viral infection | 11 |
| 12 | NF-κB | Cancer | 47 | |
| 13 | E2F-E1 | E2F transcription factor | Cancer | 48–49 |
| 14 | A30 | HER3 | Cancer | 10 |
| 15 | WT-15 | Plasminogen activator inhibitor 1 | Tumor metastasis | 50 |
The SELEX method has been further developed in combination with various other techniques, including capillary electrophoresis,23 microfluidics,24 flow cytometry,25 and fluorescence-activated cell sorting (FACS).26 It has also been modified for higher selection efficiency, including the selection of aptamers with improved biostability or bioavailability. For instance, the Ellington group developed an automated SELEX, by which RNA aptamers can be generated within a few days.27
SELEX has also been utilized to select aptamers using complex targets,28–29 including cell fragments, cell membrane fractions, red blood cell ghosts, bacteria, mammalian cancer cells, and even implanted tumors30. Recently, our group developed cell-SELEX,25 in which whole live cells were used to identify aptamers with specific recognition abilities to target cells. Aptamers have been selected using a wide range of diseased cells.14–21, 31 Cell-SELEX is also capable of generating aptamers that target diseased cells, but without having prior knowledge of particular protein signatures. This novel way to identify disease-associated biomarkers is poised to increase our understanding of molecular oncology and guide targeted therapy.
Aptamers hold many advantages over antibodies, which have been widely used as recognition elements (recently reviewed by Ellington, et al.12). Briefly, aptamers can be developed against a wide range of targets, including those toxic to organisms and therefore beyond the reach of antibody development. Thus, aptamers can be generated for toxic therapeutics and the targeted transport of toxic drugs. Moreover, aptamer identification through in vitro SELEX is usually more efficient and cost-effective than antibody development. The advancement of automated nucleic acid synthesis enables easy, cost-effective chemical synthesis and modification of functional moieties, as well as large-scale commercial production. Other advantages include high stability and long shelf-life, rapid tissue penetration based on the relatively small molecular weights, low immunogeneity,32 and ease of antidote development.33–34 These advantages makes aptamers more than attractive for clinical or biomedical applications, such as disease diagnosis and therapy.
In addition, the tertiary molecular structures of some aptamer/protein-target complexes have already been elucidated, which further indicated that noncovalent interactions formed between specific nucleic acid and target conformations, including hydrogen bonding, van der Waals contacts, stacking interactions, contributes to aptamer-target interaction at the molecular levels. 35–38 The elucidation of aptamer-target interaction is also anticipated to facilitate drug design, drug screening and discovery of new drugable sites on disease biomarkers.
This feature article summarizes recent progress in cell-SELEX to identify aptamers for specific cancer cell recognition and the application in therapeutics, targeted transport of therapeutics, biomarker identification, and aptamer-templated drug discovery.
2. Cell-SELEX
Cell-SELEX is a technique that uses living cells as targets to isolate aptamers.6, 29 In vitro cell-SELEX (and the subsequent cellular application) is usually performed in a buffer solution which does not contain nuclease, ensuring the stability of nucleic acid probes during aptamer selection. Cell-SELEX provides a unique set of capabilies: identification of aptamers that target molecules which need cofactors or post-translational modification to form functional conformations on cell surfaces; aptamer identification without prior knowledge of the molecular differences between targets and nontargets; simultaneous generation of a panel of aptamers, which may have different molecular targets; further application of the resultant aptamers for disease biomarker identification, which could, in turn, help in understanding protein expression patterns on diseased cell surfaces, unravelling the pathogenesis of complex, intractable diseases, and opening new therapeutic avenues.
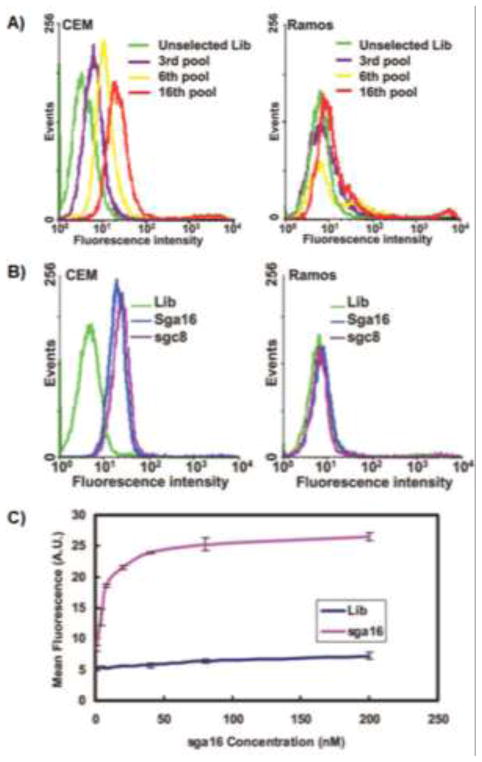
Using cell-SELEX, our group has successfully identified DNA aptamers against cells of several cancers, including acute lymphoblastic leukemia (ALL) (T cell leukemia),14 liver cancer,16 acute myeloid leukemia (AML) leukemia,17 lung cancer,18, 21 ovarian cancer,20 B cell lymphoma,42 colorectal cancer,51 and breast cancer52. Shangguan, et al. enriched DNA pools that specifically recognized CEM cells, a cultured precursor T cell ALL cell line, but did not recognize the nontarget Ramos cells, a B cell Burkitt’s lymphoma cell line (Fig. 1).14 To monitor the progress of aptamer enrichment during selection and verify the specific recognition abilities of resultant aptamers to target cells, flow cytometry was employed, in which cells bound by fluorophore-labeled DNA probes displayed larger fluorescence intensities compared to those cells not specifically bound by these DNA probes. Some aptamers identified from the enriched pools were capable of revealing the molecular differences of cancer cells from clinical smaples.53 The dissociation constants (Kds) of these aptamers range from picomolar to nanomolar range. For instance, aptamer sgc8 was shown to have a dissociation constant of 0.80 nM to CEM cells, indicating robust binding affinity.
