Abstract
CryoEM data capture the dynamic character associated with biological macromolecular assemblies by preserving the various conformations of the individual specimens at the moment of flash freezing. Regions of high variation in the data set are apparent in the image reconstruction due to the poor density that results from the lack of superposition of these regions. These observations are qualitative and, to date, only preliminary efforts have been made to quantitate the heterogeneity in the ensemble of particles that are individually imaged. We developed and tested a quantitative method for simultaneously computing a reconstruction of the particle and a map of the space-varying heterogeneity of the particle based on an entire data set. The method uses a maximum likelihood algorithm that explicitly takes into account the continuous variability from one instance to another instance of the particle. The result describes the heterogeneity of the particle as a variance to be plotted at every voxel of the reconstructed density. The test, employing time resolved data sets of virus maturation, not only recapitulated local variations obtained with difference map analysis, but revealed a remarkable time dependent reduction in the overall particle dynamics that was unobservable with classical methods of analysis.
Keywords: Time-resolved, Single particle reconstruction, Virus maturation, Protein folding, Quasi equivalence, Cryo electron microscopy, Nudaurelia Capensis Omega Virus, Maximum likelihood estimation, Expectation maximization algorithm, Heterogeneous particles, Variance map
1. Introduction
Recent success with 3-dimensional (3-D) reconstructions of biological macro molecular particles employing single-particle cryo electron microscopy (cryo EM) has been remarkable. Sub nanometer icosahedral virus structures are virtually routine and protein and nucleo-protein structures, without symmetry, are appearing more frequently at comparable resolution. Structures of icosahedral viruses at near-atomic resolution have been achieved with this technology in recent years (Liu et al., 2010; Baker et al., 2010; Zhang et al., 2008).
Cryo EM data captures biological macromolecular particles that are trapped in one of a smooth continuum of conformations at the moment of vitrification in liquid ethane. The amount of conformational change accessible to the particle is presumably space dependent, but there are limited tools available for assessing the global amount of conformational change available let alone creating a spatial map of the amount of conformational change occurring.
Cryo EM has recently been reviewed in the three volumes edited by Jensen (2010a,b,c). The idea of maximum likelihood as a method for deriving statistical estimators dates back to the early 1900s (Lehmann and Casella, Section 10.1, p. 515, 1998) and it remains an important method. Computation of a reconstruction by optimization of the fit between the images predicted by a mathematical model and the experimental images, which can be interpreted as a maximum likelihood estimator, was first done by Vogel et al. (1986) and Provencher and Vogel (1988a,b) and has recently been reviewed (Scheres, 2010; Sigworth et al., 2010). Maximum likelihood has also been used for other estimation tasks related to cryo electron microscopy, such as estimating the orientation of an image (Sigworth, 1998). Heterogeneity among a set of particles can be detected by methods such as cross-common lines residuals (Fuller et al., 1996). In this paper, maximum likelihood estimation is used not to estimate a single reconstruction or to find a homogeneous subset of particles but rather to estimate the statistics of an entire ensemble of reconstructions where the statistics of the images predicted by the statistics of the ensemble of reconstructions match the statistics of the experimental images. The most closely related work is due to Penczek et al. (2006). In this work, a space-varying variance map was constructed after the reconstruction is computed by a Monte-Carlo resampling procedure. This contrasts with the approach proposed here where the mean and covariance information are simultaneously estimated, generating not only the reconstruction but also the variance associated with every voxel of the reconstruction.
The method was used to reanalyze the time-resolved single-particle cryo EM images of Nudaurelia Capensis Omega Virus (NωV) from Matsui et al. (2010), a T = 4 icosahedral RNA virus. NωV capsid is composed of 240 copies of the same gene product, protein alpha, that in a maturation step, undergoes a autocatalytic reaction generating the major capsid protein beta and the small gamma peptide, which remains non-covalently associated with the capsid. NωV virus-like particles can be purified in the unc-leaved pro-capsid state and the maturation process can be precisely triggered by lowering the pH to 5.0. Kinetics of the cleavage is unusual with 50% of the subunits cleaved in 30 min while several hours are required for all of the subunits to cleave. Taking advantage of the slow kinetics of maturation of NωV, partially cleaved particles in intermediate stages of maturation (3 min, 30 min, and 4 h all at pH 5.0) were analyzed by cryo-EM. Because the size of the particles is the same throughout the maturation process, it was possible to use difference cryo-EM density maps. The density at each time point was subtracted from the fully mature particle. With the X-ray model as a guide, the difference density at each of the cleavage sites was evaluated. Subunits surrounding 5-fold and 3-fold icosahedral symmetry axes are quickly formed and cleave in 30 min, while the subunits not adjacent to these axes cleave slowly. Here, we show that the maximum-likelihood derived variance map can, in a single data set, reveal the same local variations that were observed with difference map analysis, and also provide an overall view of particle dynamics that was unobservable with classical methods of analysis.
2. Cryo-EM data sets and reconstructions
The data are the time-resolved single-particle cryo EM images of NωV from Matsui et al. (2010). The pixels measure 2.768 Å and the boxed image of an individual particle is 200 × 200 pixels in dimension. The reconstructions from Matsui et al. (2010) have been deposited in the EM Data Bank (EMDataBank). The times, reference numbers, and accession codes are 3 min, 25633, EMD-5426; 30 min, 25634, EMD-5427; 4 h, 25635, EMD-5428; and 3 d, 25622, EMD-5425, respectively. The original image stacks are available from J.E.J. The assumption that there is only one class of particle at each time point was sufficient to achieve resolutions of 9.3 Å, 8.6 Å, 8.3 Å, and 9.8 Å for the 3 min, 30 min, 4 h, and 3 days data sets, respectively (Matsui et al., 2010), and so the calculations described in this paper, which are at lower resolution, have continued with that assumption. The results from this paper have been deposited in the EM Data Bank (EMDataBank). Each data set results in two depositions: a mean map and a variance map. For the mean maps, the times, reference numbers, and accession codes are 3 min, 25729, EMD-5449; 30 min, 25858, EMD-5474; 4 h, 25859, EMD-5472; and 3 d, 25860, EMD-5473, respectively. For the variance maps, the times, reference numbers, and accession codes are 3 min, 25861, EMD-5468; 30 min, 25863, EMD-5469; 4 h, 25864, EMD-5471; and 3 d, 25865, EMD-5470, respectively.
