Abstract
LHX3 is a LIM-homeodomain transcription factor with critical roles in pituitary and nervous system development. Mutations in the LHX3 gene are associated with pediatric diseases featuring severe hormone deficiencies, hearing loss, developmental delay, and other symptoms. The mechanisms that govern LHX3/Lhx3 transcription are poorly understood. In this study, we examined the role of DNA methylation in the expression status of the mouse Lhx3 gene. Pituitary cells that do not normally express Lhx3 (Pit-1/0 cells) were treated with 5-aza-2’-deoxycytidine, a demethylating reagent. This treatment lead to activation of Lhx3 gene expression suggesting that methylation contributes to Lhx3 regulation. Treatment of Pit-1/0 pituitary cells with a combination of a demethylating reagent and a histone deacetylase inhibitor led to rapid activation of Lhx3 expression, suggesting possible crosstalk between DNA methylation and histone modification processes. To assess DNA methylation levels, treated and untreated Pit-1/0 genomic DNA was subjected to bisulfite conversion and sequencing. Treated Pit-1/0 cells had decreased methylation at specific sites in the Lhx3 locus compared to untreated cells. Chromatin immunoprecipitation assays demonstrated interactions between the MeCp2 methyl binding protein and Lhx3 promoter regions in the Pit-1/0 cell line. Overall, this study demonstrates that DNA methylation patterns of the Lhx3 gene are associated with its expression status.
Keywords: transcription, promoter, chromatin, LHX3
1. Introduction
The LIM-homeodomain 3 (LHX3) transcription factor plays key gene regulatory roles during pituitary gland organogenesis and nervous system development (Kelberman et al., 2009; Davis et al., 2010; Prince et al., 2011; Mollard et al., 2012). Homozygous Lhx3 knockout mice are not viable, do not develop the anterior and intermediate lobes of the pituitary, fail to differentiate four of the five anterior pituitary hormone-secreting cell types, and have impaired spinal neuron specification (Sheng et al., 1996; Sharma et al., 1998). In the anterior pituitary, LHX3 activates the transcription of hormone subunit and transcription factor genes, including the alpha glycoprotein subunit (αGSU), follicle-stimulating hormone (FSHβ), prolactin (PRL), gonadotropin-releasing hormone receptor (GnRH-R), and Pit-1 genes (reviewed in Colvin et al., 2009; Prince et al., 2011).
Recessive mutations in the human LHX3 gene are associated with complex syndromes featuring combined pituitary hormone deficiency disease (CPHD) and other symptoms (reviewed by Pfaffle and Klammt, 2011). Patients with recessive LHX3 mutations have deficiencies in growth hormone, PRL, thyroid-stimulating hormone, FSH, and luteinizing hormone, with resulting short stature, metabolic defects and delayed/failed puberty (Netchine et al., 2000; Bhangoo et al., 2006; Pfaeffle et al., 2007; Rajab et al., 2008; Kristrom et al., 2009; Bonfig et al., 2011; Colvin et al., 2011; Bechtold-Dalla Pozza et al., 2012). Other LHX3 mutation-associated symptoms may include abnormal pituitary gland morphology, rigid cervical spine, hearing loss, and adrenocorticotropic hormone deficiency.
Characterized mammalian Lhx3 genes, comprising seven exons and six introns, produce two major mRNA transcripts: Lhx3a and Lhx3b (Bach et al., 1995; Zhadanov et al., 1995; Sloop et al., 2000; Sloop et al., 2001). The Lhx3 gene contains two TATA-less, GC-rich Lhx3a and Lhx3b promoters containing conserved nuclear factor 1 (NF1) and specificity protein 1 (SP1) transcription factor binding sites (Yaden et al., 2006). The SOX2 developmental transcription factor has been shown to bind and activate the human LHX3a promoter (Rajab et al., 2008). The human LHX3 promoters are alone insufficient to guide gene expression in vivo indicating that the actions of additional elements must be required for tissue-specific transcriptional control of LHX3 protein expression (Mullen et al., 2012). A complex enhancer unit located downstream of the gene has been shown to contain modular elements conferring some aspects of pituitary and nervous system gene expression (Mullen et al., 2012) but the mechanisms by which LHX3 expression is precisely controlled over time and in specific cell types, including the roles of the two promoters, remain incompletely characterized.