Fig. 1.
Monitoring of cell-SELEX and characterization of selected aptamers. (A) Flow cytometry assay to monitor the binding of selected pools and enrichment of aptamers with CEM cells (target) and Ramos cells (control). For target CEM cells, there was an increase in binding ability of the pool as the selection was progressing, whereas there was little enhancement for the control cells. (B) Flow cytometry assay for the binding of the FITC-labeled sequences sga16 and sgc8 obtained from the above selection. (C) Flow cytometry assay to determine the binding affinity of FITC-labeled sga16 to CEM cells. The nonspecific binding was measured using an FITC-labeled random DNA sequence library. Adapted from ref.14 (Copyright (2006) National Academy of Sciences, U.S.A.)
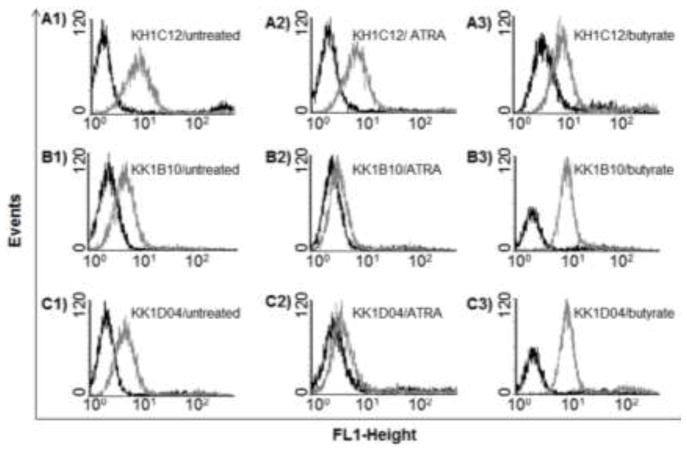
Aptamers are also able to distinguish cells that are closely related. Sefah et al. selected aptamers using three types of AML cells (HL60, K562 and NB4 cells).17 One aptamer, KH1C12, showed significant selectivity to target HL60 cells with a dissociation constant of 4.5 nM, but it did not bind to nontarget K562 or NB4 cells. KH1C12 maintained the ability to recognize the target cells in normal bone marrow aspirates and clinical samples, respectively. More importantly, this provided, for the first time, molecular probes that could distinguish two types of leukemic cells that are closely related morphologically. It is worth noting that two other aptamers, KK1B10 and KK1D04, showed different binding abilities to undifferentiated cells and chemically-induced differentiated cells. For instance, with induction by all-trans-retinoic acid (ATRA), HL60 cells differentiate in a granulocytic pathway, whereas sodium butyrate, another type of chemical inducer of HL60, induces differentiation in a monocytic pathway. Fluorophore-labeled aptamers KK1B10 and KK1D04 lost significant fluorescence signal with ATRA-treated HL60 cells, while showing an increase in fluorescence signal intensity with HL60 cells induced with sodium butyrate (Fig. 2). The ability of aptamers to probe cell differentiation stages would enable researchers to study the molecular basis of cell differentiation or cancer pathogenesis.
Fig. 2.
Flow cytometry assay showing the binding pro les of aptamers KH1C12 (A1-A3), KK1B10 (B1–B3) and KK1D04 (C1–C3) to the untreated, ATRA-treated and sodium butyrate-treated HL60 cells. The dark curves show fluorescence background using the random DNA sequence library. Adapted from ref.17
Aptamers selected using cultured cells through cell-SELEX were further shown to be capable of specific recognition of ex vivo and in vivo tumor cells (Fig. 3),54–55 and they have been widely used as recognition probes for targeted cancer cell detection and isolation,56 cell-cell interaction,57 and transport of chemotherapeutics,58–61 antibodies,62 nanostructures,60, 62 and viruses.63
Fig. 3.
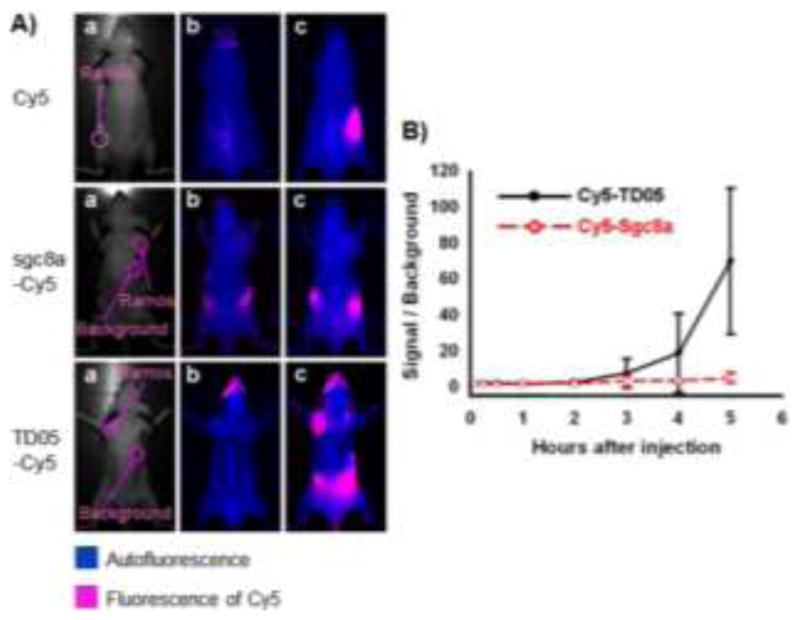
An aptamer identified through cell-SELEX for in vivo imaging of Ramos subcutaneous xenograft. (A) Optical and fluorescent imaging of tumors in mice using Cy5 dye and Cy5-labeled aptamers. Target Ramos cells were injected subcutaneously on the back of BALB/c nude mice. Cy5 dye, Cy5-labeled sgc8a (control), or Cy5-labeled TD05 was subsequently injected intravenously, and images of the dorsal side of live mice were taken at various time points after injection. Images a, b and c represent the optical photographs, the fluorescent images before injection, and 3.5 h after injection, respectively. (B) Quantification of the signal-to-background ratio for Cy5-labeled TD05 and Cy5-labeled sgc8a. The number of mice used to derive statistical information for each probe is 5. Data represent mean ± standard error. Adapted from ref.54
3. Aptamers as therapeutics
In addition to specific recognition abilities, some aptamers can bind to and further modulate the biological activities of the molecular targets which are involved in pathogenesis. As a consequence, aptamers, as therapeutics, can readily regulate biological pathways and interfere with disease development.
