Abstract
We describe a new cell-penetrating protein, B1, capable of delivering conjugated proteins and nucleic acids into mammalian cells. B1 is a 244-amino-acid product of a single-base frameshift in the gene encoding enhanced green fluorescent protein (eGFP). The molecule has a net positive charge of 43 and a very high charge-to-mass ratio of 1.5. eGFP-fused B1 potently penetrates both adherent and suspension cells with >80% of cells taking up the protein when exposed to concentrations as low as 1 μM. The protein was found to cluster in the paranuclear region of TZM-bl cells. Most importantly, we show that B1 not only facilitates cellular uptake, but allows biomolecular cargo to reach sites of biological relevance. For example, baby hamster kidney cells underwent DNA recombination when exposed to B1-tagged Cre recombinase at protein concentrations as low as 2.5 μM, indicating potent nuclear delivery of functional protein cargos. Additionally, B1 delivers non-covalently conjugated RNA and DNA across the cell membrane to cytosolic and nuclear sites accessible to the cellular translation and transcription machinery, as gauged by detection of encoded reporter functions, with efficiency comparable to commercially available cationic lipid reagents. B1 appears to utilize cell-surface glycans and multiple competing endocytic pathways to enter and traffic through cells. These studies provide both a new tool for intracellular delivery of biomolecules and insights that could aid in the design of more effective cell penetrating proteins.
Keywords: Intracellular, biopharmaceuticals, biotherapeutics, biomolecules
Introduction
Intracellular targeting of large therapeutic biomolecules poses a significant challenge. Effective delivery entails not only crossing the outer cell membrane but transport and release of therapeutic cargo to cellular loci conducive to attainment of the therapeutic effect. Several lipid-, polymeric- and inorganic-based vehicles for intracellular delivery of proteins and nucleic acids exist, including cationic lipids (1–3), polyethylenimine (PEI) (4, 5), carbon nanotubes (6–8), gold nanoparticles (9–11), supercharged green fluorescent protein (GFP) (12, 13) and nanocapsules (14, 15). However, the field of intracellular delivery of large molecules is still in its infancy and major strides need to be made both in effectiveness of delivery and our understanding of the underlying mechanisms before this approach can be used in the clinic.
Some natural and artificial proteins and peptides have the ability to move freely across the cell membrane. The first cell penetrating polypeptide (CPP) identified was the HIV Tat protein, which is able to enter cells and translocate into the nucleus (16). Subsequent studies showed that an arginine–rich motif in Tat (GRKKRRQRRR) is responsible for cell-penetration (17). This Tat peptide has been fused to many target proteins to mediate cellular delivery (18–21). Since then, a number of natural proteins have been found to have the capacity to penetrate cells, including the Antennapedia protein (Antp) from Drosophila (22, 23), VP22 protein from herpes simplex virus (24) and CaP from Brome mosaic virus(25). Similarly, several artificial CPPs have been created for protein and nucleic delivery, including highly positively charged peptides and proteins (e.g. poly arginine (26), supercharged +36 GFP (12) and related proteins (27)) and amphipathic peptides (e.g. Pep–1 (28), CADY (29)). Proteins and nucleic acids that are covalently or non–covalently conjugated to these CPPs are able to enter cells (30, 31). The cellular uptake mechanism of these CPPs is not yet fully understood, but many are believed to be internalized through endocytic pathways (32, 33).
In this paper, we describe a novel CPP, B1, discovered during a cell-based screen for genetic suppressor elements of hepatitis C virus (HCV, to be published elsewhere). B1 is 244 amino acids in length and has a very high overall positive charge of +43. This study evaluates the effectiveness of B1 as a vehicle for intracellular delivery of two types of biomolecular payloads – genetically fused proteins and non-covalently conjugated nucleic acids. We demonstrate that B1 not only potently mediates cellular uptake of large molecules, but delivers the cargo to biologically relevant cellular milieu. To our knowledge, the efficiency of mRNA delivery mediated by B1 is the highest reported for CPPs to date and is comparable to that mediated by commercially available cationic lipids. The ability of B1 to effectively deliver functional cargo to the cytosol may at least facilitate its use in in vitro stem cell engineering for which it is desirable to transiently deliver transcription factor(s) to cells to achieve cell fate reprograming(34).
This study also provides insights into the mechanism of cell penetration by B1. We show that (1) cell-surface glycans and several cellular endocytic pathways play a role in cellular entry and cargo delivery by B1, and (2) the contribution of specific endocytic pathways to cellular entry versus functional cargo delivery by B1 can be significantly different. B1 penetrates cells through a mechanism distinct from that of another high-positive-charge protein +36GFP as gauged from intracellular distribution and time-/temperature-dependent cell penetration profiles. Thus, B1 represents a complementary addition to the current toolkit for intracellular delivery.
Finally, we note that our discovery of B1 was fortuitous and not by design. It is tempting to speculate that many of the molecules that already exist in nature may, without our knowledge, already possess cell penetrating characteristics. The features that confer a molecule with cell penetrating characteristics are still somewhat mysterious, but net positive charge appears to play an important role.
Results and Discussion
B1 transduces mammalian cells and mediates cellular uptake of conjugated proteins
During a cell–based selection for a genetic suppressor element of hepatitis C virus infection, B1 emerged as a dominant species after 5 rounds of selection and enrichment (results to be published elsewhere). Sequencing analysis showed that B1 is the product of a frameshift caused by an unintended single–base insertion preceding the eGFP gene. Frameshifts in coding sequences typically yield very short polypeptides due to the concomitant introduction of new stop codons, but B1 contains 244 amino acids, making it similar in size to the original eGFP (238 amino acids). A protein database search of B1 using NCBI BLAST returned no matches, indicating no known homologs of B1. The eGFP gene is codon–optimized for expression in mammalian cells (35), and shares 71% nucleotide homology with the wild–type green fluorescent protein (GFP) from the jellyfish Aequorea Victoria (36–38) (Figure S1). Introduction of the same frameshift mutation into the original GFP sequence yields a translated sequence of only one amino acid. B1 possesses an overall charge of +43 and a charge density (+charge:mass ratio) of 1.5 at pH 7. 6xHis-tagged B1 (6H-B1, Table 1) can be expressed as a soluble protein in E. coli BL21(DE3) cells and purified via one-step immobilized-metal affinity chromatography (IMAC), although a very high concentration of imidazole (0.5–1 M) is needed to elute resin-bound B1 (Figure S2). The yield of purified 6H-B1 was ~4 mg per liter of E. coli culture, with an estimated purity > 90%. To remove excess imidazole, 6H-B1 was dialyzed in a modified PBS containing an increased concentration of NaCl (2 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4). Dialysis of purified 6H-B1 in unmodified PBS (containing 137 mM NaCl) yielded significant amounts of white precipitate in the dialysis tubing. Dialyzed 6H-B1 can be stored at 4°C for up to 2 weeks without significant loss of protein activity, or at −80°C.
Table 1.