3. Computational methodology
Using a weighted sum of basis functions to represent the electron scattering intensity function has a long history in structural biology, e.g., Fourier series in X-ray crystallography. If every instance of the object is identical, then the weights in the description of each object are the same and the goal of structure determination is to determine the numerical value of each weight. But if different instances of the object are different, then there is no unique numerical value for each weight. Different instances might differ by different stoichiometry or by different geometrical configuration, e.g., flash frozen in different vibrational conditions for single-particle cryo EM problems. If the differences can be described as statistical variation, then the goal of structure determination might be to determine the numerical values of the means and variances of each weight. If the weights are assumed to be Gaussian random variables and are grouped in a vector, then the mean vector and covariance matrix for the weight vector is a complete description of the object.
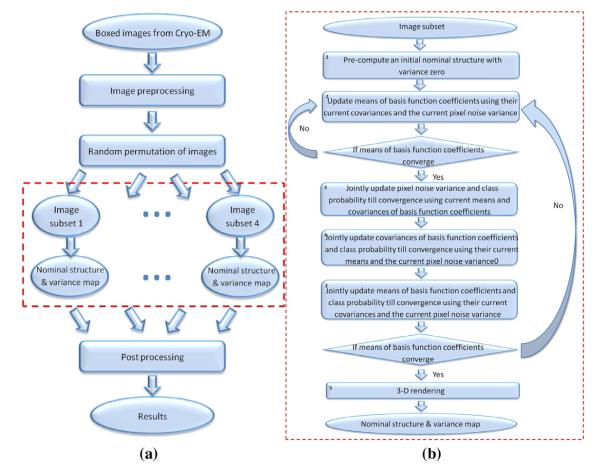
The change from describing the weights as numbers and estimating the numbers for each class of object, to describing the weights as Gaussian and estimating the statistics (the mean, corresponding to a traditional reconstruction, and the covariance, describing fluctuations around the reconstruction) for each class of object is the modeling innovation proposed in this paper. Using this new model, a maximum likelihood estimator is used to determine the means and covariances that are the solution of the reconstruction problem and the estimator is computed by a generalized expectation maximization algorithm which is an iterative algorithm which must be provided with an initial condition. The pixel noise variance and the probability that an image belongs to a particular class are also estimated. Optionally, but not used in the calculations described in this paper, the a priori probability density function on the projection orientation of the images can also be estimated. In the expectation maximization algorithm, simultaneous updates of all parameters to be estimated is a difficult optimization problem so the mean vector, the covariance matrix, and the pixel noise variance are updated sequentially (so that this is actually a generalized expectation maximization algorithm). Each update is the solution of a maximization problem. For the mean vector, the maximization problem is quadratic in the unknown vector so the new mean vector is the solution of a linear system quite similar to the situation in the homogeneous particle case (Doerschuk and Johnson, 2000; Yin et al., 2003; Lee et al., 2007; Prust et al., 2009; Lee et al., 2011). For the covariance matrix, the maximization problem is complicated because the covariance of an image is a linear combination of the covariance of the weights in the orthonormal expansion and the variance of the additive pixel noise. The linear combination is unknown and is different for each different image. Intuitively, the observed variability in the images is being partitioned into two sources which are the heterogeneity of the particle and the additive pixel noise. Because the covariance, rather than the inverse covariance, is a linear combination of the covariance of the weights in the orthonormal expansion and the variance of the additive pixel noise, this maximization problem is not convex. Formulas for first and second derivatives of the function to be maximized with respect to the covariance of the weights can be determined and, using the function and the derivatives, the maximization problem is solved numerically. Finally, for the additive pixel noise covariance, a search based on just the function to be maximized is used since the unknown is a scalar and an accurate initial condition is available by computing the sample variance of the pixels in the images in an annulus outside of the image of the particle. A flow chart of the algorithm is given in Fig. 1. Fig. 1(a) shows the entire algorithm with preprocessing, repetition of reconstruction calculations on non-overlapping sets of boxed images in order to provide the data necessary for computing sample variances, and postprocessing. Fig. 1(b) shows the reconstruction algorithm for a single set of boxed images.
Fig. 1.
Algorithm flowcharts. The four parallel computations in Panel (a) are used to determine the performance of the algorithm, e.g., the error bars in Fig. 4. The calculations contained in the red dotted-line box of Panel (a) are expanded in Panel (b) which describes the maximum likelihood estimator.
In order to compute the performance of the algorithm, at each time point the algorithm is run on each of four distinct data sets where the data sets are nonoverlapping subsets of the four image stacks of Matsui et al. (2010). Then, based on the four results, sample standard deviations can be computed which describe the performance of the algorithm. This overall computation is shown in Fig. 1(a). The algorithm is iterative and therefore requires an initial condition. At each time point, for the first of the four data sets, the algorithm is used twice: (1) the algorithm is started with means that describe a spherically-symmetric reconstruction and zero covariances and is run to the final resolution with the heterogeneity features turned off. In this case the algorithm is equivalent to the authors’ previous work (Doerschuk and Johnson, 2000; Yin et al., 2003). Alternatively, this calculation could be described as Block 2 of the flow chart in Fig. 1(b) or the third line of Algorithm 1 with the addition of the standard idea of refinement where the resolution of the reconstruction is progressively increased. (2) The algorithm is restarted with the means equal to the solution from Step (1) and the variances equal to 10% of the corresponding means and is run with the heterogeneity features turned on to determine the heterogeneous reconstruction. At each time point, for the second through fourth data sets, only Step (2) is used starting from the homogeneous reconstruction resulting from Step (1) applied to the first data set since there is no need to find a new initial condition.
Algorithm 1.
The generalized EM algorithm
| set the initial conditions on , V, q, and Q. |
| while true do |
| while not converged do |
| update q (Eq. (23)) and (Eq. (24)) (Fig. 1, Block 2) |
| end while |
| while q and Q not converged do |
| update q (Eq. (23)) and Q. (Fig. 1, Block 3) |
| end while |
| while q and V not converged do |
| update q (Eq. (23)) and V. (Fig. 1, Block 4) |
| end while |
| update q (Eq. (23)) and (Eq. (24)) (Fig. 1, Block 2) |
| if converged then |
| break |
| end if |
| end while |
Once the mean vector and covariance matrix for the weights in the orthonormal expansion have been estimated, the nominal structure can be computed from the mean vector and the variance map can be computed from the covariance matrix.
The following subsections describe the computational methods in detail.
3.1. Preprocessing
Subsequent to the steps used to create the image stacks in Matsui et al. (2010), the following procedure was carried out.