The therapeutic application of nucleic acid aptamers dates back to 1990, when Sullenger et al. reported the inhibition of HIV-1 viral infection in host cells by overexpressing a Trans-Activation Response (TAR) decoy. This nucleic acid worked as an aptamer, prevented the activation of viral gene expression, and consequently inhibited the viral replication.64 Moreover, the development of SELEX techniques by Ellington and Gold, et al. enabled the rapid generation of aptamers for a wide range of therapeutically relevant targets. The therapeutic potential and rapid in vitro identification of aptamers make them attractive as therapeutic agents.
Since then, many aptamers have been developed and have undergone clinical evaluation. One example is pegaptanib (marketed as Macugen), an anti-VEGF aptamer that can recognize the majority of human VEGFA isoforms. Pegaptanib was developed by Pfizer and Eyetech, and it was approved by the U.S. FDA in 2004 for the treatment of age-related macular degeneration (AMD). The most advanced aptamer for cancer therapy is AS1411, a G-rich aptamer previously known as AGRO001.65 AS1411 is able to bind nucleolin66 and can be internalized into target cells to interfere with intracellular pathways67. The exact mechanism of its inhibition of cell proliferation is not yet fully understood, but a few of the pathways involved include the destabilization of BCL-2 mRNA and the inhibition of NF-κB.67 AS1411 can inhibit cell proliferation in a broad range of cancers, and it is currently in Phase II clinical trials for AML. In 2010, Bates, et al. reported that cancer-selective antiproliferative activity might be generally applicable to some G-rich oligodeoxynucleotides and was associated, for example, with G-quadruplex formation, cellular internalization and protein binding.68 This may effectively lead to the isolation or design of aptamers with G-quadruplex structures and aid in elucidating the antiproliferative effect of G-quadruplex-forming aptamers. Other aptamers under clinical evaluation include MOX-A12 targeting CCL2 and NU172 targeting thrombin, among others.12
4. Aptamers for targeted delivery of therapeutics
Aptamers can work as recognition moieties and guide a variety of therapeutics to target diseased sites (Table 2). It is well known that conventional chemotherapy suffers from “off-target” effects and the resultant side effects, including hair loss, nausea, dilated cardiomyopathy or congestive heart failure.69 The targeted delivery of chemotherapeutics to diseased sites is essential to improve therapeutic efficacy. Antibodies and aptamers are the two major types of recognition moieties. The development of antibody-drug conjugates has been intensively studied in pharmaceutical companies in the past few years.70–71 Aptamers, owing to their intrinsic advantages over antibodies, are poised to play an important role in targeted delivery of therapeutics. The ability of some aptamers to recognize diseased cells, but not healthy cells, enabled them to be designed as targeting moieties and selectively transport drugs to diseased sites. The high stability of nucleic acid aptamers provides tremendous opportunities for scientists to design conjugates of aptamers and chemotherapeutics through a variety of chemical reactions or physical complexation. In addition, the ease of site-specific chemical modification of aptamers enables the conjugation of aptamers with drugs, including chemotherapeutics and photosensitizers, or nanomaterials, such as carbon nanotubes or gold nanoparticles.80–81
Table 2.