DNA methylation is a significant mediator of transcriptional regulation in the cell. DNA methyltransferases (DNMTs) catalyze the addition of a methyl group to the 5′ position of the cytosine pyrimidine ring of CpG (cytosine-phosphate bond-guanine) dinucleotides (Ramsahoye et al., 1996; Robertson and Jones, 2000). DNA methylation has been implicated in chromatin silencing, X chromosome inactivation, the imprinting of parental alleles, viral gene inactivation, and silencing of specific genes during cellular determination and differentiation (reviewed in Ramsahoye et al., 1996; Dulac, 2010). Methyl-CpG binding proteins (MBPs) bind methylated DNA and recruit regulatory proteins to modulate chromatin structure and gene activity (Buck-Koehntop and Defossez, 2013). The interactions between DNA methyl-binding proteins and histone modifying proteins are likely essential for coordinated transcriptional regulation in cells during development (Vaissiere et al., 2008). For example, the methyl-binding protein, MeCP2 can interact with histone methyltransferases to mediate repression (Lunyak et al., 2002; Fuks et al., 2003). Aberrant DNA methylation, as well as alterations to methylation machinery, is associated with many diseases (reviewed in Robertson, 2005) implicating DNA methylation as a fundamental process in regulating gene expression.
Although large-scale “methylome” data indicates that high levels of cytosine methylation occur at CpG-poor DNA and that CpG islands remain mostly unmethylated, recent studies have shown methylation of CpG islands in the promoters of important developmental genes such as Hox genes (Auclair and Weber, 2012). Like the Hox genes, Lhx3 is a developmental gene encoding a homeodomain transcription factor, but the role of DNA methylation in expression of the Lhx3 gene has yet to be characterized. To investigate the effects of methylation on Lhx3 gene transcription, we examined the methylation status of Lhx3 promoters in the mouse pituitary Pit-1/0 cell line. We describe the patterns of DNA methylation of the Lhx3 promoters, examine the effects of methylation and histone deacetylase (HDAC) inhibitors on Lhx3 gene expression and identify an interaction between the Lhx3 promoters and MeCP2 in the mouse pituitary Pit-1/0 cell line. Understanding the mechanisms involved in Lhx3 gene regulation will be important to the improved diagnosis and treatment of LHX3 gene–associated diseases.
2. Materials and Methods
2.1. Cell Culture
Mouse Pit-1/0 and Pit-1/Triple cells (Sizova et al., 2010) were a generous gift from Dr. R. Day (Indiana University; with permission from Dr. Stephen Liebhaber, University of Pennsylvania) and were maintained in Hyclone Dulbecco’s Modified Eagle’s Media/Ham’s Nutrient Mixture F-12 (1:1) with 2.5 mM L-glutamine (Hyclone, Logan, UT) supplemented with 10% heat inactivated fetal bovine serum (Biowest, Nuaillé, France) and antibiotic-antimycotic (Invitrogen, Gran Island, NY). The cells were grown at 37°C in humidified 5% CO2 atmosphere. Cells (1 × 106 cells/10 cm) were plated and 24 hours later treated with 2.5 µM 5’-aza-2’-deoxycytidine (5-aza-dc; Sigma-Aldrich, St. Louis, MO) or 2.0 µM 5-aza-dc plus 100 nM Trichostatin A (TSA; Sigma-Aldrich) in media containing 10% FBS. Cells treated with 2.0 µM 5aza-dc plus 100 nM TSA were harvested 24 hours after treatment, while cells treated with only 2 µM 5-aza-dc were harvested 48 hours after treatment. Control plates were treated with DMSO as vehicle.
2.2 Bioinformatic Analysis
EMBOSS CpGPlot (http://www.ebi.ac.uk/Tools/emboss/cpgplot/) was used to predict potential CpG islands in the mouse and human Lhx3/LHX3 genes. Gene sequences including 3.5 kb upstream and 8 kb downstream of both mouse and human Lhx3 genes (UCSC Genome Bioinformatics; http://genome.ucsc.edu/) were used as inputs in CpG Plot. The minimum length of the reported CG-rich region was 200 bp. The observed to expected ratio of C plus G to CpG in a set of 10 windows required before a CpG island is reported was 0.6.