Reconstruction algorithms can include provisions for rejecting images from the image stack because the images appear not to belong to the particle as it appears in the 3-D reconstruction being computed. Such provisions are not included in the current version of the reconstruction algorithm described in this paper. Therefore, some possibly junk images are removed from the stack before the reconstruction algorithm begins by the following mechanism: first, select the first 6000 images from the stack. Second, compute the sample mean of all the selected images. (The mean image is nearly circularly symmetric). Third, compute the difference between each particular image and the sample mean image. Fourth, compute the square of the Euclidean norms of the difference images. Fifth, from a histogram of the squared norms, decide on a threshold and remove images from the stack if their squared norm is greater than the threshold. About 16% of the images are removed. For the stack recorded at 3 min, 20 of the removed images are shown in Fig. 8 (Supplemental material).
In order to compute the performance of the algorithm, e.g., the error bars of Fig. 4, the algorithm is applied to multiple sets of images. Specifically, from those images that are not removed from the stack, we form four substacks each with 1200 images by first randomly permuting the 6000 images and then selecting subsets of 1200 images where the subsets are those images numbered 4n − 3, 4n − 2, 4n − 1, and 4n where n ∈ {1, … ,1200}.
Individually for each image in a stack, normalize the image. Specifically, ynew = a yold + b where a and b are chosen so that the sample mean and the sample variance of ynew, both evaluated outside of the image of the virus particle, have values 0 and 1, respectively.
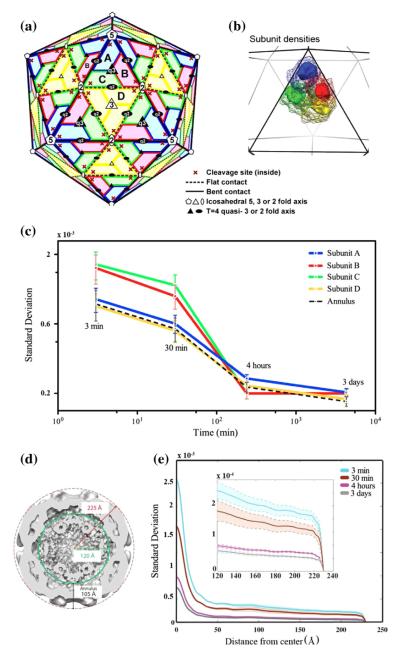
Fig. 4.
Region-specific variability analysis of the NwV protein capsid in different stages of maturation. Panels A–C: variance analysis around the cleavage sites of Subunits A, B, C and D that form the asymmetric unit of the NwV protein capsid. Panels A and B show the T = 4 surface lattice with the subunits’ locations. The total volume occupied by each subunit is rendered as a mesh in Panel B. The variance was calculated over a smaller region, enclosing the cleavage site, which is shown as a solid volume within the subunit density. As is described in Section 3.3, the smaller region is essentially the region occupied by Cα atoms within 10 Å of the active site. This is the same region analyzed by Matsui et al. (2010) using difference maps. In Panel C the standard deviation for this region is plotted log–log as a function of time for each subunit. The plot demonstrates an overall reduction of variance as a function of time after maturation is initiated, with distinct kinetics between the variances of the B and C sites (high) and the A and D sites (low). Computational methods are described by Eqs. (33)–(35). The capsid shell is defined to be the annulus with radius from 120 to 216 Å. Panels D–E: Time variation of spherical averages. A cross section perpendicular to the 2-fold axis (Panel D) shows the location of the capsid shell relative to the center of the particle. The square root of the spherically-averaged variance map versus distance from the center of the particle was computed by Eqs. (37), (34) and (35) and is plotted in Panel E. The shaded region covers plus/minus one standard deviation. The inset plot shows a zoomed version of the plot including only the capsid shell region.
3.2. Reconstruction
The electron scattering intensity of the particle is described as a weighted sum of basis functions. A standard approach is to treat the set of weights as numbers and seek to estimate the values of the numbers by a maximum likelihood estimator (Doerschuk and Johnson, 2000; Yin et al., 2003; Scheres et al., 2007; Lee et al., 2007; Prust et al., 2009; Lee et al., 2011). In contrast, in this paper we treat the set of weights as random variables where every particle is described by an independent realization of the random variables. We then seek to estimate the joint probability density function of the set of random variables. In order to simplify the task from estimating functions to estimating numbers, we assume that the joint probability density function is Gaussian so that all we must estimate is the mean vector and covariance matrix. These quantities can be estimated by a maximum likelihood estimator which is computed by an expectation–maximization algorithm where the nuisance parameters in the expectation–maximization algorithm include the unknown projection direction of the image of each particle.
3.2.1. Reconstruction: notation
If x is a random variable then x ~ p means that x is distributed with probability density function (pdf) p. is the Gaussian pdf with mean vector m and covariance matrix S evaluated at argument x. If v is a vector (which might already have multiple superscripts and subscripts) then (v)j is the jth component of the vector. Likewise, if M is a matrix then (M)j,j’ is the (j,j’) th element of the matrix.
3.2.2. Reconstruction: model
The electron scattering intensity ρ as a function of 3-D real-space coordinates x is described by a truncated orthonormal expansion with weights c and basis functions ϕ:
| (1) |
where η is the class label and there are Nη classes. In first-order image formation theory (Erickson, 1973; Lepault and Pitt, 1984; and Toyoshima and Unwin, 1988), the reciprocal-space image, denoted by Υ, parameterized by the 2-D reciprocal-space vector, denoted by κ, is the product of three factors. (1) The 2-D Fourier transform of the projection image which, by the projection slice theorem, can be computed from the 3-D Fourier transform P of the object ρ and the 3 × 3 rotation matrix R that describes the projection direction which is parameterized by the Euler angles (α, β, γ). (2) The contrast transfer function G. (3) A complex exponential of the translation χ0 of the projected location of the center of the object from the center of the reciprocal space image. The resulting equation is
| (2) |
In order to make Eqs. (1) and (2) into numerical linear algebra, the spatial frequency vector κ is discretized and Eq. (2) for each sample is one row of the resulting vector equation. In addition, the notation is augmented with an index i which indicates which of the boxed images is being described and Φ is the 3-D Fourier transform of the basis function ϕ. The resulting equation is
| (3) |
where
- yi is a vector whose jth component is the reciprocal space image evaluated at the jth sampled reciprocal space vector κj, i.e.,
(4) - zi is the Euler angles (αi, βi, γi) that describe the projection orientation of the ith image, the 2-component vector χ0,i that describes the projected location of the center of the particle in the ith image, and the class label ηi, i.e.,
all of which are unknown.(5) - c(ηi) is the vector of weights for the ith particle, i.e.,
(6) - L(zi) is the matrix that describes the transformation from weights to sampled reciprocal-space image as is given in Eq. (2), i.e., weights to 3-D cube, projection from 3-D to 2-D, the effect of the contrast transfer function, and the translation of the projected location of the center of the particle in the ith image so the (j,j’) th element of this matrix is
(7)
The statistical model used previously (Doerschuk and Johnson, 2000; Scheres et al., 2007) is that every object in the ηth class is identical and the projection image is corrupted by additive zero-mean Gaussian noise that is independent from image to image. The a priori probability that an object is from the ηth class is qη. The resulting equations are
| (8) |
| (9) |
where the goal is to estimate the vectors c(η) for η ∈ {1, … ,Nη}. This problem can be generalized to include estimating the a priori probability density function on the orientation of the projections and estimating the a priori probabilities of each class (Scheres et al., 2007).