Aptamers for targeted therapeutic delivery
| Aptamers | Therapeutic cargoes | Refs. |
|---|---|---|
| sgc8c | Chemotherapeutics, photosensitizers, nanorods, liposomes, nanogel, viral capsids, DNA origami, antibodies | 60, 62–63, 72–77 |
| TD05 | Micelle nanoparticles, photosensitizers | 78–79 |
| AS1411 | Chemotherapeutics, photosensitizers, nanoparticles, liposomes | 80–81 |
| A10 | Chemotherapeutics, siRNA, protein toxin, nanoparticles, quantum dots | 82–89 |
| APT | Radionuclides, chemotherapeutics, quantum dots | 90–92 |
4.1. Aptamer-drug conjugates for targeted drug delivery
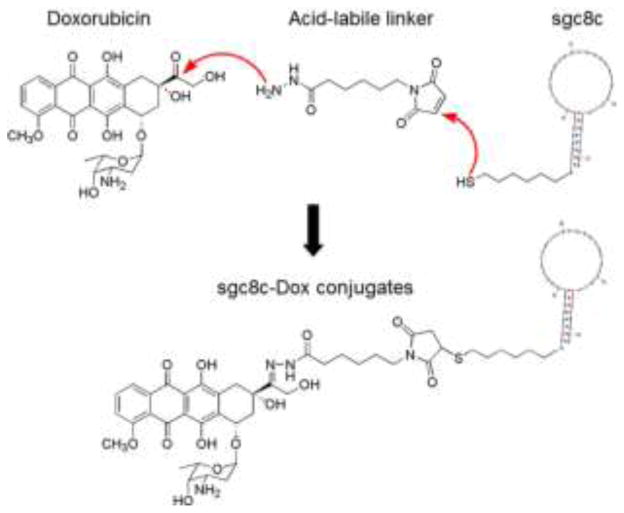
The high recognition specificity and selectivity of aptamers make them excellent molecular probes for targeted delivery of therapeutics. The chemical stability and ease of modification of aptamers enable them to be chemically conjugated with many functional moieties, including therapeutics, ranging from small molecule chemotherapeutics to photosensitizers, toxins, siRNA, and radionuclides (Table 2). Our group has conjugated an DNA aptamer, PTK7-targeting sgc8c, and Doxorubicin (Dox), one of the most widely used chemotherapeutic drugs in clinical medicine. The resultant conjugate was able to specifically deliver the drug and induce inhibition of cancer cell proliferation in target CEM cells, but not in nontarget cells, indicating high selectivity (Fig. 4). In a similar manner, a photosensitizer, Chlorin e6, was conjugated to aptamer TD05. The resultant selective cytotoxicity of this aptamer-drug conjugate demonstrated the principle of using aptamers for targeted drug delivery in photodynamic therapy (PDT). Moreover, siRNA, which is an emerging new type of nucleic acid therapeutics, has also been chemically conjugated with aptamers. The resultant chimeric aptamer-siRNA conjugates were able to specifically deliver siRNA and knock down the target genes.84, 89, 93–94
Fig. 4.
Schematic illustration of conjugation of a chemotherapeutic drug, Doxorubicin (Dox), with an DNA aptamer sgc8c for targeted drug delivery to cancer cells. Adapted from ref.72
In addition, by taking advantage of the noncovalent interactions between oligonucleotides and some therapeutics, physical conjugates of aptamers and drugs have been developed for targeted therapy. For instance, we have developed a bi-specific DNA aptamer-based drug carrier, SD, in which two different aptamers were self-assembled through a double-stranded linker with preferential Dox loading sites. Dox can be physically complexed with SD through intercalation into dsDNA linker, and transported into two distinct target cells to induce bi-specific cytotoxicity in a mixed cell environment (Fig. 5).77
Fig. 5.
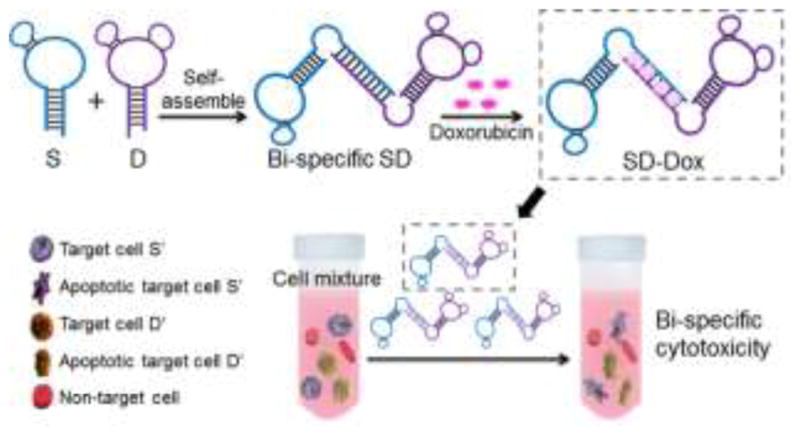
Schematic illustration of the self-assembled aptamer-based drug carrier SD for bi-specific cytotoxicity. A bi-specific aptamer-based drug carrer, SD, was self-assembled from molecularly engineered aptamers sgc8c (S) and sgc5a (D), simultaneously forming Dox intercalation sites in the dsDNA linker. Subsequently, Dox was loaded onto SD through site-specific Dox intercalation and bi-specifically delivered into target cells in a mixed cell environment, where it displayed bi-specific cytotoxicity. Adapted from ref.77
4.2. Aptamer-nanomaterial conjugates for targeted drug delivery
The past few decades have witnessed the rapid advancement in both aptamer development and nanotechnology. Suitably modified nanomaterials can be combined with aptamers for targeted drug delivery. Aptamers provide aptamer-nanomaterial conjugates with specific targeting ability, and nanomaterials provide an excellent platform for drug loading in the interior spaces or on the surface, as imaging agents via their unique optical characteristics or as therapeutic agents themselves.
As an example, our group has developed a smart multifunctional nanostructure (SMN) composed of porous hollow magnetite nanoparticles (PHMNP), PEG ligands, and aptamers.95 The PHMNP was capable of encapsulating Doxorubicin and also worked as an imaging agent for MRI. The aptamer enabled the targeted delivery of SMN-Dox complexes into target CEM cells. The Dox release was facilitated upon internalization of SMN-Dox by endocytosis, resulting in inhibition of target cell proliferation.