2.3. Bisulfite Genomic Sequencing
Genomic DNA was isolated from treated and untreated Pit-1/0 cells and Pit-1/Triple cells using a PureLink™ Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The bisulfite conversion was carried out with 500 ng of input genomic DNA with the use of EZ DNA Methylation-Gold Kit™ (Zymo Research Corp. Orange, CA) following the manufacturer’s protocol. The bisulfite converted DNA was amplified by the PCR using the Fast Start High Fidelity PCR System (Roche, Mannheim, Germany) with specific primers to the Lhx3a and Lhx3b promoters. Primers designed using the MethPrimer software (http:/ww.urogene.org/methprimer/) were as follows: mouse Lhx3a promoter forward, 5’-ggaaggttagagaaaaggttattt-3; Lhx3a promoter reverse, 5’-aaaaaacaaaaactccaaaaacac-3’; Lhx3b promoter forward, 5’-gtatttgtaggaagttttgagtggtatt-3’; Lhx3b promoter reverse, 5’-ccctaaacttcccctaaatctaac-3’. A ‘hot’ start PCR was performed by initially heating the reaction to 95°C for 2 min. prior to the addition of enzyme. The reaction then underwent 40 cycles of 95°C for 2 min, 59°C for 30 sec, 72°C for 1 min., and a final extension time of 7 min. The PCR products were gel purified using Qiaquick™ gel extraction kit (Qiagen, Valencia, CA), then cloned into pCR-4 TOPO vector using a TOPO™ TA Cloning kit (Invitrogen, Grand Island, NY). Plasmid DNA from at least 14 unique colonies per group was prepared using PureLink™ Quick Plasmid Miniprep (Invitrogen) and sequenced (ACGT Inc., Wheeling, IL) using an M13 Forward (−20) universal primer.
2.4. RNA isolation and Reverse Transcription
Pit-1/0 cells (1×106 cells/10 cm plate) and Pit-1/Triple cells (1×106cells/10cm plate) were plated and total RNA was harvested 24 hours later using an RNAeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized from 1 µg of total RNA using a High Capacity Reverse Transcription kit (Applied Biosystems, Grand Island, NY) according to the manufacturer’s protocol. The cDNA was used as a template for PCR reactions performed with GoTaq Green Master Mix (Promega, Madison, WI). Primers used were as follows: mLhx3a forward, 5'-aaccactggattagtgactg-3'; mLhx3b forward, 5'-gaagttcagggtcggaggg-3'; mLhx3a/b reverse, 5'-tggtcacagcctgcacacat-3' β-actin forward, 5'-ctacaatgagctgcgtgtgg-3'; β-actin reverse, 5'-tagctcttctccagggagga-3'. The PCR parameters were as follows: 95°C for 2 min, 40 cycles of 95°C 30 sec, 58°C 1 min, 72°C 1 min, and a final extension time of 72°C for 7 min. The PCR products were separated using 10% acrylamide/TBE electrophoresis gels (Biorad, Hercules, CA).
2.5. Protein Isolation and Western Blotting
Whole cell protein extracts of Pit-1/0 cells (1×107 cells/15 cm plate) and Pit-1/Triple cells (1×107 cells/15 cm) were isolated for western blotting. Cells were harvested in RIPA lysis buffer (150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 0.01 sodium phosphate, pH 7.2, 50 mM NaFl) plus one Complete Mini protease inhibitor cocktail tablet (Roche). BCA™ Protein Assay kit (Thermo Scientific, Rockford, IL) was used to determine protein concentration, according to the manufacture’s protocol. The harvested protein (25 µg) was separated on a 12% acrylamide gel and immunoblotted to check expression of LHX3 and MeCP2. Protein was transferred to PVDF membranes and incubated with anti-LHX3 (Chemicon, Temecula, CA; 1:2,500) or anti-MeCP2 primary antibodies (Abcam, Cambridge, MA; 2 µg/µl) in TBST (154 mM NaCl, 52 mM Tris, 1% Tween 20) overnight at 4°C. The membrane was washed and then incubated with the appropriate peroxidase-conjugated secondary antibody (1:25,000) in TBST for 1 hour. The membrane was incubated in SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, Rockford, IL) and then documented using a G-Box Imaging System (Syngene, Frederick, MD).