In this paper it is proposed to allow each instance of an object in the ηth class to have a different structure where the variability is described statistically by assuming that the weights for the orthonormal expansion (Eq. (1)) collected into a vector (Eq. (6)) are Gaussian random vectors with mean vector and covariance matrix Vη. The resulting equations are
| (10) |
| (11) |
| (12) |
where the ci random vectors are nuisance parameters, that is, they are not known but instead of estimating them, a pdf for them is provided. Since linear transformations of Gaussian random vectors are Gaussian random vectors, rewrite Eqs. (10)–(12) with a single Gaussian random vector v’ rather than two Gaussian random vectors c and v. The resulting equations are
| (13) |
| (14) |
Eqs. (10)–(14) differ in two important ways. First, the ci random vectors are gone leading to simpler estimator equations. Second, v’ has a structured covariance matrix, specifically, L(zi)VηiLT (zi)+Q. The goal is to estimate Q, qη, , Vη for η ∈ {1, … ,Nη}. In addition, though it is not done in this paper, it is possible to estimate the a priori pdf on the orientation of the projections.
3.2.3. Reconstruction: estimator
Using the notation of Section 3.2.2, it follows from Eqs. (13) and (14) that the conditional mean, denoted by , and the conditional covariance, denoted by Ξi(θi, ηi, Vηi, Qi, of the ith image, denoted by yi, are
| (15) |
| (16) |
| (17) |
| (18) |
where the operators E and Cov are expectation and covariance, respectively, and that the conditional probability density function (pdf) on yi is
| (19) |
The absence of a subscript or superscript implies that the variable is the collection of variables with the subscript or superscript, e.g., . In this abbreviated notation, the log likelihood function for the maximum likelihood estimator is
| (20) |
where Nv is the number of particles that are imaged, p(yi∣θi, ηi, , Vηi, Qi) is given in Eq. (19), and p(θi) is the a priori pdf on θi and the definition of the estimator is
| (21) |
where the ^ indicates that the variable is an estimate.
The method used for computing the maximization is a generalized expectation–maximization algorithm. The idea in expectation–maximization algorithms is that there is a set of so-called nuisance parameters which, if their values were measured, would greatly simplify the computation of the maximum. However, the values are not measurable. The iterative nature of the algorithm results from repeating a pair of steps: average over the possible values of the nuisance parameters (the so-called expectation step) and compute new values for the parameters being estimated by maximizing the result of the averaging with respect to the parameters. For this problem, the natural nuisance parameters are the variables θi,ηi (i ∈ {1, … ,Nv}).
The conditional pdf on the nuisance parameters is
| (22) |
which uses Eq. (19). Using Eq. (22) repeatedly and following the calculation of Doerschuk and Johnson (2000), the update equations for the generalized expectation maximization algorithm are described in the following paragraphs. In all the following equations, variables with a leading subscript of 0, e.g., 0V, are the result of the previous iteration and variables without a leading subscript of 0, e.g., V, are the variables being computed in the current iteration.
- For each class (equivalently, each value of η’ in the set {1, … ,Nη}), the new value of the a priori class probability, denoted by qη’ as a function of , 0V, 0q, and 0Q is
where the computation of p(θi, η’∣yi, , 0Vη’, 0q, 0Qi) is from Eq. (22). The primary computational expense is to compute the integrals in Eqs. (23), (25), (26), and (28). (Fig. 1, Blocks 2, 3, and 4).(23) - The new value of as a function of V, Q, , 0V, 0Q is determined by solving the following linear system for each η’ ∈ {1, … ,Nη} to compute the corresponding vectors:
where(24) (25)
(Fig. 1, Block 2).(26) The new value of Q as a function of V, , , 0V, 0Q is only considered for the case where the pixel noise is independent and identically distributed at all pixels of all images. Then Q is just a scalar covariance which is denoted by λ and which must be determined by nonlinear programming to maximize the value of Eq. (28). (Fig. 1, Block 3).
3.2.4. Reconstruction: algorithm
These four steps of Section 3.2.3 can be combined in many ways to yield valid expectation maximization algorithms. Focusing on the importance of the mean vector, which is the traditional reconstruction, the calculations described in this paper use the algorithm described in Algorithm 1.
Several aspects of Algorithm 1 need additional explanation. The algorithm is an ab initio algorithm but it has not often been used in that mode. Instead, it has typically been used based on a traditional homogeneous reconstruction which provides a high-quality estimate of mean for each value of η ∈ {1, … ,Nη} and this estimate is used as the initial condition. Because the optimization problem is for the covariance Vη and not, for instance, the Cholesky factor of Vη, it is necessary to impose the constraint that Vη be semi positive definite. Therefore, the biologically-natural initial condition of Vη = 0 is on the boundary of the feasible set and the nonlinear programming algorithms that have been used do not behave well in this situation. Therefore, the initial condition that has been used is a diagonal initial condition where the jth element is 10% of the jth element of the initial condition. The initial condition for Q, the pixel noise, is the sample variance in an annulus of the image surrounding the portion of the image that displays the virus particle, averaged over all the images in the calculation. The initial condition for qη, the class probability, is uniform, i.e., qη = 1/Nη for each value of η ∈ {1, … ,Nη}.