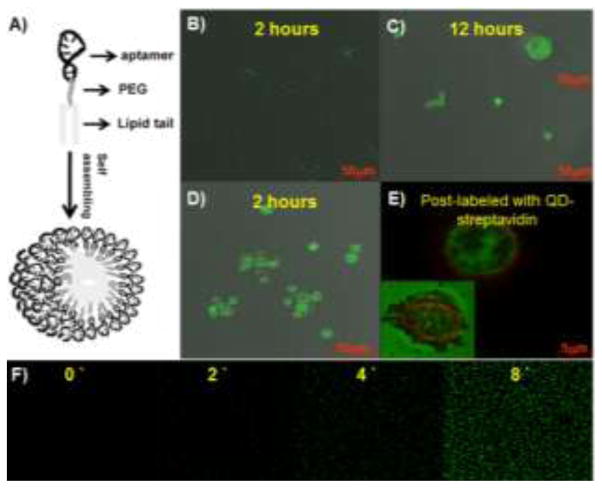
Other than metallic nanomaterials, polymeric nanomaterials in combination with aptamers have also been investigated for targeted transport of therapeutics. For instance, a diacyl moiety was tethered at one end of an aptamer sequence by automated solid-phase DNA synthesis. The resultant chimeric lipid-aptamers self-assembled into monodispersed lipid nanoparticles, termed micelles (Fig. 6A).96 The aptamer-coated lipid micelles were further studied as a vector for targeted delivery.78 In this study, aptamer TD05 was conjugated with micelles, and the resultant TD05-micelles showed improved binding ability at physiological temperature and enhanced aptamer binding affinity by approximately 750-fold as a consequence of multivalency. These aptamer-tethered micelles were able to integrate with the cell membrane and facilitate the delivery of a dye doped into cells (Fig. 6B, C). Strikingly, the authors also demonstrated that these aptamer micelles showed dynamic specificity in flow channel systems mimicking drug delivery in a circulation system. The specific recognition and facilitated transport of loaded model molecules make aptamer-micelles promising as vectors for targeted drug delivery.
Fig. 6.
Aptamer-micelles for targeted delivery of doped dye. (A) Schematic illustration of aptamer micelle formation. Bright field and fluorescent images of Ramos cells after incubation with free CellTracker Green BODIPY for 2 h (B) and 12 h (C), or incubation with biotin-TD05-micelle doped with CellTracker Green BODIPY for 2 h (D). Image E is the enlarged fluorescent image after postlabeling the biotinylated TD05 aptamer with QD705 streptavidin. The inset in image C is the enlarged individual cell image. The inset in image E is the fluorescent image of the dead cell. (F) Real-time monitoring of doped CellTracker™ released from the core of the micelles. The numbers indicated the time (minutes) of incubation at room temperature. Adapted from ref.78
In addition to serving as drug carriers, nanomaterials conjugated with aptamers can also work as therapeutics themselves. Huang et al. have conjugated multiple copies of aptamer sgc8 onto the surfaces of gold nanorods (AuNRs), which have high absorption efficiency of near-infrared light for hyperthermia effect.74 The resultant sgc8-NRs were then capable of selectively binding to target CEM cells and induce cytotoxicity by hyperthermia. In another example, an acrydite phosphoramidite was conjugated to the 5′-end of aptamers by automated solid-phase DNA synthesis, and the resultant acrydite-aptamer chimera were able to polymerize in the presence of ammonium persulfate (APS) and tetramethylethylenediamine (TEMED) (Fig. 7).97 The preformed polymeric aptamers were able to specifically bind and be internalized into target cells. More importantly, the internalized aptamer-tethered polymers led to cytotoxicity in target cancer cells, including those exhibiting multidrug resistance.
Fig. 7.
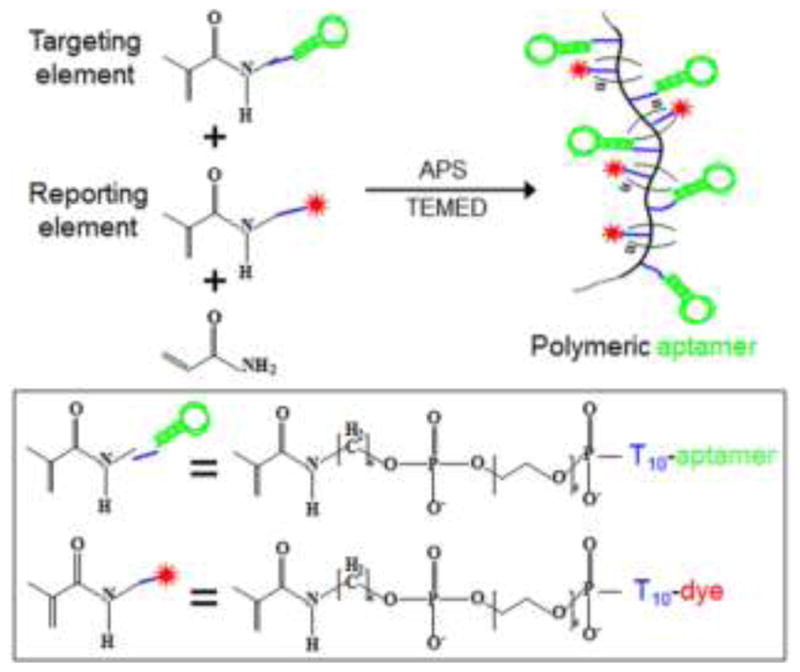
Schematic illustration of polymeric aptamer construction. Polymeric aptamers, which work as flexible molecular probes, were engineered from multiple dye-labeled reporting elements and aptamer-labeled targeting elements. Adapted from ref.97
5. Aptamers for biomarker identification and drug discovery
In the past decade or so, intensive study in genome sequencing and proteomics has produced a wealth of information at the genomic, genetic, and proteomic levels. However, disease biomarkers remain scarce, especially those that can be used as drug targets. In parallel, the limited number of drugable biological activity sites on molecular biomarkers has hampered the discovery of new drugs that could otherwise target those sites. Aptamer selection against targets without prior knowledge at the molecular level provides an excellent platform to identify biomarkers and drugable sites on biomarkers.