Constructs used in this study.
| Number | Short Name | Short Sequence | Molecular Weight (kDa) |
|---|---|---|---|
| 1 | 6H-B1 | HHHHHH-B1 | 30.3 |
| 2 | GFP-L-B1 | HHHHHH-GFP-(GGGGS)2-B1 | 60.2 |
| 3 | GFP-L-B1-F | HHHHHH-GFP-(GGGGS)2-B1(F) | 41.5 |
| 4 | GFP-L-B1-M | HHHHHH-GFP-(GGGGS)2-B1(M) | 41.2 |
| 5 | GFP-L-B1-R | HHHHHH-GFP-(GGGGS)2-B1(R) | 38.7 |
| 6 | mCherry-L-B1 | HHHHHH-mCherry-(GGS)9-B1 | 58.9 |
| 7 | mCherry-L-R10 | HHHHHH-mCherry-(GGS)9-(R)10 | 32.4 |
| 8 | mCherry-L-TAT | HHHHHH-mCherry-(GGS)9-GRKKRRQRRR | 32.2 |
| 9 | Cre-L-B1 | HHHHHH-Cre-(GGS)9-B1 | 72.2 |
| 10 | Cre-L-R10 | HHHHHH-Cre-(GGS)9- (R) 10 | 43.4 |
| 11 | +36GFP | HHHHHH-+36GFP | 28.5 |
| 12 | +36GFP-L-mCherry | +36GFP-(GGS)9-mCherry-HHHHHH | 56.9 |
| 13 | +36GFP-L-Cre | +36GFP-(GGS)9-Cre-HHHHHH | 70.2 |
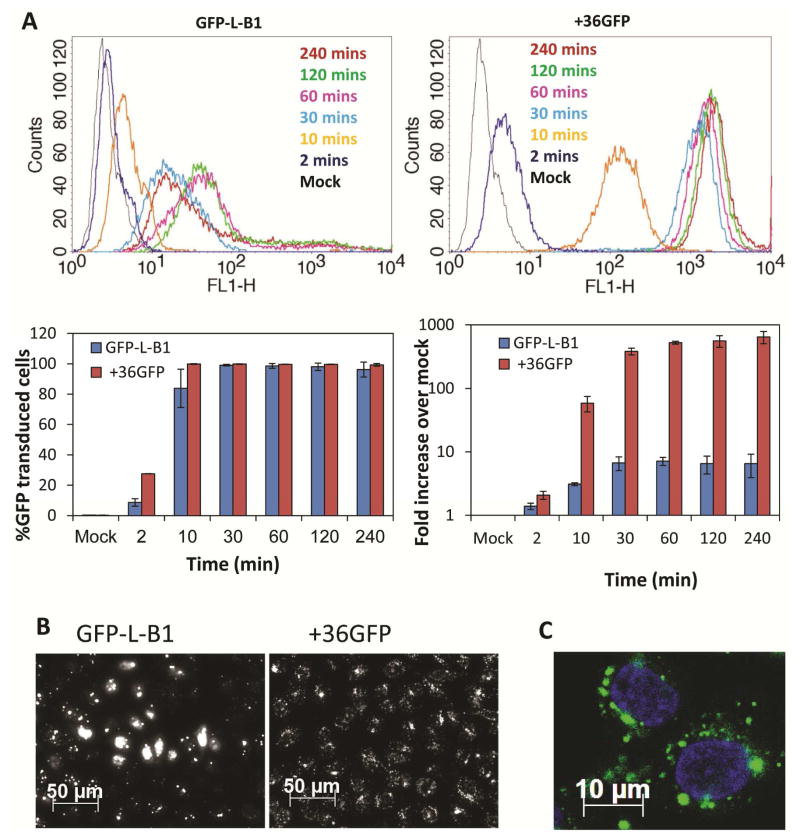
To enable detection of intracellular B1, we fused the globular protein eGFP to the N-terminus of B1 via a flexible linker to form GFP-L-B1 (Table 1). We purified GFP-L-B1 by one-step IMAC, akin to our purification of B1 (Figure S2). Purified GFP-L-B1 exhibited greatly improved stability compared to B1. The purified protein can be easily dialyzed with minimal precipitation and the dialyzed protein can be stored at 4°C for a few months without loss of activity. GFP-L-B1 is non-toxic to mammalian cells (Figure S3). TZM-bl cells were incubated with 2 μM GFP-L-B1 at 37°C and 5% CO2 for different amounts of time. After incubation, cells were treated with 0.04% Trypan Blue for two minutes to completely quench the signal from extracellular GFP (39), digested with 0.25% trypsin-EDTA for five minutes to completely remove the extracellular GFP and analyzed by flow cytometry (Figure 1A). GFP-L-B1 was found to rapidly and efficiently enter TZM-bl cells.
Figure 1. B1 penetrates cells.
(A) TZM-bl cells were incubated with GFP-L-B1 (2 μM) or +36GFP (2 μM) at 37°C/5% CO2 for up to 4 hours, washed with DPBS containing 0.04% Trypan Blue to quench extracellular GFP (39) and analyzed via flow cytometry. Error bars represent the standard deviation of two independent experiments. (B) Fluorescence microscopic images of TZM-bl cells treated with 2 μM of GFP-L-B1 or +36GFP at 37°C/5% CO2 for 1 hour. Cells were washed with PBS containing 0.04% Trypan Blue prior to imaging. (C) Confocal microscopic image of TZM cells treated with 2 μM GFP-L-B1 at 37°C and 5% CO2 for 1 hour. Cell nuclei were stained with DAPI (blue).
Two major differences were noticed between cells that had taken up GFP-L-B1 (+charge:MW ratio of 1.5) and those that had taken up the supercharged +36GFP (+charge:MW ratio of 1.3): (1) Fluorescence intensity. +36GFP-positive cells exhibited a much higher (up to 50-fold) fluorescence intensity on average than GFP-L-B1-positive cells (Figure 1A). (2) Intracellular distribution. Unlike +36GFP, which accumulated evenly in a large number of tiny, endosomal compartments, as gauged by the appearance of finely spotted cells, intracellular GFP-L-B1 was found to accumulate in much larger intracellular clusters in one subpopulation of cells while exhibiting an even diffuse distribution in another subpopulation (Figure 1B). Confocal microscopy confirmed that large GFP-L-B1 clusters formed in the paranuclear region of cells (Figure 1C).
