Abstract
Obesity, and in particular central adiposity, is a key feature of metabolic syndrome, which includes trends toward increased triglycerides, insulin resistance, high blood pressure, hypercholesterolemia, and heart disease. It has a prevalence of 25% or more and is a dominant component of the health care budgets in Western societies. In addition to genetic causes, high-fat diets and disrupted sleep patterns have major influences on the development of metabolic syndrome. Recent studies have demonstrated active roles for the nuclear receptor superfamily and the energy-sensing kinase adenosine monophosphate (AMP)-activated protein kinase (AMPK) in regulating metabolism and circadian rhythm. In this chapter, we review these findings and attempt to develop a better understanding of the interplay between metabolism and circadian rhythm and their coordinated regulation by nuclear receptors and AMPK. This supraregulatory network may be considered a target for novel therapeutic applications against metabolic syndrome.
Nuclear receptors (NRs) form a large family of ligand-modulated transcription factors (TFs) that respond to a variety of lipophilic molecules, including lipids, steroids, retinoids, hormones, and xenobiotics. To date, a total of 48 human NRs and 49 mouse NRs have been identified, including classic endocrine receptors that mediate the actions of steroid hormones, fatty acids, thyroid hormones, and fat-soluble vitamins A and D, as well as a large number of orphan NRs, whose ligands, target genes, and physiological functions are still largely unknown (Mangelsdorf and Evans 1995; Yang et al. 2007).
All NRs comprise functional modular domains. The highly conserved DNA binding domain (DBD) consists of a well-characterized zinc finger motif that recognizes a degenerate six to eight nucleotide sequence on target DNAs. The ligand binding domain (LBD) resides in the carboxyl terminus. Ligands bind to LBDs and induce conformational changes that affect the NR-associated protein complexes, resulting in both epigenetic chromatin changes and transcriptional regulation (Wurtz et al. 1996; Norris et al. 2009). This unique property of NRs to directly modulate transcriptional activity upon ligand binding provides the body with essential machinery to monitor and respond to complex metabolic and environmental cues (such as diet and exercise) and may also explain why NRs are mainly present in metazoans that interact with the environment (Laudet 1997).
The adenosine monophosphate (AMP)-activated protein kinase (AMPK) is a central mediator of metabolism. AMPK is a heterotrimeric kinase composed of a catalytic α subunit and two regulatory subunits, β and γ. AMP binds to the γ subunit, leading to a conformational change and the subsequent phosphorylation of the α subunit by upstream kinases, which activates the enzyme. This effect of AMP is antagonized by ATP, giving AMPK the capability to monitor and respond to nutrient and energy fluctuations. Upon activation, AMPK phosphorylates its targets, either promoting catabolic energy-producing pathways or inhibiting anabolic energy-consuming pathways (Hardie 2007).
The diurnal light–dark cycle regulates the interaction between living organisms and the environment. In mammals, such circadian rhythms are controlled by a central clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus as well as peripheral clocks that reside in most tissues (Dibner et al. 2010). The central clock in the SCN is directly entrained by light, which also synchronizes the peripheral clocks by delivering synaptic or paracrine signals. In addition to light entrainment, circadian clocks are also entrained by metabolic and environmental cues such as feeding and resting.
To maintain metabolic homeostasis, the body relies on metabolic sensors such as NRs and AMPK to coordinate cellular energy usage with the fluctuations in metabolites that occur owing to daily fed/fasting cycles. NRs sense hormones and metabolites such as lipids, steroids, retinoids, and xenobiotics, whereas conversely, AMPK senses the energy state of the body through its ability to respond to changes in the AMP:ATP ratio that take place during rest and exercise. These characteristics make them ideal candidates to transmit metabolic input signals to other aspects of physiology, including circadian rhythm. On the other hand, the ability of NRs and AMPK to directly regulate metabolism also makes it possible for their activities to feed back circadian signals to metabolism.
In this chapter, we discuss our recent findings on the roles of NRs and AMPK in regulating metabolism and circadian rhythm. By doing so, we hope to expand the current understanding of the interactions between the circadian and metabolic networks and thus bring new insight to the prevention and treatment of metabolic diseases.