5.1 Aptamers for disease biomarker identification
As described above, aptamer selection does not require prior knowledge of, for instance, a particular protein conformation. This property, combined with the ability of aptamers to distinguish diseased cells and healthy cells, makes it conceivable to identify previously unknown disease biomarkers. Towards this goal, Tan and colleagues have successfully identified target biomarkers of aptamers through a chemical biology approach. In one example, a biotinylated aptamer, sgc814, was generated to bind target CEM cells with subsequent crosslinkage to cell membrane proteins. The resultant cells were then lysed, and the crosslinked aptamer-protein complexes were extracted using magnetic beads, purified using gel electrophoresis, and subjected to mass spectrometric analysis. MS data subsequently verified that the target biomarker was PTK7 (protein tyrosine kinase 7), a pseudokinase without detectable catalytic tyrosine kinase activity.41, 98 Although PTK7 was reported to be upregulated in many types of cancers, the biomedical relevance of this upregulation remains unclear, and PTK7 still has no clinical application.99 Our further investigation demonstrated that silencing PTK7 genes through RNAi induced apoptosis in colon cancer HCT116 cells via a mitochondrial pathway.99 The successful identification and important biological role of PTK7 in cancer cell proliferation and apoptosis suggest that aptamers are promising in the discovery of biomarkers of therapeutic interest.
5.2 Aptamers for rational drug screening and drugable site discovery
The generally strong binding affinities of aptamers to targets and the high stability of aptamer-target interactions have enabled the elucidation of the crystal structures of these complexes, including aptamers with thrombin,35–36 HIV TAR,37 and NF-κB38. These structures shed light on the loci of aptamer/target interaction, which could be further applied to rational structure-based drug design and screening. To this end, aptamers have been used as both templates and substitutes in high-throughput drug screening. That is, small molecule drugs that compete with aptamers for target binding were identified as drug candidates.100–109 In 2007, Famulok, et al. reported the identification of a small molecule inhibitor, SY-3E4, against HIV reverse transcriptase, which plays a vital role in viral replication.102 This inhibitor was identified through the displacement of aptamers from reverse transcriptase with small molecule drug candidates and further verified to inhibit viral replication. More importantly, the structure analysis of this target protein revealed that this drug binding site had not been previously considered as a drugable site for viral therapy.102 Thus, the structural analysis of aptamer-target complexes is a new avenue for investigating aptamers as templates for drug design, high-throughput drug screening and discovery of alternative drugable sites.
6. Concluding remarks
During the two decades since aptamers were first described, many challenges for therapeutic aptamer application have come to the forefront, particularly issues of biostability, bioavailability, therapeutic formulation and pharmacokinetics, and cost of large-scale DNA production. These properties differ between antibodies or small molecule drugs and aptamers, which represent a new class of therapeutics, thus requiring thorough investigation in advance of clinical application. Nonetheless, the advantages of nucleic acid aptamers offer tremendous opportunities. For instance, antibody-drug conjugates (ADC)110 and bi-specific antibodies111–112 are two areas of tremendous interest and opportunities in pharmaceutical research and development. However, while the use of antibodies is always associated with complicated and time-consuming genetic engineering, limited stability, and high cost, In contrast, bi-specific aptamer or aptamer-drug conjugate engineering is fairly straightforward as a consequence of facile modification, high stability and programmability.57, 61–62, 72, 79, 113–114 With further understanding and improvement of aptamers, as well as the advancement of DNA synthesis technology, nucleic acid aptamers are poised to revolutionize the next generation of therapeutics in the context of disease diagnosis and prevention.
Supplementary Material
Acknowledgments
This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086), by the National Key Scientific Program of China (2011CB911000) and China National Instrumentation Program 2011YQ03012412.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
This article is part of the ‘Nucleic acids: new life, new materials’ web themed issue.
Notes and references
- 1.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 4.Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 5.Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W. Chem Commun. 2006:3229–3231. doi: 10.1039/b604778e. [DOI] [PubMed] [Google Scholar]
- 6.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc Natl Acad Sci U S A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parekh P, Tang Z, Turner PC, Moyer RW, Tan W. Anal Chem. 2010;82:8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 9.Hu J, Wu J, Li C, Zhu L, Zhang WY, Kong G, Lu Z, Yang CJ. Chembiochem. 2011;12:424–430. doi: 10.1002/cbic.201000470. [DOI] [PubMed] [Google Scholar]
- 10.Chen C-hB, Chernis GA, Hoang VQ, Landgraf R. Proc Natl Acad Sci U S A. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khati M, Schüman M, Ibrahim J, Sattentau Q, Gordon S, James W. J of Virology. 2003;77:12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keefe AD, Pai S, Ellington A. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo KT, Schäfer R, Paul A, Gerber A, Ziemer G, Wendel HP. Stem Cells. 2006;24:2220–2231. doi: 10.1634/stemcells.2006-0015. [DOI] [PubMed] [Google Scholar]