3.2.5. Reconstruction: software
The theory of earlier subsections applies for any choice of basis functions. However, the software uses the specific basis functions described in Yin et al. (2003) where each basis function is the product of an icosahedral harmonic and a spherical Bessel function. A software implementation of the method was written that is suitable for execution in either the proprietary Matlab (Mathworks) or open source Octave (Octave) engine on a shared-memory computer. The update of Q is done by fminbnd in both Matlab and Octave. The update of V is implemented only for the case where V is a diagonal matrix and is done by fmincon in Matlab and SQP in Octave. The limits of the software are partly memory requirements and partly use of simple numerical linear algebra algorithms. As an example of algorithmic limitations, Eq. (24) is solved by LU decomposition where the F matrix (Eq. (25)) and g vector (Eq. (26)) are each computed without taking advantage of the fact that orientations that are close to each other lead to similar contributions to the integrals in Eqs. (25) and (26). Relative to memory requirements, in order to make efficient Matlab code, the data is treated as matrix with dimensions that are the number of pixels per image by the number of images. This is the largest data structure in the runs described in this paper. For the results described in this paper, the software was run using the Matlab engine on a dual-cpu quad-core Xeon (E5430 at 2.66 GHz) with 16 GB memory. In order to fit a computation into this hardware–software system, using more images implies using fewer basis functions or visa versa. For the results described in this paper, all calculations used 1200 images and 720 basis functions (the so-called Step 7 of Yin et al. (2003)) and each reconstruction takes approximately 2 days. Please contact the corresponding author for a copy of the software.
3.2.6. Reconstruction: comments
In the work of Penczek et al. (2006), a space-varying variance map is constructed by a Monte-Carlo resampling procedure after the reconstruction is computed while in the approach proposed in this paper, the mean and covariance are simultaneously estimated. Potentially, though not demonstrated in the example of Section 4, the simultaneous estimation will allow for a better reconstruction since the reconstruction algorithm is allowed the additional degrees of freedom of assigning high variance to a part of the structure rather than allowing the somewhat disordered state of a part of the structure to contaminate better ordered parts of the structure. A second contrast is that the information estimated in this paper is sufficient to construct the complete second-order statistics of the reconstruction, i.e., a space-varying mean (the reconstruction) and a space-varying autocorrelation function. The autocorrelation function is the covariance between the electron scattering intensity at two different locations and therefore is a function of 6 independent variables (two 3-D spatial positions). It would be very challenging to estimate such a large amount of information by resampling. An advantage of resampling is that very little must be assumed about the probability density functions. However, the assumptions that are made in this paper have a long history (dating back to at least 1984 (Redner and Walker, 1984)) in the pattern recognition and machine learning communities as assumptions that are still useful even if there is no underlying physical model to motivate them.
The Gaussian assumption used in the homogeneous case (Doerschuk and Johnson, 2000; Yin et al., 2003; Scheres et al., 2007; Lee et al., 2007; Prust et al., 2009; Lee et al., 2011) (Eq. (9)) greatly simplifies the maximization step of the expectation–maximization algorithms but may not have a more fundamental motivation. Here, however, the joint Gaussian assumption on the pixel noise and weights (Eqs. (11) and (12)) is important because it allows the combination of these two sources of variability into a single equivalent source (Eq. (14)).
For the reconstruction of homogeneous particles (Eqs. (8) and (9)), a fast algorithm exists (Lee et al., 2007) that takes advantage of the fact that one of the Euler angles corresponds to a rotation of the image in the plane of the image. However, no corresponding algorithm appears to be possible for reconstruction of heterogeneous particles (Eqs. (13) and (14)).
3.3. Postprocessing
In order to interpret the results, estimates of the statistics of the weights in the orthonormal expansion are not as intuitive as estimates of the statistics of the electron scattering intensity function. Conditional on a particular class, the spatial mean function (which depends on position in 3-D space) and the spatial variance function (which depends on position in 3-D space) of the electron scattering intensity are
| (29) |
| (30) |
and
| (31) |
| (32) |
respectively. Let and be Eqs. (30) and (32) evaluated at the estimated values of and V rather than the true values. For biological purposes, the natural quantities to visualize are and , especially the standard deviation .
The unit of the electron scattering intensity in the reconstruction at 3 days is set by the scaling described in Section 3.1 Item 3. The standard deviation has the same unit. The reconstructions at different time points, denoted by , are scaled to the reconstruction at 3 days, denoted by , by the following algorithm. First, compute the optimal gain g* by where ∥f∥ = ∫∣f(x)∣dx. Second, the scaled reconstruction is .
Fig. 4 concerns variability versus time. Variability is described by averaged standard deviation and is computed as follows. Let νη’,δ(x) be the variance for the δth repetition of the calculation. Define
| (33) |
Then the plotted value is
| (34) |
and the sample standard deviation marks are at ±μη’ where
| (35) |
For the capsid calculation, the volume Υ is the annulus with inner radius 120 Å and outer radius 216 Å. For the four subunit calculations, the volume Υ is described implicitly by the following algorithm: (1) compute a cube of the space-varying variance map with sampling interval 2.768 Å. (2) Rotate the cube from the coordinate system of Zheng and Doerschuk (2000) to the coordinate system of VIPERdb. (3) In the refined crystal structure for NωV (1OHF) (Munshi et al., 1996; Helgstrand et al., 2004), locate all amino acids for which the α carbon is within 10 Å of the α carbon of the asparagine at the cleavage site (Asn570). (4) Locate a 3 × 3 × 3 cube of voxels around each voxel containing a α carbon in Step (3). This collection of voxels is the volume denoted by Υ.
Spherical averages of the variance map are used in Fig. 4(D–E). Each spherical average is computed using a formula analogous to Eqs. (22–25) of Yin et al. (2003), specifically,
| (36) |
| (37) |
where is the radial basis function (Yin et al., 2003), ∮ dΩ is integration over the sphere, and
| (38) |
where Il,n(·,·) is the (l,n) th icosahedral harmonic (Zheng and Doerschuk, 2000) and Yl,m(·,·) is the (l,m) th spherical harmonic. Applying Eq. (37) to the results of multiple calculations on different data sets indexed by δ ∈ {1, … ,Δ} gives . Then, . Finally, the sample mean and sample standard deviations of the spherical averages are computed by Eqs. (34) and (35).