THE NUCLEAR RECEPTOR RING OF PHYSIOLOGY
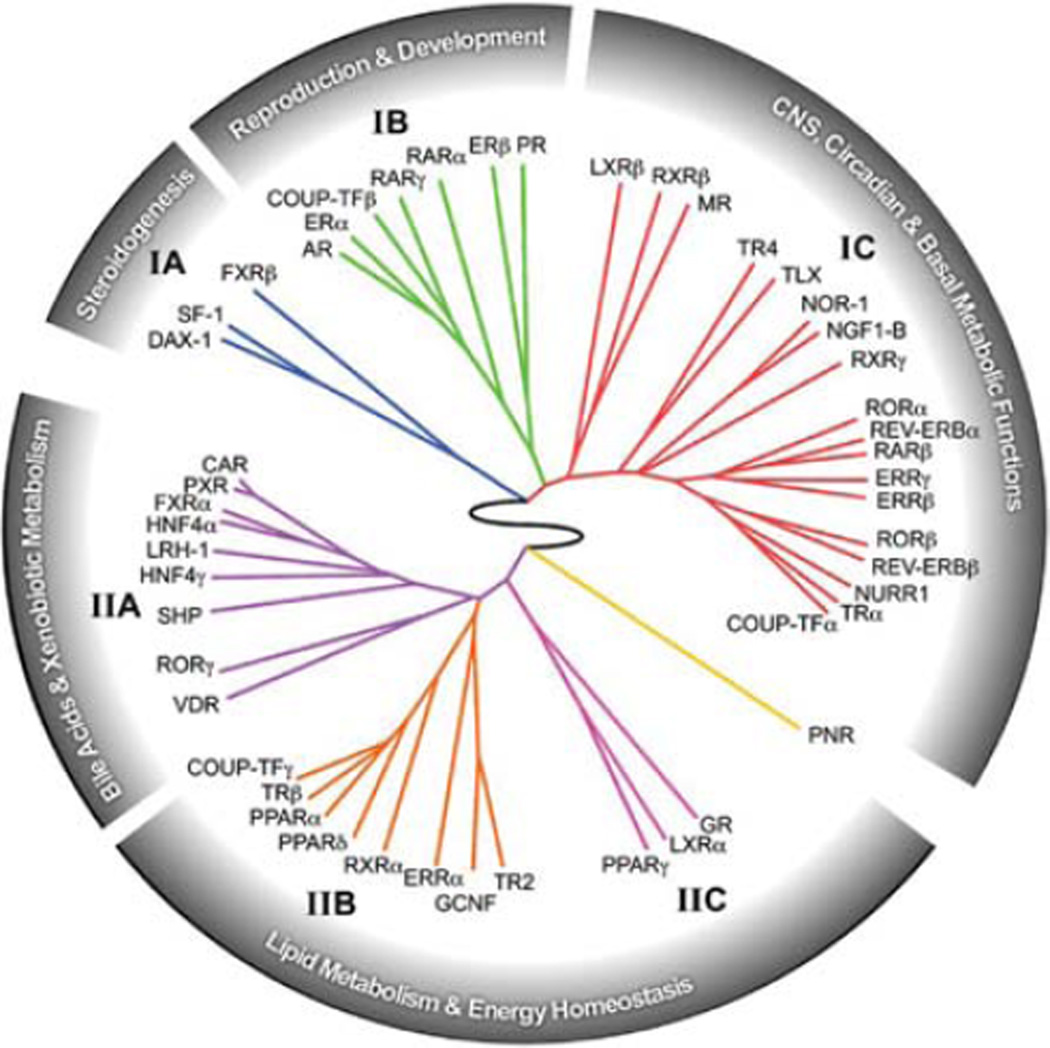
NRs are metazoan innovations to mediate their physiological functions such as reproduction, development, metabolism, and immune response. The members can be grouped into clusters that correspond to different physiological functions, best depicted as the NR ring of physiology (Fig. 1). In a previous study, we examined the tissue-specific expression profiles of all 49 mouse NRs in 39 different tissues, which served as the basis for a non-supervised hierarchical analysis to group the NRs into six main clusters (Bookout et al. 2006).
Figure 1.
The NR ring of physiology. Based on their tissue-specific expression patterns, the NR family is divided into six clusters: cluster I, steroidogenesis; cluster II, reproduction and development; cluster III, central nervous system (CNS), circadian, and basal metabolic functions; cluster IV, bile acids and xenobiotic metabolism; and clusters V and VI, lipid metabolism and energy homeostasis.