- 14.Shangguan D, Li Y, Tang Z, Cao Z, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikaratchy P, Chen HW, Li Y, Tan W. Anal Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 16.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Anal Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 17.Sefah K, Tang Z, Shangguan D, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim Y, Tan W. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W. Chemmedchem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W. ACS Chem Neuroscience. 2011;2:175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simaeys DV, López-Colón D, Sefah K, Sutphen R, Jimenez E, Tan W. Plos One. 2010:5. doi: 10.1371/journal.pone.0013770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- 22.O’Donoghue M, Shi X, Fang X, Tan W. Anal and BioAnal Chem. 2012;402:3205–3209. doi: 10.1007/s00216-011-5667-y. [DOI] [PubMed] [Google Scholar]
- 23.Mosing RK, Mendonsa SD, Bowser MT. Anal Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 24.Mosing RK, Bowser MT. J of Separation Science. 2007;30:1420–1426. doi: 10.1002/jssc.200600483. [DOI] [PubMed] [Google Scholar]
- 25.Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Nat Protocols. 2009;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 26.Mayer G, Ahmed MSL, Dolf A, Endl E, Knolle PA, Famulok M. Nat Protocols. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 27.Cox JC, Rudolph P, Ellington AD. Biotechnology Progress. 1998;14:845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 28.Shamah SM, Healy JM, Cload ST. Accounts of Chemical Research. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 29.Fang X, Tan W. Accounts of Chemical Research. 2009;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. Nat Chem Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z, Parekh P, Turner P, Moyer RW, Tan W. Clin Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Group TES. Retina. 2002;2:143–152. [Google Scholar]
- 33.Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Scardino E, Fay WP, Sullenger BA. Nat Biotechnol. 2004;22:1423–1428. doi: 10.1038/nbt1023. [DOI] [PubMed] [Google Scholar]
- 34.Oney S, Lam RTS, Bompiani KM, Blake CM, Quick G, Heidel JD, Liu JYC, Mack BC, Davis ME, Leong KW, Sullenger BA. Nat Medicine. 2009;15:1224–1228. doi: 10.1038/nm.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The J of biological chemistry. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 36.Long SB, Long MB, White RR, Sullenger BA. RNA-Publ RNA Soc. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebars I, Legrand P, Aime A, Pinaud N, Fribourg S, Di Primo C. Nucleic Acids Research. 2008;36:7146–7156. doi: 10.1093/nar/gkn831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, Ghosh G. Proc Natl Acad Sci U S A. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR. New England J of Medicine. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 40.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Experimental and Molecular Pathology. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J of Proteome Research. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Molecular & Cellular Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Cancer Research. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 44.Ferreira CSM, Matthews CS, Missailidis S. Tumor Biology. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- 45.Wiegand T, Williams P, Dreskin S, Jouvin M, Kinet J, Tasset D. The J of Immunology. 1996;157:221–230. [PubMed] [Google Scholar]
- 46.Mi J, Zhang X, Giangrande PH, McNamara JO, II, Nimjee SM, Sarraf-Yazdi S, Sullenger BA, Clary BM. Biochemical and Biophysical Research Communications. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 47.Lebruska LL, Maher LJ. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 48.Martell RE, Nevins JR, Sullenger BA. Mol Ther. 2002;6:30–34. doi: 10.1006/mthe.2002.0624. [DOI] [PubMed] [Google Scholar]
- 49.Ishizaki J, Nevins JR, Sullenger BA. Nat Medicine. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- 50.Blake CM, Sullenger BA, Lawrence DA, Fortenberry YM. Oligonucleotides. 2009;19:117–128. doi: 10.1089/oli.2008.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan W. Plos One. 2010:5. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K, Sefah K, Tang L, Zhao Z, Zhu G, Ye M, Sun W, Goodison S, Tan W. Chemmedchem. 2012;7:79–84. doi: 10.1002/cmdc.201100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shangguan D, Cao Z, Li Y, Tan W. Clin Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 54.Shi H, Tang Z, Kim Y, Nie H, Huang YF, He X, Deng K, Wang K, Tan W. Chemistry-an Asian J. 2010;5:2209–2213. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 55.Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B. Proc Natl Acad Sci U S A. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Anal Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Yan H, Liu Y, Chang Y. Small. 2011;7:1673–1682. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 58.Tan W, Wang H, Chen Y, Zhang X, Zhu H, Yang C, Yang R, Liu C. Trends in Biotechnol. 2011;29:634–640. doi: 10.1016/j.tibtech.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang YF, Kim Y, Meng L, Tan W. Chimica Oggi-Chemistry Today. 2009;27:52–54. [Google Scholar]
- 60.Luo YL, Shiao YS, Huang YF. ACS Nano. 2011;5:7796–7804. doi: 10.1021/nn201592s. [DOI] [PubMed] [Google Scholar]
- 61.Mallikaratchy P, Liu H, Huang YF, Wang H, Lopez-Colon D, Tan W. Chem Commun. 2009:3056–3058. doi: 10.1039/b823258j. [DOI] [PubMed] [Google Scholar]
- 62.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 63.Tong GJ, Hsiao SC, Carrico ZM, Francis MB. J Am Chem Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 65.Ireson CR, Kelland LR. Molecular Cancer Therapeutics. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 66.Soundararajan S, Wang L, Sridharan V, Chen W, Courtenay-Luck N, Jones D, Spicer EK, Fernandes DJ. Molecular Pharmacology. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. Cancer Research. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 68.Choi EW, Nayak LV, Bates PJ. Nucleic Acids Research. 2010;38:1623–1635. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Pharmacological Reviews. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 70.Hughes B. Nat Rev Drug Discov. 2010;9:665–667. doi: 10.1038/nrd3270. [DOI] [PubMed] [Google Scholar]