2.6. Chromatin Immunoprecipitation Assays
Pit-1/0 chromatin was isolated for chromatin immunoprecipitation (ChIP) analyses. Approximately 1–2 × 107 Pit-1/0 cells were grown in 15 cm culture dishes and treated with 2.5 µM 5-aza-dc, 100 nM TSA, 2.0 µM 5-aza-dc plus 100 nM TSA or DMSO vehicle. Twenty-four hours after treatment, chromatin was isolated from the 100 nM TSA cells and 2.0 µM 5-aza-dc plus 100 nM TSA treated cells. Cells from the 2.5 µM 5-aza-dc only treatment were harvested 48 hours post treatment. The ChIP assay was carried out with EZ ChIP Kit (Millipore, Billerica, MA) following the manufacturer’s protocol. DNA shearing efficiency was determined via electrophoresis. After the crosslinks were reversed lysates were sheared in a water bath sonicator (Diagenode, Denville, NJ) with 30 × 30 seconds sonication, 30 seconds off. The sheared samples were separated on 1.5% agarose gel to check for DNA fragmentation below 1 kb. Pit-1/0 chromatin was mixed with cold dilution buffer and protease inhibitor cocktail. The samples were pre-cleared for 1 hour with Salmon Sperm-Blocked Protein A agarose beads and centrifuged at 10K for 30 seconds. An input control sample (1%) was saved for downstream PCR. The supernatants were incubated with 5 µg of antibody overnight with gentle shaking. The following antibodies were used: anti-acetyl Histone 3 (Millipore), MeCP2 (Millipore), RNA Polymerase II (Millipore) as a positive control, and rabbit IgG as a negative control. Then, protein A agarose beads were added and incubated at 4°C for 2–4 hours. According to the manufacturer’s instructions, the beads were washed with 1 mL of each solution in the order listed for 5 minutes at 4°C: low salt buffer, high salt buffer, LiCl buffer and TE.