Cluster I contains the following NRs: FXRβ, SF1, and DAX-1. FXRβ is not present in human and its function in mouse remains unclear (Otte et al. 2003). SF1 and DAX-1 are key TFs controlling sexual differentiation and steroidogenesis (Beuschlein et al. 2002; Parker and Schimmer 2002). Consistently, their expression is highly restricted in the central nervous system (CNS), reproductive organs, and adrenals (Fig. 1).
NRs in cluster II govern the sex-steroid transcriptional cascade. This cluster contains all of the endocrine steroid hormone receptors that control gender determination and sexual reproduction, including androgen receptor (AR), estrogen receptor α and β (ERα and β), and progesterone receptor (PR). The cluster also includes COUP-TFβ and two of the retinoic acid receptors, RARα and RARγ. COUP-TFβ is believed to modulate retinoic acid signaling, and RARα and RARγ are known for their important roles in development (Cupp et al. 1999; Parker and Schimmer 2002; Chung et al. 2004). Consistent with their clustering with the sex hormone receptors, retinoid signaling is known to be important for reproduction (Fig. 1).
Cluster III comprises NRs that are predominantly or mainly expressed in the CNS. Such NRs include TLX, COUP-TFα, TR4, and the NR4A orphan receptors NGFI-B, NOR1, and NURR1, all known for their roles in differentiation of neural cells and peripheral tissues (Park et al. 2003; Perlmann and Wallen-Mackenzie 2004; Ponnio and Conneely 2004; Shi et al. 2004). This cluster also contains NRs important for circadian rhythm and metabolism, including REV-ERBα and β, RORα and β, ERRβ and γ, and the NR4A subfamily (Yang 2010). Other NRs in this cluster, such as TRα, MR, and LXRβ, regulate cardiovascular function (Tontonoz and Mangelsdorf 2003; Le Menuet et al. 2004; Marrif et al. 2005). This cluster also includes the retinoid X receptors, RXRβ and RXRγ, which serve as heterodimeric partners for endocrine and lipid sensing receptors (Chawla et al. 2001). Together, this cluster appears to define a clade of NRs involved in CNS functions, circadian rhythm, and metabolism (Fig. 1).
NRs in cluster IV control nutrient uptake and dietary toxin clearance. In this cluster, HNF4α is required for gastro/enterohepatic development and HNF4γ is crucial for carbohydrate metabolism and insulin action (Chen et al. 1994; Hayhurst et al. 2001). FXRα, LRH-1, and SHP are required for bile acid metabolism, which is critical for the uptake of fat-soluble nutrients (Lu et al. 2001; Edwards et al. 2002; Downes et al. 2003). This cluster also contains the xenobiotic receptors PXR and CAR, which regulate the clearance of steroids, dietary xenobiotic lipids, and harmful bioactive lipids (Dixit et al. 2005). VDR is another functionally related NR in this group, required for calcium absorption and metabolism (Kato 2000). It is also involved in sensing and clearing toxic bile acids in the colon (Makishima et al. 2002). RORγ, the last NR in this cluster, is required for lymph node development and thymopoiesis (Winoto and Littman 2002); however, its relationship with other members in this cluster remains unclear (Fig. 1).
Cluster V contains NRs that function at the next level of the nutrient regulatory network to modulate energy metabolism. In this cluster, TRβ mediates thyroid hormone responses to promote thermogenesis as well as fatty acid and cholesterol metabolism (Gullberg et al. 2000). The peroxisome proliferator-activated receptors PPARα and PPARδ are activated by lipid metabolites and positively regulate fatty acid oxidation (Evans et al. 2004). The orphan receptor ERRα regulates oxidative gene expression, lipid metabolism, and mitochondrial biogenesis (Schreiber et al. 2004). Three other orphan receptors, COUP-TFγ, TR2, and GCNF, are also included in this cluster; however, their functions and relationships with other members in this cluster remain unclear. The last receptor in this category, RXRα, serves as the predominant heterodimeric partner for other receptors in this cluster (Fig. 1) (Chawla et al. 2001).