4. Results
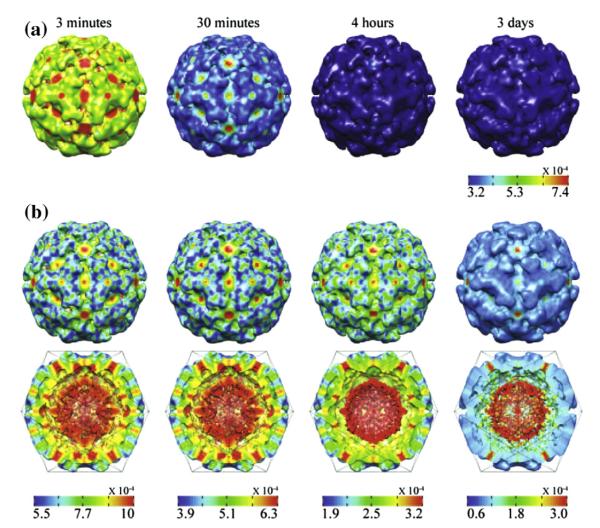
Fig. 2 shows the four time-resolved reconstructions as surface and cross section plots. These plots are colored by the square root of the variance map (i.e., the standard deviation map). The overall impression from the capsid surfaces shown in Fig. 2(A) is that the variability decreases in amplitude as time passes and the particle matures. The gradual stabilization of the capsid can be easily appreciated by comparing the variance at 3 min, 30 min and 4 h time points. However, if individual scales are used to plot the variance map, it became apparent that the stabilization process is still incomplete 4 h after the initiation of maturation (Fig. 2(B)). Because the variance is computed for each voxel of the reconstruction, we can analyze the stabilization process for the entire structure, as demonstrated by the cross-section view in Fig. 2(B). It can be seen that even 3 days after maturation the internal density continues to have high variance, which is expected if the RNA core of the particle is not highly ordered and does not obey icosahedral symmetry. However, the protein shell of the completely cleaved particle, i.e., the infectious particle, still retains a region of relative high variance in the center of the five fold axes.
Fig. 2.
The four time-resolved reconstructions. Panel A: surface of each of the four reconstructions colored by the square root of the variance map (i.e., the standard deviation map) and displayed using the VIPERdb convention. The same color map is used in all images. Panel B: the surface and a cross section perpendicular to a 2-fold axis of each of the four reconstructions colored by the standard deviation map. The surface and cross section visualizations at a particular time point share the same color map. Different color maps are used at different time points. Visualization by UCSF Chimera (Pettersen et al., 2004).
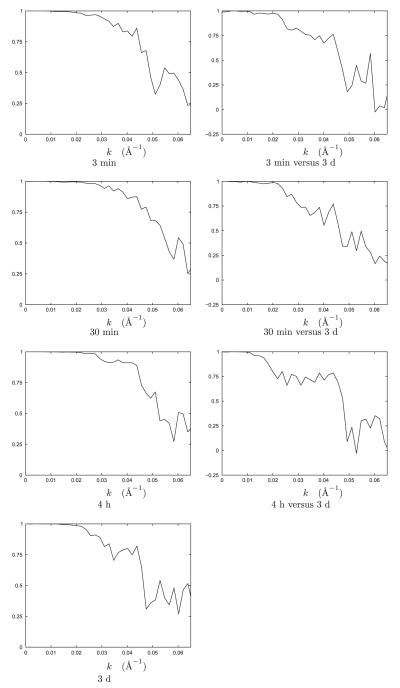
Fig. 3 provides information about resolution as Fourier Shell Correlation (FSC) plots for the reconstructions. In the first column of Fig. 3, the FSC plots show that the achieved resolutions for the reconstructions are 22, 21, 21, and 20 Å for 3 min, 30 min, 4 h, and 3 days, respectively. In the second column of Fig. 3, the FSC plots show that the reconstructions at times 3 min, 30 min, and 4 h agree with the reconstruction at time 3 days within resolutions of 27, 24, and 33 Å, respectively. In all cases, resolution is defined to be the inverse spatial frequency when the curve first interSections 0.5. Resolution is also described in Figs. 6 and 7 (Supplemental material). Fig. 7 (Supplemental material) shows the 3 min reconstruction at reduced resolution, specifically, using only 180 coefficients (the so-called Step 5 of Yin et al. (2003)) and Fig. 7 (Supplemental material) shows cross sections of the four reconstructions colored by the mean map or colored by the square root of the variance map (i.e., the standard deviation map).
Fig. 3.
Resolution of the four time-resolved reconstructions as a function of k, which is the magnitude of the reciprocal-space frequency vector measured in Å−1. First column: Fourier Shell Correlation (FSC) curves for comparing reconstructions from non-overlapping subsets containing 1200 images from the same data set. Based on these curves, the resolution of the four structures are approximately 21 Å. Second column: Fourier Shell Correlation (FSC) curves between the 3 days reconstruction and each of the 3 min, 30 min, and 4 h reconstructions for the nominal structures. Based on these difference curves, all the early structures agree with the capsid structure to approximately 24–33 Å. All FSC curves were computed using command procʒd in EMAN.
Next, we used the reconstructions generated in this work to analyze the variance around the cleavage site in each of the four quasi-equivalent subunits. Fig. 4(A) shows the position of subunits A, B, C, and D in the T = 4 surface lattice with the location of the autocatalytic site indicated by a red cross. The voxels covering the region occupied by amino acid residues within 10 Å of the active site were used to quantify the variance around the autocatalytic site of each subunit (Fig. 4(B)). This is the same region analyzed by Matsui et al. (2010) with difference maps. Fig. 4(C) shows that the variance in volumes encompassing the B and C active sites is clearly higher than the variance in A and D active sites at early time points and they all converge to the same variance at later time points when all the subunits have cleaved. We assume that positions of higher variance are still changing and that these cleavage sites have not yet formed. This is consistent with the position-specific active site formation observed by Matsui et al. (2010), validating the maximum-likelihood derived variance maps as a quantitative tool to address protein dynamics. An unexpected feature observed in this new analysis is that not only the active sites but also the average of the annulus containing the majority of the capsid protein densities reduce in variance by at least a factor of 3 as all of the subunits undergo cleavage. This important result could not be determined from the analysis of difference maps, but emerges naturally from the time resolved data sets when analyzed with the maximum likelihood algorithm that explicitly takes into account the continuous variability from one instance to another instance of the particle. Fig. 4(D–E) shows the radial dependence of the average of the variance map for each of the four time points, which decreases by a factor of 4 from time 3 min to time 3 days independent of the radial position from the center of the particle. All standard deviations plotted in Fig. 4 are computed by Eq. (35) with Δ = 4 repeats of the reconstruction based on nonover-lapping subsets of the image stack at a particular time point.