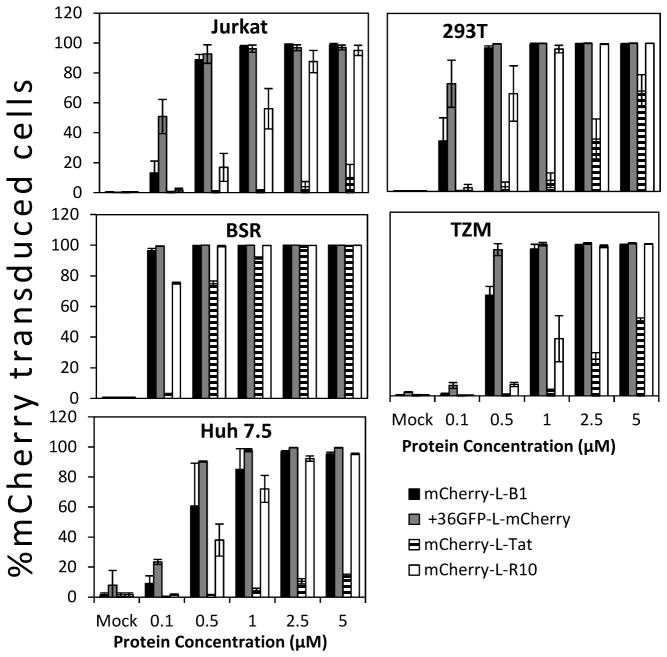
We also determined the ability of B1 to deliver covalently attached mCherry protein (32 kDa) (40) into different cell types and evaluated B1 against other known cell-penetrating proteins/peptides. mCherry was attached to the N-terminus of B1, supercharged +36GFP, Tat and R10 (deca-arginine) via a flexible linker (Table 1). As shown in Figure 2, 80–100% of all the target cells take up mCherry-L-B1 when exposed to a protein concentration as low as 1 μM. The potency of B1-mediated delivery of mCherry into cells was comparable to R10 and +36GFP and significantly higher than Tat.
Figure 2. B1-mediated mCherry delivery into selected cell lines.
Jurkat, 293T, BSR, TZM-bl, or Huh-7.5 cells were exposed to the indicated mCherry-protein fusions for 4 hours at 37°C/5% CO2, digested with trypsin to remove cell-surface-bound proteins and analyzed via flow cytometry. Error bars represent the standard deviation of two independent experiments.
Abbreviated forms of B1 yield significantly weakened cell transduction
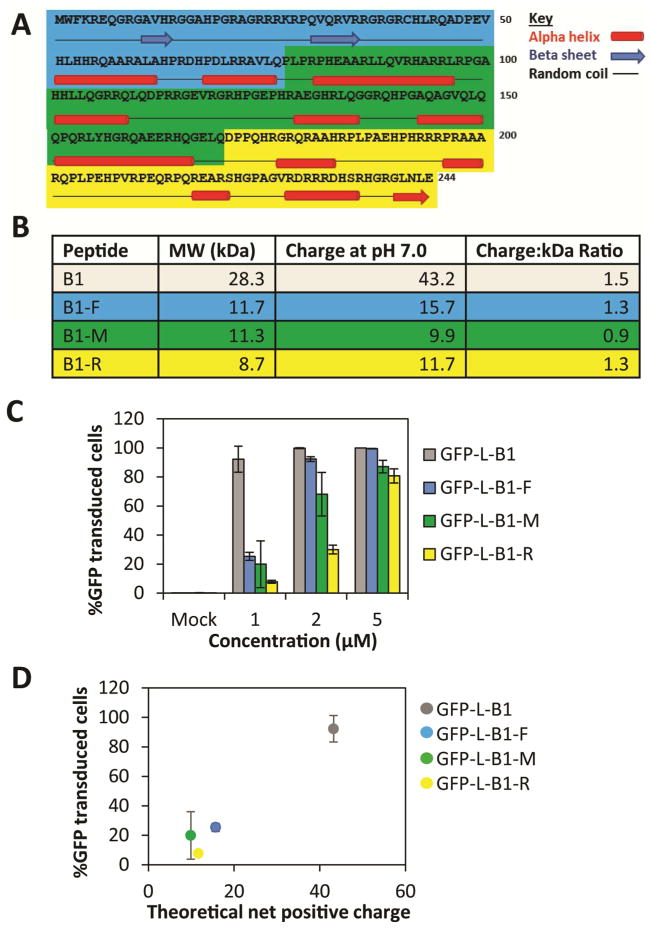
We sought to determine whether B1’s cell transduction functionality could be traced to a specific region of the protein. B1 was segmented into three non-overlapping fragments that avoid the disruption of predicted structural motifs, named B1-F, B1-M, and B1-R (Figure 3A). As shown in the table at the bottom of Figure 3A, each segment comprises approximately one third of the mass of B1 and possesses a charge:MW ratio similar to the parental B1 (Figure 3B). Each segment was fused to the C-terminus of eGFP via a flexible linker (Table 1) and the ability of TZM-bl cells to take up these constructs was measured. All three segments of B1 mediated cell transduction, with >80% of cells taking up the proteins at 5 μM exposure (Figure 3C). However, the cell transduction potency of these abbreviated forms of B1 is significantly lower than that of full-length B1. It is noteworthy that, compared to B1-R, B1-M possesses a lower charge:kDa ratio but exhibits a higher cell transduction potency (Figure 3C, D).
Figure 3. Abbreviated forms of B1 transduce cells, but not as potently as full-length B1.
(A) B1 protein sequence with secondary structure predicted by the GOR4 algorithm (57). B1-F (blue), B1-M (green), B1-R (yellow). (B) Table showing the molecular weight and the predicted charge of truncated B1. (C) TZM-bl cells were exposed to the indicated GFP-fusions for 4 hours at 37°C/5% CO2 prior to quenching with 0.04% Trypan Blue, trypsin digestion and flow cytometric analysis. (D) A plot of % GFP-transduced cells vs. theoretical net positive charge. Error bars represent the standard deviation of two independent experiments.
B1-delivered proteins are able to access the cytosol and nucleus
Many cellular functions require proteins to be able to access the cytosol or nucleus. The punctate subcellular distribution of GFP-L-B1 (Figure 1C) raised the concern that B1 may be trapped in subcellular compartments, without access to sites critical for cellular functions. To address this concern, we performed two major studies. The first study evaluated B1-mediated nuclear transport of protein cargo by measuring the ability of a B1-delivered recombinase to mediate DNA recombination. The second major study evaluated the ability of two types of nucleic acids – mRNA and DNA – to undergo processing by the cell’s transcription/translation machinery in the cytosol (mRNA) and nucleus (DNA).
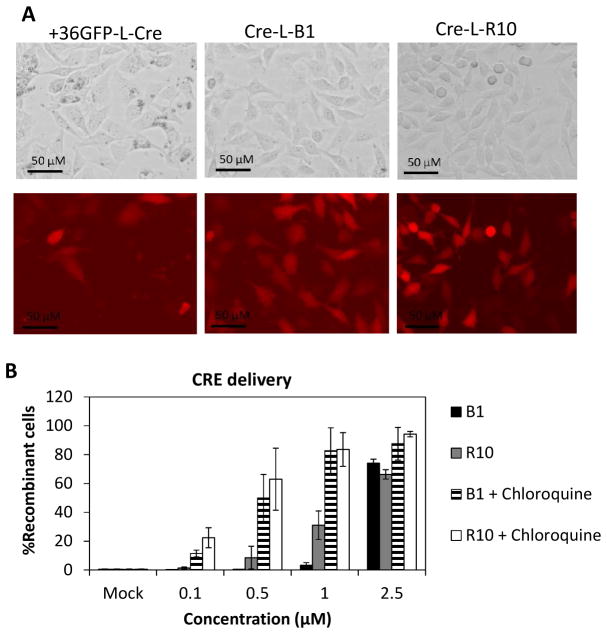
For the nuclear transport study, B1 was used to deliver covalently conjugated Cre recombinase into BSR.LNL.tdTomato cells (41). BSR.LNL.tdTomato cells contain two loxP recombination sites upstream of the reporter gene tdTomato, causing the gene to be silenced (40). Reporter gene expression is triggered by Cre-mediated DNA recombination (13). Cre fusions to B1 and R10 used in this study are depicted in Table 1. A comparable extent of transfection was observed with +36GFP-L-Cre, Cre-L-B1 and Cre-L-R10 based on fluorescence microscopic analysis (Figure 4A). We were unable to quantify the percentage of activated (tdTomato-expressing) cells in the +36GFP-L-Cre-exposed cell population via flow cytometry due to significant overlap in the fluorescence spectra of GFP and tdTomato (40, 42). Over 70% of cells were activated after exposure to 2.5 μM Cre-L-B1 or Cre-L-R10 (Figure 4B). As with +36GFP (13), chloroquine significantly enhanced Cre-induced recombination, particularly at low protein concentrations (Figure 4B). Chloroquine is a known inhibitor of lysosomal protein degradation (43) and likely extends the residence time of Cre fusion proteins in endosomes, allowing for increased endosomal escape.