NRs in cluster VI, including PPARγ, LXRα, and GR, control specialized aspects of energy metabolism. PPARγ and LXRα sense the fed state by responding to dietary cholesterol and fat. Their activation promotes insulin signaling and fat storage in adipose tissues (Evans et al. 2004; Kalaany et al. 2005). On the other hand, GR is activated by glucocorticoids in the fasted state and promotes lipolysis and gluconeogenesis (Fig. 1) (Wintermantel et al. 2004).
Owing to its unique expression pattern, PNR is not grouped into any of the above clusters. It is selectively expressed in the eye and is required for photoreceptor cell development and function (Fig. 1) (Kobayashi et al. 1999).
The above clustering of NRs based on their expression patterns rather than on the traditional structural homology can help us to better understand the roles of NRs in physiology. It demonstrates that NRs work in cooperative networks to modulate different aspects of animal physiology—reproduction, development, and metabolism.
NUCLEAR RECEPTORS COUPLE METABOLISM WITH CIRCADIAN RHYTHM
Physiology is controlled by circadian rhythm. At the molecular level, circadian clocks are governed by a cell-autonomous transcriptional feedback loop containing two transcriptional activators, CLOCK and BMAL1, and their target genes Period (PER) and Cryptochrome (CRY). CLOCK and BMAL1 form heterodimers and activate the transcription of PER and CRY. Consequently, PER and CRY proteins accumulate and form a repressor complex, which in turn can interact with CLOCK and BMAL1 and block their own transcription (Dibner et al. 2010).
NRs are directly involved in the regulation of circadian clocks. In addition to the primary feedback loop described above, peripheral clocks are also regulated by a secondary feedback loop consisting of the orphan NRs REV-ERBs and RORs (Liu et al. 2008). The REV-ERB and ROR proteins recognize similar cis response elements (ROREs) on target genes (Forman et al. 1994). REV-ERBs act as constitutive transcriptional repressors, whereas RORs are constitutive activators. In this feedback loop, the CLOCK/BMAL1 heterodimer governs the expression of REV-ERBs, which in turn compete with RORs at ROREs within the BMAL1 promoter and thereby inhibit the expression of BMAL1. Although this secondary feedback loop is not required for circadian rhythm generation, it is thought to enhance and allow fine tuning of the circadian oscillation (Yang et al. 2007). In fact, the PPAR coactivator PGC1α regulates circadian rhythm through the interactions with RORs and REV-ERBs, which integrate metabolic signals into the clock (Liu et al. 2007).
In addition to REV-ERBs and RORs, other NRs may also regulate circadian rhythm by sensing rhythmic metabolic signals. Circulating glucocorticoids display a strong diurnal rhythm that is controlled by both the central clock in SCN and the peripheral clock in the adrenal gland (Ishida et al. 2005; Oster et al. 2006). In response to glucocorticoids, GR binds to GREs in PER1 and PER2 genes to regulate their rhythmic expression (So et al. 2009). The levels of circulating thyroid hormones are also rhythmic (Allan and Czeisler 1994). In rodents, surgical removal of the thyroid gland alters the diurnal expression of PER2 and disrupts their circadian rhythm (Amir and Robinson 2006). Another rhythmic NR ligand is oleoylethanolamide (OEA), an endogenous ligand for PPARα whose level is diurnally regulated by food intake (Fu et al. 2007). PPARα is known to regulate the transcription of both BMAL1 and REV-ERBα, possibly by interacting with the peroxisome proliferator response elements (PPREs) at their promoters (Canaple et al. 2006). PPARγ directly regulates BMAL1 expression by interacting with cis elements at its promoter. Knockout of vascular Pparγ significantly reduces the circadian rhythms in blood pressure and heart rate and is accompanied by impaired rhythmicity of clock genes (Wang et al. 2008). The retinoic acid receptors RAR and RXR directly interact with CLOCK in a ligand-dependent manner to block its transactivation activity (McNamara et al. 2001). In fact, the administration of retinoids in mice causes a phase shift in rhythm and reduced amplitudes of circadian gene expression (Sherman et al. 2011). Other NRs such as HNF4, TRα, NURR1, and RORs have also been shown to physically interact with PER2, although their direct involvement in the regulation of circadian rhythm has not been established (Schmutz et al. 2010).