The relative abundance of regions of interest in precipitated DNA was measured by quantitative (q) PCR (ABI 7900 PRISM, Applied Biosystems, Foster City, CA) using SYBR green (Roche, Indianapolis, IN). The isolated ChIP input DNA was diluted 1:10 and primers to the Lhx3 promoters were as follows: Lhx3a forward, 5'-tgaaagaggtccagcacttcc-3'; Lhx3a reverse, 5'-tggcaatcgagttctgcttc-3'; Lhx3b forward, 5'-tctgtaggaagccttgagtgg-3’; Lhx3b reverse, 5'-cgtctggcttgcacttcc-3'. The PCR parameters were as follows: 95°C 15 min. for one cycle, 95°C 15 sec, 60°C 60 sec for 40 cycles and 95°C 15 sec, 60°C 1 min., 95°C 15 sec for one cycle. Fold enrichment was calculated using the following formula: 2^[Ct (IgG)−Ct(experimental)]. The Student’s t-test was used to determine differences between groups. Values were considered significantly different when P < 0.05.
3. Results
3.1. Analysis of Lhx3 mRNA and protein expression
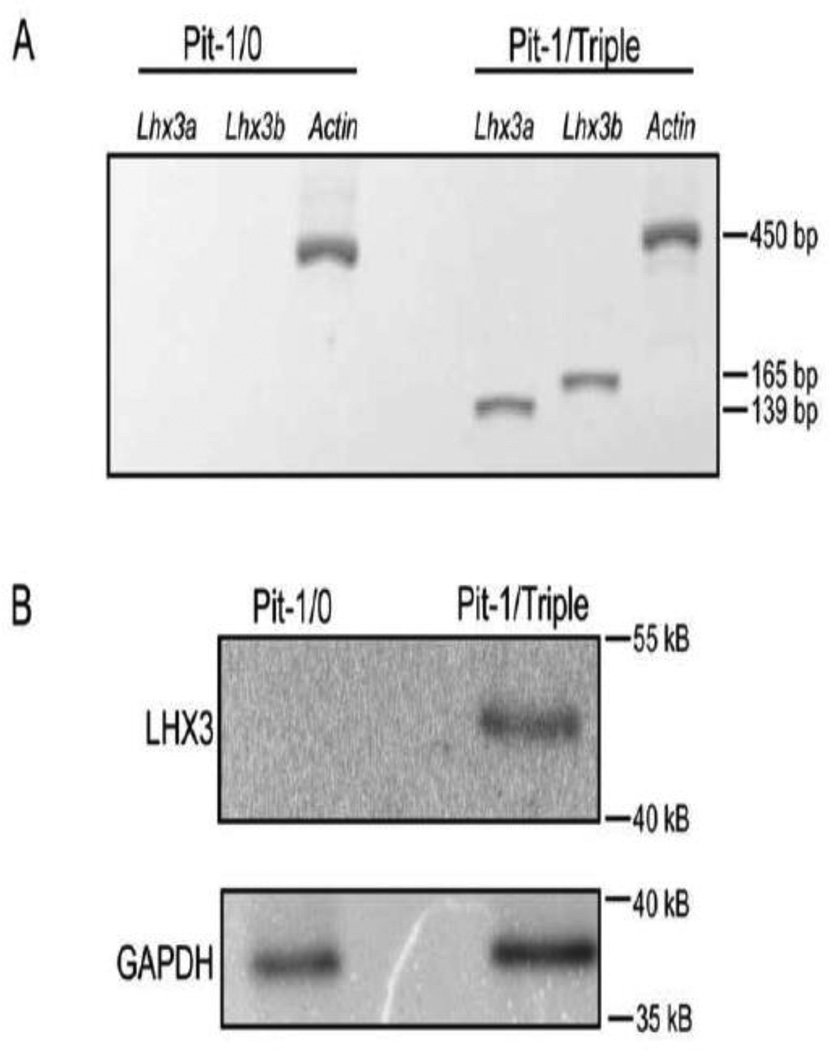
Recently, mouse cell lines representing different stages of anterior pituitary cell development have been generated using pituitary promoters to target immortalizing genes in transgenic animals (Sizova et al., 2010). Of these, the Pit-1/0 and Pit-1/Triple lines represent earlier and later stages, respectively. Unfortunately, similar reagents