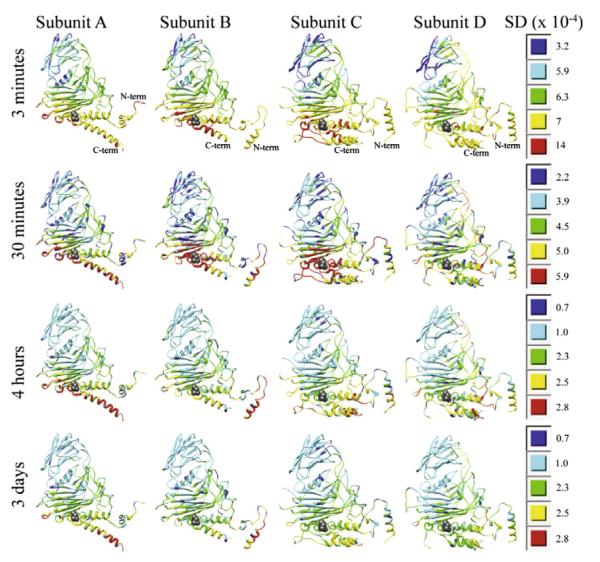
Fig. 5 shows ribbon diagrams of each of the subunits at each of the time points colored by the standard deviation map. The standard deviation tends to be largest in the helical region of the capsid protein near the autocatalytic site (Asn 570). These diagrams emulate diagrams used to display the Debye–Waller temperature factor in crystallography, where similar plots are made with the temperature factor displayed using color for each alpha carbon position of the peptide. The coordinates shown in the ribbon diagrams are from the refined X-ray crystallographic structure of NωV (1OHF) (Munshi et al., 1996; Helgstrand et al., 2004). The standard deviation of the pixel position closest to a given Cα coordinate was used to color code the ribbons.
Fig. 5.
Ribbon diagrams of the four subunits at the four times colored by the square root of the variance map (i.e., the standard deviation map) with the asparagine at the self-catalytic site (Asn 570) shown as a ball-and-stick model. Each time point has its own color map analogous to the second row of Fig. 2. For instance, red at the 3 min time point is 14 × 10−4/5.9 × 10−4 = 2.4 times higher than red at the 30 min time point.
5. Discussion
We showed that the maximum likelihood derived variance maps calculated for NωV in different stages of maturation successfully captured the same subunit-specific dynamics features previously observed with difference maps by Matsui et al. (2010). However, while the difference map analysis was technically limited to a small portion of the structure, this new approach allowed us to observe an overall reduction in structural variability as the particle matures. This data correlates with the increase in particle stabilization as a function of cleavage, as previously demonstrated biochemically (Taylor et al., 2002). Moreover, this new analysis afforded the identification of highly dynamic regions in the fully mature capsid that were not obvious in the crystal structure. At the 5-fold symmetry axes, the high variance region (Fig. 2) encompasses a central channel formed by the C-terminal gamma peptide of Subunit A and the N-terminal helix of Subunit B (Fig. 5). Recently, it was demonstrated that NωV membrane disruption activity is promoted by gamma peptides specifically derived from Subunit A cleavage (Domitrovic et al., 2012). In the crystal structure the fivefold central channel is protected from the solvent, however, the increased mobility observed in this analysis agrees with the high dynamics required to expose gamma peptide to the external environment, where it would be accessible to protease activity, as already demonstrated by Bothner et al. (2005), and could interact with cellular membranes. Therefore, the maximum likelihood derived variance maps can possibly provide information about putative biding sites and regulatory regions in cryo-EM structures. Another important advantage of the approach proposed here is that no difference maps are involved so the method would still be applicable if the overall structure underwent large changes.
The new method is based on simultaneously computing a nominal reconstruction and a map of the space-varying heterogeneity of a biological particle from single-particle cryo EM data. The method depends on describing the heterogeneity probabilistically and estimating the statistics of the heterogeneity from the image data. This is a generalization of previous work (Doerschuk and Johnson, 2000; Yin et al., 2003; Scheres et al., 2007; Lee et al., 2007; Prust et al., 2009; Lee et al., 2011) to the case where the particle is described probabilistically rather than deterministically. The method can be extended from maximum likelihood estimation to maximum a posteriori estimation (a form of Bayesian estimation (Scheres, 2012)) as is described for the homogeneous case in Section VII of Doerschuk and Johnson (2000). In this paper, resolution is measured by the standard FSC method of comparing two reconstructions computed from non-overlapping sets of images. This can be done rapidly using previously published formulas (Yin et al., 2003, Eqs. (22)–(25)). A more statistical approach that is natural for maximum likelihood estimators has been described (Prust et al., 2009, Section 4). In the approach proposed here for heterogeneous particles, both of these methods measure resolution in terms of the nominal structure not the variance map.
The resolutions of the maps presented here are moderate compared to the sub nanometer reconstructions in Matsui et al. (2010) due to the computationally intensive nature of the algorithm and current limited computing capability. In the NωV example of Section 4, the new method produced variance maps that agreed closely with the difference maps computed at the higher resolution emphasizing the power of the method and motivating the use of high performance computers that will allow calculations to be performed at the resolutions dictated by the data. Potentially, though not demonstrated in the example of this paper, the simultaneous estimation will lead to a better reconstruction since the reconstruction algorithm is allowed the additional degrees of freedom of assigning high variance to a part of the structure rather than allowing the somewhat disordered state of a segment of the structure to contaminate better ordered parts of the structure. The method presented in the present paper should have broad application to existing EM data sets that can be reanalyzed with explicit spatial variance maps that may well provide added value for relating these structures to the function of the macromolecules.
Supplementary Material
Acknowledgments
Q.W., Y.Z., and P.C.D. are grateful for support from the National Science Foundation under Grant Nos. 0836656 and 0735297. T.D. is supported by PEW Latin American Fellowship Program and by Conselho Nacional de Desenvolvimento Cientifico (CNPq) Brasil. J.E.J. and T.M. are supported by NIH Grant R01 GM054076. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2012.11.005.
References
- Baker Matthew L., Zhang Junjie, Ludtke Steven J., Chiu Wah. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat. Protoc. 2010;5(10):1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothner B, Taylor D, Jun B, Lee KK, Siuzdak G, et al. Maturation of a tetravirus capsid alters the dynamic properties and creates a metastable complex. Virology. 2005;334:17–27. doi: 10.1016/j.virol.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Doerschuk Peter C., Johnson John E. Ab initio reconstruction and experimental design for cryo electron microscopy. IEEE Trans. Info. Theory. 2000;46(5):1714–1729. [Google Scholar]
- Domitrovic Tatiana, Matsui Tsutomu, Johnson John E. Dissecting quasi-equivalence in non-enveloped viruses: membrane disruption is promoted by lytic peptides released from subunit pentamers, not hexamers. J. Virol. 2012 doi: 10.1128/JVI.01089-12. (VI Accepts, published online ahead of print on 3 July 2012). http://dx.doi.org/10.1128/JVI.01089-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EM Data Bank Available at: < http://www.emdatabank.org/>.