Figure 4. B1-mediated nuclear delivery of Cre recombinase.
(A) BSR.LNL.td.Tomato cells were exposed to 2.5 μM of Cre-L-B1, Cre-L-R10 or +36GFP-L-Cre at 37°C/5% CO2 for 6 hours. The expression of tdTomato was visualized by fluorescence microscopy 24 hours later. (B) BSR.LNL.td.Tomato cells were exposed to Cre-L-B1 or Cre-L-R10 in the absence or presence of chloroquine (100 μM) at 37°C/5% CO2 for 6 hours. The percentage of cells expressing tdTomato was determined via flow cytometry 24 hours later. The error bars represent the standard deviation of two independent experiments.
The potency of B1-mediated delivery of Cre recombinase as gauged by nuclear recombination activity is significantly lower than that of B1–mediated delivery of mCherry as reported by cell fluorescence (compare chart for BSR cells in Figure 2 with Figure 4B). This result indicates that the vast majority of B1-conjugated proteins are not accessible to the cytosol and nucleus. These inaccessible proteins likely remain trapped in endosomes, consistent with the observation that GFP-L-B1 was found to accumulate in large intracellular patches (Figure 1B).
Presumably, any cargo protein can be covalently attached to B1 for cell delivery. Given that B1 is highly positively charged, a negatively charged cargo protein could potentially be non-covalently conjugated to B1 for cellular delivery. This strategy was successfully used by Waugh et al. to deliver negatively charged BOTOX into the skin of mice through conjugation with a TAT-polylysine fusion peptide (30). Alternatively, a disulfide bond could be inserted between the cargo protein and B1 through the introduction of a pair of cysteine substitutions in both proteins. Upon delivery to the reducing environment of the cytosol, the disulfide linkage is likely to break. Wiyagi et al. used this approach to deliver a δPKC inhibitor peptide into mice through using TAT as the CPP (44).
B1 can efficiently deliver nucleic acids
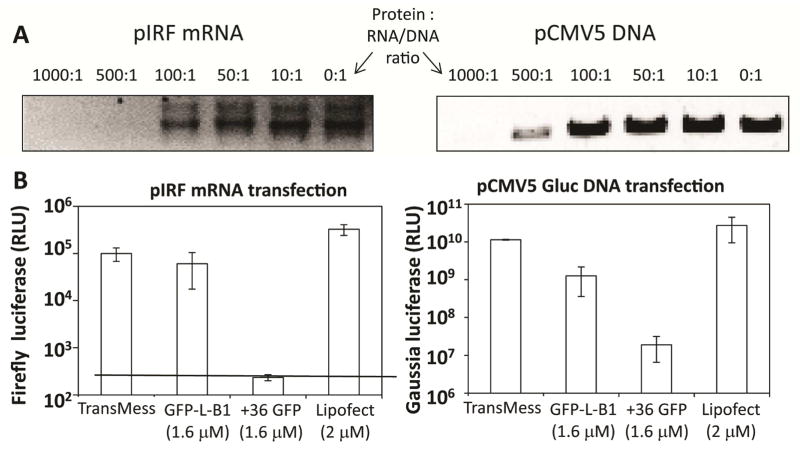
Our next study sought to determine the ability of B1 to transport conjugated nucleic acids to cytosolic and nuclear sites accessible to the cell’s transcription/translation machinery, as assessed by the measurement of encoded reporter functions. To do this, we used two reporter constructs as payloads – the mRNA pIRF (3 kb) and the plasmid DNA pCMV5-Gluc (5.2 kbp). pIRF is a bicistronic mRNA containing a firefly luciferase (Fluc) gene expressed via cap-dependent translation and a Renilla luciferase (Rluc) gene expressed via the incorporated internal ribosome entry site (IRES) of hepatitis C virus (45). Only the Fluc reporter was quantified in this study. pCMV5-Gluc encodes a Gaussia luciferase gene under the control of a CMV promoter (46). In this experiment and other studies described in this paper, GFP-L-B1 and not B1 was used as the CPP because of difficulties in obtaining large amounts of stable B1 protein.
As seen in Figure 5A, GFP-L-B1 is able to bind pIRF mRNA, as gauged by the decreasing intensity of the mRNA band at increasing protein:mRNA molar ratios. At B1:pIRF molar ratios at and above 500, the mRNA was not able to migrate into the gel at all, indicating supershift of the mRNA. Similarly, B1 forms complexes with linearized plasmid DNA, although at a higher B1:nucleic acids ratio (Figure 5A). The higher molar ratio of B1 needed for DNA gel-shift likely derives from the larger molecular weight of the DNA. Each pCMV5-Gluc DNA molecule has approximately 3 times the number of nucleotides as pIRF mRNA. It is also possible that B1 interacts more strongly with single-stranded RNA than double-stranded DNA.
Figure 5. B1-mediated cytosolic delivery of RNA and nuclear delivery of DNA.
(A) Gel-shift assays of GFP-L-B1-conjugated DNA and RNA. pIRF RNA (43 femtomoles) or linearized pCMV5 DNA (14.5 femto moles) were incubated with B1 protein at the indicated molar ratios in EMSA buffer for 10 minutes at room temperature analyzed on an agarose gel. (B) 293T cells were transfected with 1.6 nM of pIRF mRNA or linearized pCMV5-Gluc plasmid DNA using GFP-L-B1 (1.6 μM), +36GFP (1.6 μM) or the indicated commercial reagents according to the manufacturer’s directions. The dotted line indicates the limit of detection of the Fluc activity assay. TransMess: TransMessenger. Lipofec: Lipofectamine 2000. Error bars represent the standard deviation from two independent experiments.