modeling human pituitary cell types are not available. To examine whether the Lhx3a and Lhx3b mRNAs are expressed in Pit-1/0 and Pit-1/Triple cells, total RNA was obtained and Lhx3a, Lhx3b and actin (positive control) transcripts were detected by RT-PCR. The Pit-1/Triple cells expressed both Lhx3a and Lhx3b, while neither transcript was expressed in the Pit-1/0 cells (Figure 1A). Correspondingly, LHX3 protein was detected in the Pit-1/Triple cells, but was not observed in the Pit-1/0 cells using western assays of whole cell protein extracts (Figure 1B).
Figure 1. Lhx3 mRNA and protein are expressed in mouse pituitary Pit-1/Triple but not Pit-1/0 cells.
(A) Lhx3a or Lhx3b mRNA was amplified by RT-PCR in Pit-1/0 and Pit-0/Triple cells. Beta actin primers were used in a positive control to test cDNA integrity. (B) Protein levels of LHX3 were determined by western blotting of whole cell protein extracts of Pit-1/0 and Pit-1/Triple cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies were used as positive controls. Each experiment was performed three times with similar results.
3.2. Identification and investigation of methylated regions in Lhx3 promoters
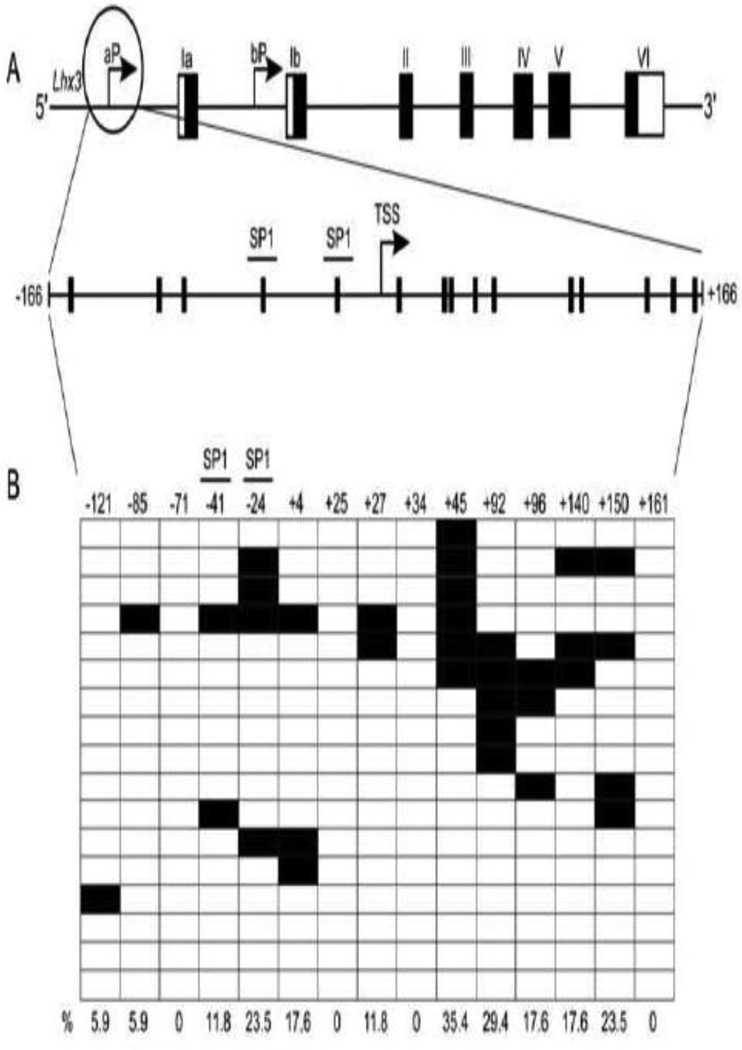
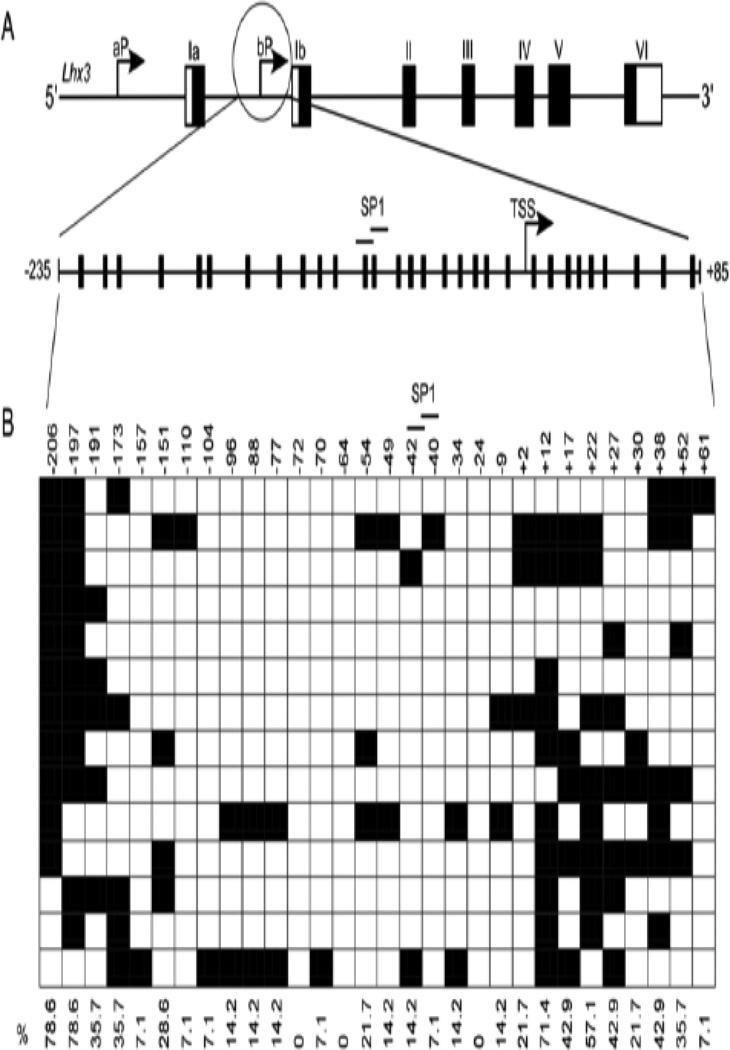
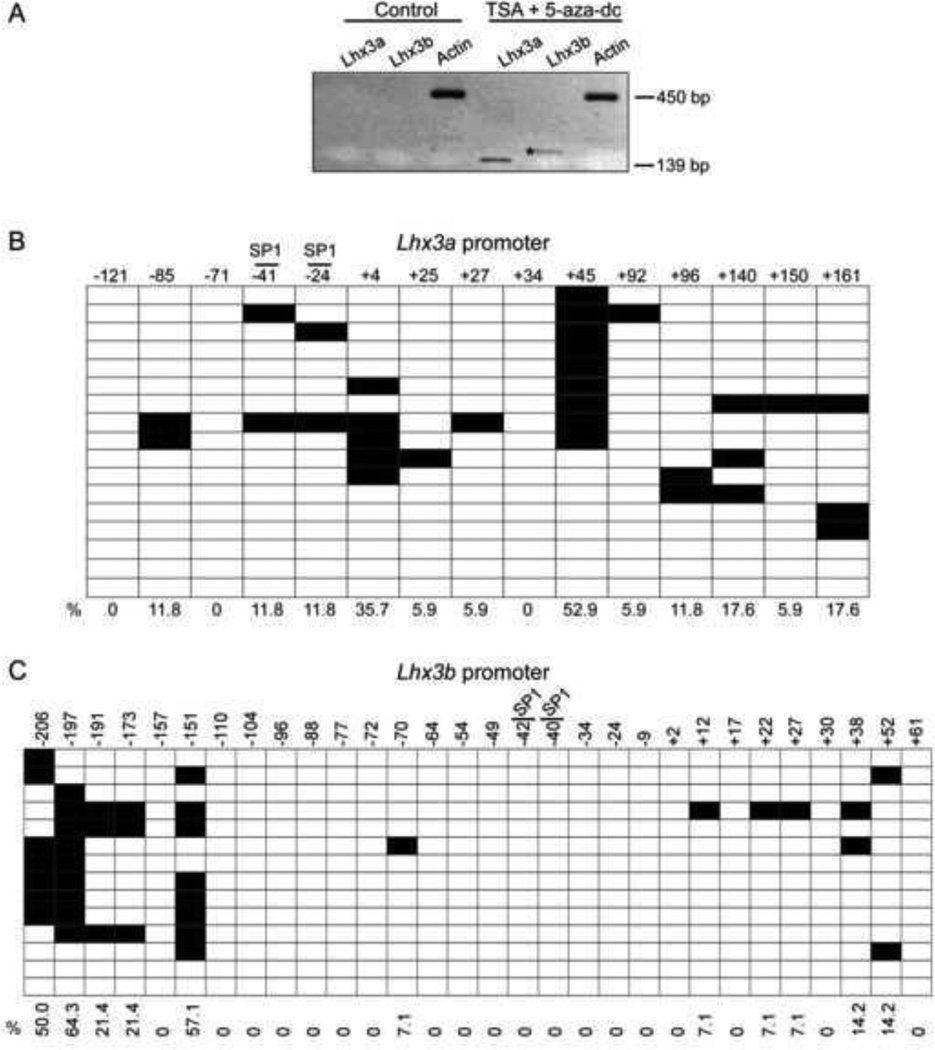