Not only do NRs regulate circadian clocks, but they also serve in transmitting circadian signals from the clock to metabolic outputs. In a comprehensive survey of the diurnal expression profiles of all 49 mouse NRs in major metabolic tissues, we have shown that of the 45 NRs expressed in these tissues, 25 exhibit rhythmic expression patterns (Yang et al. 2006). Some of these rhythmic NRs are directly regulated by the core clock genes. PPARα expression is regulated by CLOCK and BMAL1 proteins via intronic cis elements (Oishi et al. 2005; Canaple et al. 2006) and that of PPARγ is regulated by two clock-controlled TFs, DBP and E4BP4, at a novel promoter in its first exon (Takahashi et al. 2010). CAR expression is also regulated by the circadian TFs DBP, HLF, and TEF. Ablation of these three genes results in the loss of rhythmic expression of CAR and its target genes, which are associated with defects in xenobiotic detoxification (Gachon et al. 2006).
As discussed above, circadian rhythm and metabolism are connected by NR signaling pathways. The circadian clock directly governs the rhythmic expression of NRs to control metabolism. NRs in turn respond to metabolic signals and regulate circadian rhythm, either by regulating the transcription of clock genes or interacting with clock proteins to regulate their downstream targets.
AMPK COUPLES METABOLISM WITH CIRCADIAN RHYTHM
In addition to NRs, the nutrient sensor AMPK is another regulator that couples metabolism with circadian rhythm. AMPK is activated when energy supplies are low, such as during fasting or exercising. When activated, AMPK promotes catabolic pathways to burn glucose or fatty acids to produce ATP or inhibits anabolic pathways to limit ATP consumption. It functions by either directly phosphorylating metabolic enzymes or regulating gene transcription. The targets of AMPK cover many aspects of metabolism including glucose uptake, glycolysis, gluconeogenesis, glycogen synthesis, fatty acid oxidation and synthesis, steroid synthesis, protein synthesis, and mitochondrial biogenesis (Hardie 2007).
AMPK activity is regulated by circadian rhythm. Phosphorylation of the AMPK substrate acetyl-CoA carboxylase (ACC) by AMPK follows a diurnal rhythm (Davies et al. 1992). Our studies in mouse liver have revealed the circadian expression of the AMPK β2 subunit, rhythmic phosphorylation of the AMPK substrates Raptor and ACC, and oscillating nuclear localization of the AMPK α1 subunit (Lamia et al. 2009). On the other hand, AMPK also directly regulates circadian rhythm through the degradation of PER2 or CRY1 (Um et al. 2007; Lamia et al. 2009). PER2 degradation is induced by its phosphorylation by casein kinase ε (CKε) (Eide et al. 2005). When activated by metformin, AMPK phosphorylates CKε, thus inducing PER2 degradation, which alters the circadian period length (Um et al. 2007). We have also shown that activation of AMPK results in direct phosphorylation of CRY1 and induces its degradation, resulting in an increase in circadian period and reduced amplitude of BMAL1 expression (Lamia et al. 2009).
Therefore, the circadian clock regulates AMPK activity to fulfill the body’s different energy requirements during the diurnal physiological cycles. In addition, various metabolic energy states also regulate AMPK activity and influence the circadian rhythm by controlling the degradation of PER2 and CRY1.
CROSS TALK BETWEEN NUCLEAR RECEPTORS AND AMPK IN METABOLISM
NRs and AMPK form a highly interactive network to regulate metabolism. In liver, the orphan NR HNF4α regulates gluconeogenesis. AMPK can directly phosphorylate HNF4α to reduce its transcriptional activation activity and promote its protein degradation (Hong et al. 2003). AMPK activation also induces the expression of SHP, another hepatic orphan NR, resulting in inhibition of gluconeogenesis (Kim et al. 2008; Lee et al. 2010). The xenobiotic receptor CAR plays an essential role in xenobiotic detoxification, which requires a large amount of energy. In primary rodent hepatocytes, activation of AMPK blocks the nuclear translocation of CAR and inhibits its transactivation of drug-metabolizing genes, thereby reducing energy consumption (Kanno et al. 2010). AMPK also interacts with the glucocorticoid signaling pathway via the phosphorylation of GR to regulate its transcriptional activity and promote energy production (Nader et al. 2010). In muscle, AMPK is actively involved in regulating energy metabolism, in cooperation with NRs PPARδ and ERRγ (Narkar et al. 2008, 2011). AMPK activation in muscle induces running endurance, oxidative fiber composition, mitochondrial biogenesis, and oxidative metabolism, which is associated with its direct interaction with PPARδ (Narkar et al. 2008). We have also found that AMPK is constitutively activated in mice that overexpress muscle-specific ERRγ, suggesting potential interactions between AMPK and ERRγ (Narkar et al. 2011).