To determine the ability of B1 to transport nucleic acids to cytosolic sites accessible to the cell’s transcription/translation machinery, 1000:1 mixtures of either GFP-L-B1:pIRF or GFP-L-B1:pCMV5-Gluc were incubated with 293T cells for 6 hours. Our rationale for using a higher ratio of protein to nucleic acids than the minimum amount required for gel-shift was that each RNA/DNA molecule needs to form a complex with multiple B1 proteins in order to be functionally delivered into cells (47). The size of the protein:DNA/RNA complexes is presumably 100–1000-fold larger than GFP-L-B1. The activities of Fluc and Gluc deriving from intracellularly processed pIRF RNA (cytosol) and pCMV5-Gluc DNA (nucleus) were quantified several hours following exposure to cell transduction constructs. As a control, the same amounts of mRNA or DNA were transfected into 293T cells using commercially available cationic lipid reagents according to the manufacturers’ protocols. As shown in Figure 5B, the efficiency of B1-mediated mRNA transfection is comparable to that obtained with the commercially available reagents TransMessenger and Lipofectamine 2000. Although +36GFP has been reported to support RNA interference by efficient delivery of siRNA (48), we found that exposure of 293T cells to +36GFP-conjugated pIRF reporter RNA did not yield a detectable Fluc reporter signal despite multiple attempts using different batches of purified proteins. The reason for this apparent discrepancy is unclear, but may derive from the much larger size of pIRF RNA (3 kb) relative to siRNA (22 bp). Another possible explanation is that pIRF RNA may interact with +36GFP in a way that inhibits its translation. DNA transfection mediated by GFP-L-B1 is 10–20-fold weaker than that mediated by TransIT and Lipofectamine 2000, and ~100-fold stronger than that mediated by unmodified +36GFP (Figure 5C). It has previously been reported that the ability of free +36GFP to transport DNA into the nucleus is poor (48). However, +36GFP attached to a fusion peptide derived from hemagglutinin (12)can transfect DNA with similar efficiency as Lipofectamine (48). It is possible that an even higher DNA transfection efficiency can be achieved when B1 is fused to the same fusion peptide.
B1 enters/traffics through cells via multiple endocytic pathways
We attempted to elucidate the mechanism through which B1 enters cells. We first quantified cell penetration by GFP-L-B1 at different temperatures. As seen in Figure 6A, although cell penetration by GFP-L-B1 is significantly weaker at lower temperatures all cells are able to take up GFP-L-B1 even at 4°C. This result indicates that B1 can penetrate cells in a temperature-independent manner. To evaluate the permeability of B1, we treated TZM-bl cells with the membrane-impermeable dye propidium iodide in the presence of GFP–L–B1 (Figure S4)(49). Intracellular propidium iodide was not detected in the presence of B1, indicating that B1 does not permeabilize cell membranes.
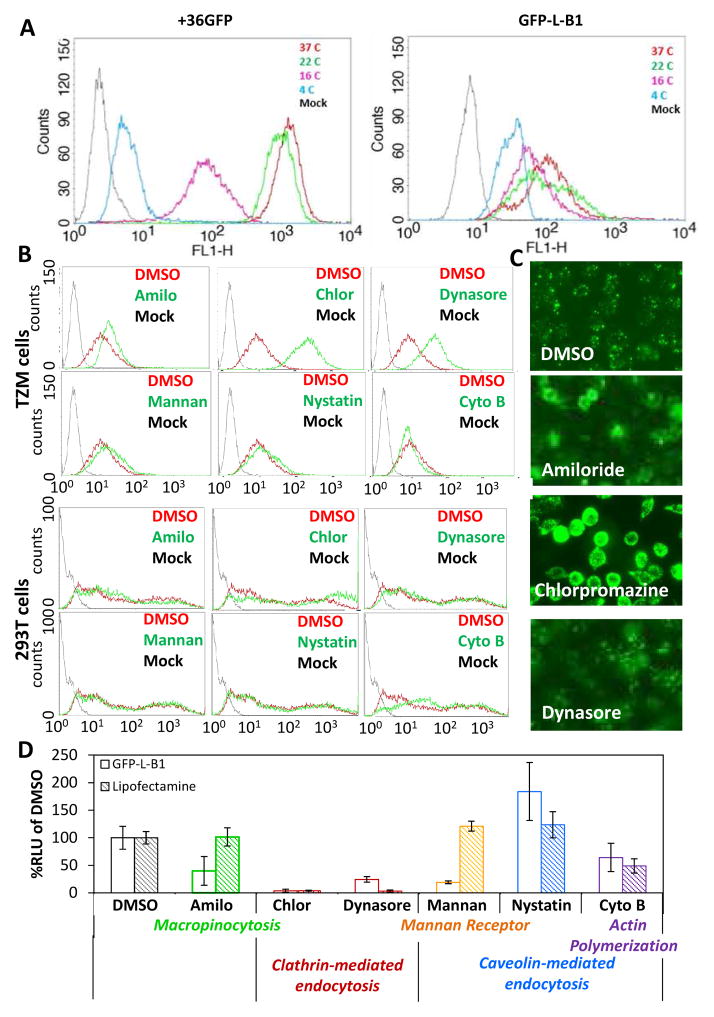
Figure 6. B1 enters/traffics through cells using multiple endocytic pathways, and the ability of these pathways to mediate cellular uptake does not correlate with the ability to support payload delivery to sites where bioactivity can be realized.
(A) Effect of temperature on cellular internalization of B1. TZM-bl cells were incubated with 2 μM GFP-L-B1 or 2 μM +36GFP at 4, 16, 22 or 37°C for one hour, washed with DPBS containing 0.04% Trypan Blue to quench extracellular GFP and analyzed by flow cytometry. (B) Role of different endocytic pathways in cellular uptake of B1. TZM-bl or 293T cells were pretreated with the endocytic inhibitors amiloride (5 mM) (50), chlorpromazine (55 μM) (52), dynasore (50 μM) (51), mannan (100 μg/ml) (53), nystatin (50 nM) (58) or cytochalasin B (4 μM) for one hour prior to the addition of GFP-L-B1 (2 μM). Part (D) indicates the endocytic pathways inhibited by these small molecules. One hour later, these cells were washed with DPBS containing 0.04% Trypan Blue and analyzed by flow cytometry. (C) Fluorescence microscopic images of TZM-bl cells transfected with GFP-L-B1 in the presence of the indicated inhibitors. Data is representative of 4 independent experiments. (D) Role of different endocytic pathways in supporting B1-mediated functional delivery of a RNA payload. 293T cells were pretreated with the indicated inhibitors for 1 hour prior to pIRF mRNA transfection using GFP-L-B1 or Lipofectamine 2000. The activity of Fluc deriving from translation of the delivered RNA was measured 6 hours post transfection. Error bars represent the standard deviation of two independent experiments.