- Erickson Harold P. The Fourier transform of an electron micrograph—first order and second order theory of image formation. In: Barer R, Cosslett VE, editors. Advances in Optical and Electron Microscopy. vol. 5. Academic Press; London, New York: 1973. pp. 163–199. [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles – the uncommon line. J. Struct. Biol. 1996;116(1):48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Helgstrand Charlotte, Munshi Sanjeev, Johnson John E., Liljas Lars. The refined structure of Nudaurelia capensis ω Virus reveals control elements for a T = 4 capsid maturation. Virology. 2004;318:192–203. doi: 10.1016/j.virol.2003.08.045. [DOI] [PubMed] [Google Scholar]
- Jensen Grant J., editor. Methods in Enzymology. vol. 481. Elsevier Inc; 2010a. Cryo-EM, Part A: Sample Preparation and Data Collection. [DOI] [PubMed] [Google Scholar]
- Jensen Grant J., editor. Methods in Enzymology. vol. 482. Elsevier Inc; 2010b. Cryo-EM, Part B: 3-D Reconstruction. [DOI] [PubMed] [Google Scholar]
- Jensen Grant J., editor. Methods in Enzymology. vol. 483. Elsevier Inc; 2010c. Cryo-EM, Part C: Analyses, Interpretation, and Case Studies. [Google Scholar]
- Lee Junghoon, Doerschuk Peter C., Johnson John E. Exact reduced-complexity maximum likelihood reconstruction of multiple 3-D objects from unlabeled unoriented 2-D projections and electron microscopy of viruses. IEEE Trans. Image Proc. 2007;16(11):2865–2878. doi: 10.1109/tip.2007.908298. [DOI] [PubMed] [Google Scholar]
- Lee Seunghee, Doerschuk Peter C., Johnson John E. Multi-class maximum likelihood symmetry determination and motif reconstruction of 3-D helical objects from projection images for electron microscopy. IEEE Trans. Image Proc. 2011;20(7):1962–1976. doi: 10.1109/TIP.2011.2107329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann EL, Casella George. Theory of Point Estimation. 2 ed Springer-Verlag; New York: 1998. [Google Scholar]
- Lepault J, Pitt T. Projected structure of unstained, frozen-hydrated T-layer of Bacillus brevis. EMBO J. 1984;3(1):101–105. doi: 10.1002/j.1460-2075.1984.tb01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Hongrong, Jin Lei, Koh Sok Boon S., Atanasov Ivo, Schein Stan, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathworks Available at: < http://www.mathworks.com/>.
- Matsui Tsutomu, Lander Gabriel C., Khayat Reza, Johnson John E. Subunits fold at position-dependent rates during maturation of a eukaryotic RNA virus. Proc. Natl. Acad. Sci. USA. 2010;107(32):14111–14115. doi: 10.1073/pnas.1004221107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi S, Liljas L, Cavarelli J, Bomu W, McKinney B, et al. The 2.8 Å structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 1996;261(1):1–10. doi: 10.1006/jmbi.1996.0437. [DOI] [PubMed] [Google Scholar]
- Octave Available at: < http://www.gnu.org/software/octave/, http://octave.sourceforge.net/>.
- Penczek Pawel A., Yang Chao, Frank Joachim, Spahn Christian M.T. Estimation of variance in single-particle reconstruction using the bootstrap technique. J. Struct. Biol. 2006;154(2):168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Provencher Stephen W., Vogel Robert H. Three-dimensional reconstructions from electron micrographs of disordered specimens: I. Method. Ultramicroscopy. 1988a;25:209–222. doi: 10.1016/0304-3991(88)90016-2. [DOI] [PubMed] [Google Scholar]
- Prust Cory J., Doerschuk Peter C., Lander Gabriel C., Johnson John E. Ab initio maximum likelihood reconstruction from cryo electron microscopy images of an infectious virion of the tailed bacteriophage P22 and maximum likelihood versions of Fourier Shell Correlation appropriate for measuring resolution of spherical or cylindrical objects. J. Struct. Biol. 2009;167:185–199. doi: 10.1016/j.jsb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redner Richard A., Walker Homer F. Mixture densities, maximum likelihood and the EM algorithm. SIAM Rev. 1984;26(2):195–239. [Google Scholar]
- Scheres Sjors H.W. Classification of structural heterogeneity by maximum-likelihood methods. In: Jensen Grant J., editor. Cryo-EM, Part B: 3-D Reconstruction. vol. 482. Elsevier Inc.; 2010. pp. 295–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres Sjors H.W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres Sjors H.W., Gao Haixiao, Valle Mikel, Herman Gabor T., Eggermont Paul P.B., et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods. 2007;4(1):27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ. A maximum-likelihood approach to single-particle image refinement. J. Struct. Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- Sigworth Fred J., Doerschuk Peter C., Carazo Jose-Maria., Scheres Sjors H.W. An introduction to maximum-likelihood methods in cryo-EM. In: Jensen Grant J., editor. Cryo-EM, Part B: 3-D Reconstruction. vol. 482. Elsevier Inc.; 2010. pp. 263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Krishna NK, Canady MA, Schneemann A, Johnson JE. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J. Virol. 2002;76:9972–9980. doi: 10.1128/JVI.76.19.9972-9980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Chikashi., Unwin Nigel. Contrast transfer for frozen-hydrated specimens: determination from pairs of defocused images. Ultramicroscopy. 1988;25(4):279–291. doi: 10.1016/0304-3991(88)90003-4. [DOI] [PubMed] [Google Scholar]
- VIPERdb Available at: < http://viperdb.scripps.edu/>.
- Vogel Robert H., Provencher Stephen W. Three-dimensional reconstructions from electron micrographs of disordered specimens: II. Implementation and results. Ultramicroscopy. 1988b;25:223–240. doi: 10.1016/0304-3991(88)90017-4. [DOI] [PubMed] [Google Scholar]
- Vogel RH, Provencher SW, von Bonsdorff C-H, Adrian M, Dubochet J. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature. 1986;320:533–535. doi: 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- Yin Zhye, Zheng Yili, Doerschuk Peter C., Natarajan Padmaja, Johnson John E. A statistical approach to computer processing of cryo electron microscope images: virion classification and 3-D reconstruction. J. Struct. Biol. 2003;144(1/2):24–50. doi: 10.1016/j.jsb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Zhang Xing, Settembre Ethan, Wu Chen, Dormitzer Philip R., Bellamy Richard, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA. 2008;105(6):1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Yibin, Doerschuk Peter C. Explicit computation of orthonormal symmetrized harmonics with application to the identity representation of the icosahedral group. SIAM J. Math. Anal. 2000;32(3):538–554. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.