- 71.Webb S. Nat Biotechnol. 2011;29:297–298. doi: 10.1038/nbt0411-297. [DOI] [PubMed] [Google Scholar]
- 72.Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. Chembiochem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang H, O’Donoghue MB, Liu H, Tan W. Chem Commun. 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- 74.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan W. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 75.Wang K, You M, Chen Y, Han D, Zhu Z, Huang J, Williams K, Yang CJ, Tan W. Angew Chem, Int Ed. 2011;50:6098–6101. doi: 10.1002/anie.201008053. [DOI] [PubMed] [Google Scholar]
- 76.Kang H, Trondoli AC, Zhu G, Chen Y, Chang YJ, Liu H, Huang YF, Zhang X, Tan W. ACS Nano. 2011;5:5094–5099. doi: 10.1021/nn201171r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu G, Meng L, Ye M, Yang L, Sefah K, O’Donoghue MB, Chen Y, Xiong X, Huang J, Song E, Tan W. Chemistry An Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Sefah K, Liu H, Wang R, Tan W. Proc Natl Acad Sci U S A. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mallikaratchy P, Tang Z, Tan W. Chemmedchem. 2008;3:425–428. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shieh YA, Yang SJ, Wei MF, Shieh MJ. ACS Nano. 2010;4:1433–1442. doi: 10.1021/nn901374b. [DOI] [PubMed] [Google Scholar]
- 81.Cao Z, Tong R, Mishra A, Xu W, Wong GCL, Cheng J, Lu Y. Angew Chem, Int Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 82.Bagalkot V, Farokhzad OC, Langer R, Jon S. Angew Chem, Int Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 83.Chu TC, Marks JW, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Cancer Research. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 84.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 85.Kim D, Jeong YY, Jon S. ACS Nano. 2010;4:3689–3696. doi: 10.1021/nn901877h. [DOI] [PubMed] [Google Scholar]
- 86.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Letters. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 87.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Proc Natl Acad Sci U S A. 2011;108:1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Proc Natl Acad Sci U S A. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dassie JP, Liu X-y, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH. Nat Biotechnol. 2009;27:839–846. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pieve CD, Perkins AC, Missailidis S. Nuclear Medicine and Biology. 2009;36:703–710. doi: 10.1016/j.nucmedbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 91.Savla R, Taratula O, Garbuzenko O, Minko T. J of Controlled Release. 2011;153:16–22. doi: 10.1016/j.jconrel.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 92.Tan L, Gee Neoh K, Kang E-T, Choe W-S, Su X. J of Pharmaceutical Sciences. 2012;101:1672–1677. doi: 10.1002/jps.23101. [DOI] [PubMed] [Google Scholar]
- 93.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Nucleic Acids Research. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen T, Shukoor MI, Wang R, Zhao Z, Yuan Q, Bamrungsap S, Xiong X, Tan W. ACS Nano. 2011;5:7866–7873. doi: 10.1021/nn202073m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu H, Zhu Z, Kang H, Wu Y, Sefan K, Tan W. Chemistry-a European J. 2010;16:3791–3797. doi: 10.1002/chem.200901546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L, Meng L, Zhang X, Chen Y, Zhu G, Liu H, Xiong X, Sefah K, Tan W. J Am Chem Soc. 2011;133:13380–13386. doi: 10.1021/ja201285y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shin WS, Maeng YS, Jung JW, Min JK, Kwon YG, Lee ST. Biochem and Biophys Res Communi. 2008;371:793–798. doi: 10.1016/j.bbrc.2008.04.168. [DOI] [PubMed] [Google Scholar]
- 99.Meng L, Sefah K, O’Donoghue MB, Zhu G, Shangguan D, Noorali A, Chen Y, Zhou L, Tan W. Plos One. 2010:5. doi: 10.1371/journal.pone.0014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Green LS, Bell C, Janjic N. Biotechniques. 2001;30:1094. doi: 10.2144/01305dd02. [DOI] [PubMed] [Google Scholar]
- 101.Srivatsan SG, Famulok M. Combinatorial Chem & High Throughput Screening. 2007;10:698–705. doi: 10.2174/138620707782507359. [DOI] [PubMed] [Google Scholar]
- 102.Yamazaki S, Tan L, Mayer G, Hartig JS, Song JN, Reuter S, Restle T, Laufer SD, Grohmann D, Kraeusslich HG, Bajorath J, Famulok M. Chemistry & Biology. 2007;14:804–812. doi: 10.1016/j.chembiol.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 103.Hafner M, Vianini E, Albertoni B, Marchetti L, Gruene I, Gloeckner C, Famulok M. Nat Protocols. 2008;3:579–587. doi: 10.1038/nprot.2008.15. [DOI] [PubMed] [Google Scholar]
- 104.Famulok M. J of Medicinal Chem. 2009;52:6951–6957. doi: 10.1021/jm9014789. [DOI] [PubMed] [Google Scholar]
- 105.Mayer G, Faulhammer D, Graettinger M, Fessele S, Blind M. Chembiochem. 2009;10:1993–1996. doi: 10.1002/cbic.200900325. [DOI] [PubMed] [Google Scholar]
- 106.Yamazaki S, Famulok M. In: Methods in Molecular Biology. Mayer G, editor. 2009. pp. 187–199. [DOI] [PubMed] [Google Scholar]
- 107.Niebel B, Lentz C, Pofahl M, Mayer G, Hoerauf A, Pfarr KM, Famulok M. Chemistry-a European J. 2010;16:11100–11107. doi: 10.1002/chem.201001192. [DOI] [PubMed] [Google Scholar]
- 108.Auslaender D, Wieland M, Auslaender S, Tigges M, Fussenegger M. Nucleic Acids Research. 2011:39. doi: 10.1093/nar/gkr829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mueller M, Ackermann D, Famulok M. Comptes Rendus Chimie. 2011;14:819–825. [Google Scholar]
- 110.Ducry L, Stump B. Bioconjugate Chem. 2009;21:5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 111.Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA. Drug Discovery Today. 2005;10:1237–1244. doi: 10.1016/S1359-6446(05)03554-3. [DOI] [PubMed] [Google Scholar]
- 112.Beck A, Wurch T, Bailly C, Corvaia N. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 113.Boltz A, Piater B, Toleikis L, Guenther R, Kolmar H, Hock B. J of Biol Chem. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA. Nucleic Acids Research. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.