To determine the role of different cellular endocytic pathways in cell transduction by B1, we evaluated cellular uptake of GFP-L-B1 in the presence of selected inhibitors of macropinocytosis, clathrin-mediated endocytosis, mannan receptor-dependent phagocytosis, and caveolin-mediated endocytosis. No significant reduction in GFP-L-B1 uptake was observed in the presence of any of the inhibitors in either 293T or TZM-b1 cell line (Figure 6B), suggesting that any one of these pathways alone is not critical for GFP-L-B1 uptake by cells. In fact, GFP-L-B1 uptake by TZM-bl cells appeared to be somewhat enhanced by the macropinocytosis inhibitor amiloride (50) and clathrin-mediated endocytosis inhibitor dynasore (51), and dramatically enhanced by chlorpromazine, another inhibitor of clathrin-mediated endocytosis (52) (Figure 6B, C). This result indicates that macropinocytosis and clathrin–mediated endocytosis may be “weak links” for cell transduction by B1 that obstruct B1–mediated protein entry through other, more energetically favorable pathways.
The monitoring of green fluorescent cells following exposure of cells to GFP-L-B1 provides a measure of cellular uptake of protein from the surroundings but does not provide information on its intracellular fate. We therefore sought to investigate the ability of cellular endocytic pathways to support the B1-mediated transport of an mRNA payload to sites accessible to the cell’s translation machinery. 293T cells were exposed to pIRF-mRNA-conjugated GFP-L-B1 in the presence of endocytic inhibitors. In contrast to the data shown in Figure 6B, RNA transfection efficiency was significantly reduced by inhibitors of clathrin-mediated endocytosis (chlorpromazine, dynasore) and mannan receptor-dependent phagocytosis (100 μg/ml mannan (53)) (Figure 6D). In contrast, nystatin, an inhibitor of caveolin–mediated endocytosis, was found to enhance the delivery of mRNA pointing to a possible inhibitory role of caveolin–mediated endocytosis in mRNA cargo delivery.
The discrepancy in the roles of endocytic pathways in cellular uptake versus successful processing of an mRNA cargo underscores the lack of correlation between B1’s cell-transduction ability and its access to cellular milieu conducive to cargo bioactivity. Clathrin-mediated endocytosis is an example of a pathway that is dispensable for cellular uptake of B1 (Figure 6B) but critical for providing cytosolic access to B1–conjugated cargo (Figure 6D). In this example, it is possible that blockage of clathrin–mediated endocytosis forces B1 to enter cells through other endocytic pathways whose endosomal compartments are less permissive to cytosolic access (33).
An endocytic pathway for B1’s cell-transduction is presumed, since fluorescence imaging indicates that the GFP–fused protein appears to be agglomerated in punctate regions presumed to be endosomes (Figure 1B, C). Any form of direct membrane translocation is not expected, since B1 is composed of mainly hydrophilic residues, and since the cell membrane remains intact during transduction (Figure S4). Such pathways would presumably be responsible for the GFP transduction observed at 4 °C, and would as such possess a relatively limited energy dependence. Since the known mechanisms of cellular transport require considerable amounts of energy and cellular resources, such a pathway must therefore use a currently undescribed mechanism. It is likely that other cell transducing proteins also make use of this mechanism. Elucidating the details of this mechanism, however, will prove a challenge. This is because B1, and other cell transducing proteins, are capable of delivering their cargo via multiple mechanisms(54, 55).
Finally, we explored the role of negatively charged cell–surface glycans in cellular uptake/processing of B1. 293T cells and TZM-bl cells were treated for 24 hours with sodium chlorate (80 mM), an inhibitor of glycan synthesis (56), prior to exposure to GFP-L-B1. B1-mediated cell uptake and RNA transfection were both reduced by sodium chlorate treatment (Figure 7), indicating that the ability of GFP-L-B1 to bind and delivery functional cargo into cells is affected by the cell-surface negative charge. Previously, Cronican et al. observed that highly positively charged proteins can efficiently transduce mammalian cells and predicted that proteins with a +charge:MW ratio greater than 0.75 represent candidate CPPs (27, 48). B1 possesses a +charge:MW ratio of 1.5, further supporting a role of ionic interactions in at least partially mediating its cell transduction functionality.
Figure 7. B1-mediated cell entry/trafficking utilizes cell-surface glycans.
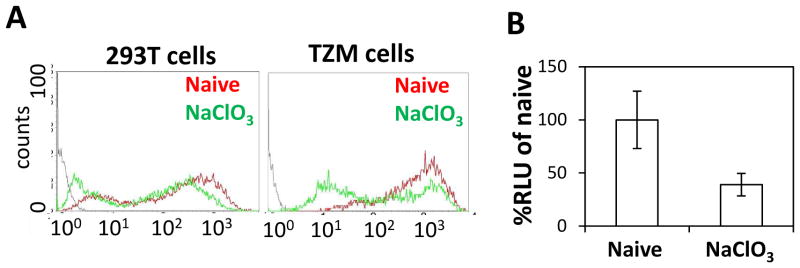
(A) 293T or TZM-bl cells were incubated with 80 mM of the glycan synthesis inhibitor sodium chlorate (NaClO3) for 24 hours at 37°C/5% CO2. GFP-L-B1 (2 μM) was then added to the wells. 1 hour later, the cells were washed with DPBS containing 0.04% Trypan Blue and analyzed via flow cytometry. Representative histograms from two independent experiments are shown. (B) 293T cells were exposed to 80 mM NaClO3 for 24 hours and then incubated with a mixture containing 5 ng (0.1 nM) pIRF mRNA and 1.6 μM GFP-L-B1 for 6 hours in the presence of NaClO3. The cells were lysed 6 hours later and the intracellular Fluc signal was measured. The error bars represent the standard deviation of two independent experiments.
In conclusion we identified a new cell-penetrating protein, B1, which is able to potently deliver conjugated biomolecular cargo to several mammalian cell lines. Most importantly, we show that B1 not only mediates cellular uptake, but effectively transports cargo to intracellular sites of biological relevance. B1 appears to utilize cell-surface glycans for cellular binding and transports its cargo via multiple competing cellular endocytic pathways. Interestingly, we found that the contribution of specific endocytic pathways to cellular entry versus functional cargo delivery by B1 can be significantly different. B1 provides a new tool for intracellular delivery of biomolecules. Additionally, insights garnered from these studies could aid in the design of more effective vehicles for the cellular delivery of therapeutic payloads.
Methods
For complete materials and methods including chemicals, cell lines, protein purification and gel shift assays, please see Supporting Information.
Cellular uptake determination of fluorescent protein-fused CPPs
On the day prior to CPP exposure, the following cells were seeded in 24-well plates at the indicated densities: 293T cells (2 × 105 cells/well), Huh-7.5 and TZM cells (1.9 × 105 cells/well), BSR cells (1 × 105 cells/well). These seeding densities were chosen such that the cells reach ~70% confluency the following day. On the day of CPP exposure, Jurkat cells were transferred to V-bottom 96-well plates at 1 × 105 cells/well. All cells were washed once with OptiMEM to remove residual serum. Protein solutions were diluted in cold OptiMEM to the desired concentrations and equilibrated to room temperature before being added to cells. Cells were incubated at 37°C/5% CO2 unless otherwise indicated. After incubation, these cells were washed once with cold DPBS.