CONCLUSION
In the past few years, significant progress has been made in understanding the roles of the nuclear receptor superfamily and AMPK in regulating metabolism and circadian rhythm. NRs function in a supraregulatory network to cooperatively regulate many aspects of physiology, including metabolism and circadian rhythm. This regulatory network is further enriched by interaction with AMPK. However, our current knowledge is just the tip of the iceberg, because many parts of this supraregulatory network still remain unclear and much future effort will be required to find the final pieces of this physiological puzzle.
REFERENCES
- Allan JS, Czeisler CA. Persistence of the circadian thyrotropin rhythm under constant conditions and after light-induced shifts of circadian phase. J Clin Endocrinol Metab. 1994;79:508–512. doi: 10.1210/jcem.79.2.8045970. [DOI] [PubMed] [Google Scholar]
- Amir S, Robinson B. Thyroidectomy alters the daily pattern of expression of the clock protein, PER2, in the oval nucleus of the bed nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neurosci Lett. 2006;407:254–257. doi: 10.1016/j.neulet.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Beuschlein F, Keegan CE, Bavers DL, Mutch C, Hutz JE, Shah S, Ulrich-Lai YM, Engeland WC, Jeffs B, Jameson JL, et al. SF-1, DAX-1, and acd: Molecular determinants of adrenocortical growth and steroidogenesis. Endocr Res. 2002;28:597–607. doi: 10.1081/erc-120016972. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Chung SS, Sung W, Wang X, Wolgemuth DJ. Retinoic acid receptor α is required for synchronization of spermatogenic cycles and its absence results in progressive breakdown of the spermatogenic process. Dev Dyn. 2004;230:754–766. doi: 10.1002/dvdy.20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp AS, Dufour JM, Kim G, Skinner MK, Kim KH. Action of retinoids on embryonic and early postnatal testis development. Endocrinology. 1999;140:2343–2352. doi: 10.1210/endo.140.5.6720. [DOI] [PubMed] [Google Scholar]
- Davies SP, Carling D, Munday MR, Hardie DG. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem. 1992;203:615–623. doi: 10.1111/j.1432-1033.1992.tb16591.x. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dixit SG, Tirona RG, Kim RB. Beyond CAR and PXR. Curr Drug Metab. 2005;6:385–397. doi: 10.2174/1389200054633907. [DOI] [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, et al. A chemical, genetic, and structural analysis of the bile acid receptor FXR. Mol Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA, Kast HR, Anisfeld AM. BAREing it all: The adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIε-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM. Cross-talk among ROR α 1 and the Reverb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, Piomelli D. Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. J Biol Chem. 2007;282:1518–1528. doi: 10.1074/jbc.M607809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gullberg H, Rudling M, Forrest D, Angelin B, Vennstrom B. Thyroid hormone receptor β-deficient mice show complete loss of the normal cholesterol 7α-hydroxylase (CYP7A) response to thyroid hormone but display enhanced resistance to dietary cholesterol. Mol Endocrinol. 2000;14:1739–1749. doi: 10.1210/mend.14.11.0548. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YH, Varanasi US, Yang W, Leff T. AMP-activated protein kinase regulates HNF4α transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem. 2003;278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- Ishida A, Mutoh T, Ueyama T, Bando H, Masubuchi S, Nakahara D, Tsujimoto G, Okamura H. Light activates the adrenal gland: Timing of gene expression and glucocorticoid release. Cell Metab. 2005;2:297–307. doi: 10.1016/j.cmet.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Inoue Y, Inouye Y. 5-Aminoimidazole-4-carboxamide-1-β-ribofuranoside (AICAR) prevents nuclear translocation of constitutive androstane receptor by AMP-activated protein kinase (AMPK) independent manner. J Toxicol Sci. 2010;35:571–576. doi: 10.2131/jts.35.571. [DOI] [PubMed] [Google Scholar]
- Kato S. The function of vitamin D receptor in vitamin D action. J Biochem. 2000;127:717–722. doi: 10.1093/oxfordjournals.jbchem.a022662. [DOI] [PubMed] [Google Scholar]