For uptake determination of GFP-L-B1 or +36 GFP, cells were incubated with 0.04% Trypan Blue (in DPBS) for 2 minutes at room temperature to quench the fluorescence of extracellular GFP (39), washed again with cold DPBS and then imaged using a Zeiss Axiovert 200M fluorescent microscope (Carl Zeiss Microscopy, NY). For confocal microscopy, TZM cells (seeded on a glass chamber slide) that had been exposed to 2 μM GFP-L-B1 were incubated with 300 nM DAPI solution (Invitrogen, NY) for 5 minutes at room temperature and imaged using a Zeiss 510 Meta NLO Mulitphoton microscope (Carl Zeiss Microscopy). For flow cytometric analysis, cells that had been exposed to CPPs were incubated with 0.25% trypsin-EDTA at room temperature for 5 minutes and resuspended in complete growth medium (DMEM supplemented with 10% FBS and 1X NEAA) and analyzed on a BD FACScan flow cytometer (BD Biosciences) gated for GFP fluorescence (excitation/emission 488/530). At least 1 × 104 cells were analyzed for each sample.
For cellular uptake analysis of CPPs containing mCherry-fusion proteins, CPP-exposed cells were detached by incubation with 0.25% trypsin-EDTA at room temperature for five minutes, resuspended in complete growth medium and then analyzed by flow cytometry (excitation/emission 488/650). At least 1 × 104 cells were analyzed for each sample.
Cre recombinase delivery
BSR.LNL.tdTomato reporter cells were seeded in 24-well plates at 5 × 104 cells per well. The following day, the cells were washed once with OptiMEM to remove residual serum, exposed to Cre-fused CPPs in the presence or absence of 100 μM chloroquine and incubated at 37°C/5% CO2 for 6 hours. Following CPP exposure, cells were washed once with OptiMEM to remove excess proteins, the supernatants were replaced with fresh, complete growth medium, and the cells were incubated at 37°C/5% CO2 for another 24 hours followed by fluorescence microscopic analysis (Zeiss Axiovert 200M) to quantify Cre delivery. After imaging, cells were trypsinized, resuspended in complete growth medium and then analyzed by flow cytometry (excitation/emission 488/650). Cells were gated for at least 1 × 104 live cells in each sample.
Quantification of luciferase activity deriving from DNA/RNA delivery
293T cells were seeded in 96-well plates at 4 × 104 cells per well. The following day, 10Xconcentrated protein samples were prepared in OptiMEM. Linearized pCMV5-Gluc DNA (46 ng/μL) and pIRF mRNA (10 ng/μL) were prepared in OptiMEM. 10 μL of 10X protein samples were mixed with 10 μl of the nucleic acids and the mixtures were incubated at room temperature for 10 minutes to allow protein/nucleic acids association. In the meantime, the cells were washed twice with OptiMEM to remove residual serum and incubated in 40 μL OptiMEM. 10 μL of the protein/nucleic acids mixtures were then added to cells followed by incubation at 37°C/5% CO2 for 6 hours. The amount of RNA and DNA in individual wells was 50 ng (50.5 fmol) and 230 ng (67 fmol), respectively. For control experiments utilizing commercial reagents for nucleic acids delivery, linearized pCMV5-Gluc (230 ng in 10 μL) was mixed with 0.8 μL Lipofectamine 2000 or TransIT reagent and pIRF mRNA (50 ng in 10 μL) was mixed with 0.4 μL Lipofectamine 2000 or TransMessenger reagent. The mixtures were incubated at room temperature for 20 minutes, diluted in 40 μL OptiMEM and added to 293T cells. 6 hours later, the cells were washed and the supernatants were replaced with the complete growth medium. DNA transfection was quantified by measuring the Gaussia luciferase (Gluc) activity in supernatants 24 hours post medium change using the Biolux Gaussia Luciferase Assay Kit (New England Biolabs). RNA transfection was quantified by measuring the intracellular firefly luciferase (Fluc) levels measured 6 hours post medium change using the Luciferase Assay System (Promega).
Supplementary Material
Acknowledgments
We thank D. Liu (Harvard University) for graciously providing the BSR.LNL.tdTomato cell line and the plasmids expressing +36GFP, +36GFP-L-mCherry and +36GFP-L-Cre; C. Rice (Rockefeller University) for providing the Huh-7.5 cell line; and P. Lindahl (Texas A&M University) for providing the Jurkat cell line. We also thank K. Chockalingam for critical reading and editing the manuscript, and Y. Shi for assisting with protein purification. This work was supported by the Artie McFerrin Department of Chemical Engineering, Texas A&M University, and by the National Institutes of Health (R21A1083965-01).
Abbreviations
- CPP
cell-penetrating polypeptide
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proceedings of the National Academy of Sciences. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose JK, Buonocore L, Whitt MA. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520. [PubMed] [Google Scholar]
- 3.Vigneron J-P, Oudrhiri N, Fauquet M, Vergely L, Bradley J-C, Basseville M, Lehn P, Lehn J-M. Guanidinium-cholesterol cationic lipids: efficient vectors for the transfection of eukaryotic cells. Proceedings of the National Academy of Sciences. 1996;93:9682–9686. doi: 10.1073/pnas.93.18.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E, Schluep T, Wen S, Engler H, Heidel J. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjugate chemistry. 2004;15:831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kam NWS, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. Journal of the American Chemical Society. 2005;127:12492–12493. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 7.Pantarotto D, Singh R, McCarthy D, Erhardt M, Briand JP, Prato M, Kostarelos K, Bianco A. Functionalized carbon nanotubes for plasmid DNA gene delivery. Angewandte Chemie. 2004;116:5354–5358. doi: 10.1002/anie.200460437. [DOI] [PubMed] [Google Scholar]
- 8.Bianco A, Kostarelos K, Partidos CD, Prato M. Biomedical applications of functionalised carbon nanotubes. Chem Commun. 2005:571–577. doi: 10.1039/b410943k. [DOI] [PubMed] [Google Scholar]
- 9.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 10.Sandhu KK, McIntosh CM, Simard JM, Smith SW, Rotello VM. Gold nanoparticle-mediated transfection of mammalian cells. Bioconjugate chemistry. 2002;13:3–6. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]
- 11.Han G, Ghosh P, De M, Rotello VM. Drug and gene delivery using gold nanoparticles. Nanobiotechnology. 2007;3:40–45. [Google Scholar]
- 12.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proceedings of the National Academy of Sciences. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS chemical biology. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Biswas A, Zhao M, Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 15.Biswas A, Joo KI, Liu J, Zhao M, Fan G, Wang P, Gu Z, Tang Y. Endoprotease-mediated intracellular protein delivery using nanocapsules. ACS nano. 2011;5:1385–1394. doi: 10.1021/nn1031005. [DOI] [PubMed] [Google Scholar]
- 16.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 17.Vives E. Present and future of cell-penetrating peptide mediated delivery systems: “Is the Trojan horse too wild to go only to Troy? J Control Release. 2005;109:77–85. doi: 10.1016/j.jconrel.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tatmediated delivery of heterologous proteins into cells. Proceedings of the National Academy of Sciences. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker-Hapak M, McAllister SS, Dowdy SF. TAT-Mediated Protein Transduction into Mammalian Cells. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- 20.Xia H, Mao Q, Davidson BL. The HIV Tat protein transduction domain improves the biodistribution of [beta]-glucuronidase expressed from recombinant viral vectors. Nat Biotech. 2001;19:640–644. doi: 10.1038/90242. [DOI] [PubMed] [Google Scholar]
- 21.Wadia JS, Dowdy SF. Modulation of cellular function by TAT mediated transduction of full length proteins. Current Protein & Peptide Science. 2003;4:97–104. doi: 10.2174/1389203033487289. [DOI] [PubMed] [Google Scholar]
- 22.Thorén PEG, Persson D, Karlsson M, Nordén B. The Antennapedia peptide penetratin translocates across lipid bilayers – the first direct observation. FEBS Letters. 2000;482:265–268. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 23.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc Natl Acad Sci U S A. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kueltzo LA, Normand N, O’Hare P, Middaugh CR. Conformational lability of herpesvirus protein VP22. J Biol Chem. 2000;275:33213–33221. doi: 10.1074/jbc.M002476200. [DOI] [PubMed] [Google Scholar]
- 25.Qi X, Droste T, Kao CC. Cell-Penetrating Peptides Derived from Viral Capsid Proteins. Molecular Plant-Microbe Interactions. 2010;24:25–36. doi: 10.1094/MPMI-07-10-0147. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell DJ, Steinman L, Kim DT, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. The Journal of Peptide Research. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 27.Cronican James J, Beier Kevin T, Davis Tina N, Tseng J-C, Li W, Thompson David B, Shih Allen F, May Erin M, Cepko Constance L, Kung Andrew L, Zhou Q, Liu David R. A Class of Human Proteins that Deliver Functional Proteins into Mammalian Cells In Vitro and In Vivo. Chem Biol. 2011;18:833–838. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 29.Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R, Heitz F, Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waugh J, Lee J, Dake M, Browne D. Nonclinical and Clinical Experiences with CPP-Based Self-Assembling Peptide Systems in Topical Drug Development. In: Langel Ü, editor. Cell-Penetrating Peptides. Humana Press; 2011. pp. 553–572. [DOI] [PubMed] [Google Scholar]
- 31.Lopes LB, Furnish EJ, Komalavilas P, Flynn CR, Ashby P, Hansen A, Ly DP, Yang GP, Longaker MT, Panitch A, Brophy CM. Cell permeant peptide analogues of the small heat shock protein, HSP20, reduce TGF-beta1-induced CTGF expression in keloid fibroblasts. J Invest Dermatol. 2009;129:590–598. doi: 10.1038/jid.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson David B, Villaseñor R, Dorr Brent M, Zerial M, Liu David R. Cellular Uptake Mechanisms and Endosomal Trafficking of Supercharged Proteins. Chemistry & Biology. 2012;19:831–843. doi: 10.1016/j.chembiol.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mäger I, Langel K, Lehto T, Eiríksdótir E, Langel Ü. The role of endocytosis on the uptake kinetics of luciferin-conjugated cell-penetrating peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818:502–511. doi: 10.1016/j.bbamem.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell stem cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang TT, Cheng L, Kain SR. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 37.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 38.Yang T-T, Cheng L, Kain SR. Optimized Codon Usage and Chromophore Mutations Provide Enhanced Sensitivity with the Green Fluorescent Protein. Nucleic Acids Research. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL. CHIP Suppresses Polyglutamine Aggregation and Toxicity In Vitro and In Vivo. The Journal of Neuroscience. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 41.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson G, Day RN, Piston D. Fluorescent protein spectra. Journal of cell science. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 43.Lammers G, Jamieson JC. Studies on the effect of lysosomotropic agents on the release of Gal beta 1-4GlcNAc alpha-2,6-sialytransferase from rat liver slices during the acutephase response. Biochem J. 1989;261:389–393. doi: 10.1042/bj2610389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyaji Y, Walter S, Chen L, Kurihara A, Ishizuka T, Saito M, Kawai K, Okazaki O. Distribution of KAI-9803, a Novel δ-Protein Kinase C Inhibitor, after Intravenous Administration to Rats. Drug Metabolism and Disposition. 2011;39:1946–1953. doi: 10.1124/dmd.111.040725. [DOI] [PubMed] [Google Scholar]
- 45.Laporte J, Malet I, Andrieu T, Thibault V, Toulme J-J, Wychowski C, Pawlotsky J-M, Huraux J-M, Agut H, Cahour A. Comparative Analysis of Translation Efficiencies of Hepatitis C Virus 5′ Untranslated Regions among Intraindividual Quasispecies Present in Chronic Infection: Opposite Behaviors Depending on Cell Type. Journal of virology. 2000;74:10827–10833. doi: 10.1128/jvi.74.22.10827-10833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Katzenellenbogen BS, Katzenellenbogen JA, Zhao H. Directed evolution of human estrogen receptor variants with significantly enhanced androgen specificity and affinity. J Biol Chem. 2004;279:33855–33864. doi: 10.1074/jbc.M402118200. [DOI] [PubMed] [Google Scholar]
- 47.Gao S, Simon MJ, Morrison B, 3rd, Banta S. Bifunctional chimeric fusion proteins engineered for DNA delivery: optimization of the protein to DNA ratio. Biochim Biophys Acta. 2009;1790:198–207. doi: 10.1016/j.bbagen.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. P Natl Acad Sci USA. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macklis JD, Madison RD. Progressive incorporation of propidium iodide in cultured mouse neurons correlates with declining electrophysiological status: a fluorescence scale of membrane integrity. Journal of neuroscience methods. 1990;31:43–46. doi: 10.1016/0165-0270(90)90007-3. [DOI] [PubMed] [Google Scholar]
- 50.Dangoria NS, Breau WC, Anderson HA, Cishek DM, Norkin LC. Extracellular simian virus 40 induces an ERK/MAP kinase-independent signalling pathway that activates primary response genes and promotes virus entry. Journal of General Virology. 1996;77:2173–2182. doi: 10.1099/0022-1317-77-9-2173. [DOI] [PubMed] [Google Scholar]
- 51.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Developmental cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Subtil A, Hèmar A, Dautry-Varsat A. Rapid endocytosis of interleukin 2 receptors when clathrin-coated pit endocytosis is inhibited. Journal of cell science. 1994;107:3461–3468. doi: 10.1242/jcs.107.12.3461. [DOI] [PubMed] [Google Scholar]
- 53.Shibata Y, Metzger WJ, Myrvik QN. Chitin particle-induced cell-mediated immunity is inhibited by soluble mannan: mannose receptor-mediated phagocytosis initiates IL-12 production. The Journal of Immunology. 1997;159:2462–2467. [PubMed] [Google Scholar]
- 54.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. British Journal of Pharmacology. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Advanced Drug Delivery Reviews. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J Biol Chem. 1999;274:36267–36273. doi: 10.1074/jbc.274.51.36267. [DOI] [PubMed] [Google Scholar]
- 57.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 58.Rothberg KG, Ying YS, Kamen BA, Anderson R. Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate. The Journal of Cell Biology. 1990;111:2931–2938. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.