- Kim YD, Park KG, Lee YS, Park YY, Kim DK, Nedumaran B, Jang WG, Cho WJ, Ha J, Lee IK, et al. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase-dependent regulation of the orphan nuclear receptor SHP. Diabetes. 2008;57:306–314. doi: 10.2337/db07-0381. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: Early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- Lee JM, Seo WY, Song KH, Chanda D, Kim YD, Kim DK, Lee MW, Ryu D, Kim YH, Noh JR, et al. AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB.CRTC2 complex by orphan nuclear receptor small heterodimer partner. J Biol Chem. 2010;285:32182–32191. doi: 10.1074/jbc.M110.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Menuet D, Viengchareun S, Muffat-Joly M, Zennaro MC, Lombes M. Expression and function of the human mineralocorticoid receptor: Lessons from transgenic mouse models. Mol Cell Endocrinol. 2004;217:127–136. doi: 10.1016/j.mce.2003.10.045. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBα and β and nonessential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Repa JJ, Mangelsdorf DJ. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J Biol Chem. 2001;276:37735–37738. doi: 10.1074/jbc.R100035200. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Marrif H, Schifman A, Stepanyan Z, Gillis MA, Calderone A, Weiss RE, Samarut J, Silva JE. Temperature homeostasis in transgenic mice lacking thyroid hormone receptor-α gene products. Endocrinology. 2005;146:2872–2884. doi: 10.1210/en.2004-1544. [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Nader N, Ng SS, Lambrou GI, Pervanidou P, Wang Y, Chrousos GP, Kino T. AMPK regulates metabolic actions of glucocorticoids by phosphorylating the glucocorticoid receptor through p38 MAPK. Mol Endocrinol. 2010;24:1748–1764. doi: 10.1210/me.2010-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JD, Joseph JD, Sherk AB, Juzumiene D, Turnbull PS, Rafferty SW, Cui H, Anderson E, Fan D, Dye DA, et al. Differential presentation of protein interaction surfaces on the androgen receptor defines the pharmacological actions of bound ligands. Chem Biol. 2009;16:452–460. doi: 10.1016/j.chembiol.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α (PPARα) in mice. Biochem J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, et al. Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Mol Cell Biol. 2003;23:864–872. doi: 10.1128/MCB.23.3.864-872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Tsai SY, Tsai MJ. Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J Med. 2003;52:174–181. doi: 10.2302/kjm.52.174. [DOI] [PubMed] [Google Scholar]
- Parker KL, Schimmer BP. Genes essential for early events in gonadal development. Ann Med. 2002;34:171–178. [PubMed] [Google Scholar]
- Perlmann T, Wallen-Mackenzie A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004;318:45–52. doi: 10.1007/s00441-004-0974-7. [DOI] [PubMed] [Google Scholar]
- Ponnio T, Conneely OM. Nor-1 regulates hippocampal axon guidance, pyramidal cell survival, and seizure susceptibility. Mol Cell Biol. 2004;24:9070–9078. doi: 10.1128/MCB.24.20.9070-9078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman H, Gutman R, Chapnik N, Meylan J, leCoutre J, Froy O. All-trans retinoic acid modifies the expression of clock and disease marker genes. J Nutr Biochem. 2012;23:209–217. doi: 10.1016/j.jnutbio.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci. 2009;106:17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Inoue I, Nakajima Y, Seo M, Nakano T, Yang F, Kumagai M, Komoda T, Awata T, Ikeda M, et al. A promoter in the novel exon of hPPARγ directs the circadian expression of PPARγ. J Atheroscler Thromb. 2010;17:73–83. doi: 10.5551/jat.2410. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iε (CKIε)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, et al. Vascular PPARγ controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8:482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(suppl.):S57–S66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Berger S, Greiner EF, Schutz G. Genetic dissection of corticosteroid receptor function in mice. Horm Metab Res. 2004;36:387–391. doi: 10.1055/s-2004-814567. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- Yang X. A wheel of time: The circadian clock, nuclear receptors, and physiology. Genes Dev. 2010;24:741–747. doi: 10.1101/gad.1920710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]