To determine if methylation might play a role in Lhx3 gene expression, bioinformatics was used to identify possible CpG rich regions in the Lhx3 promoters. The EMBOSS CpGPlot online tool predicted a CpG-rich region encompassing the Lhx3b promoter of both the mouse and human genes. A CpG-rich region also was noted in the human LHX3a promoter, but the corresponding region in the mouse Lhx3a gene promoter does not have quite as high a CpG density. To further probe the role of methylation of the two mouse Lhx3 promoters, a 333 bp region of the mouse Lhx3a promoter (−166 to +166 relative to the major transcription start site) containing SP1 binding sites and 15 CG sites was identified for further analysis (Figure 2A). Similarly, the CpG-rich region corresponding to the mouse Lhx3b promoter was included in a 321 bp region (−235 to +85) containing 30 CG sites and SP1 binding sites A (Figure 3A).
Figure 2. Lhx3a promoter and its methylation pattern.
(A) The upper diagram represents the mouse Lhx3 gene, with black rectangles signifying protein-coding exons and white areas denoting untranslated exonic regions. Two upstream promoters (aP and bP) produce the Lhx3a and Lhx3b mRNAs. The lower diagram below shows potential sites of methylation in the Lhx3a promoter, spanning from position −166 to +166 with the major transcription start site (TSS) set to position 0. Each vertical black bar indicates a CG base pair. The positions of SP1 binding sites (Yaden et al., 2006) are shown. (B) The methylation pattern of the Lhx3a promoter in mouse pituitary Pit-1/0 cells was determined by bisulfite sequencing (see materials and methods). Black boxes indicate methylated CpGs and open boxes indicate non methylated CpGs. Horizontal rows in each diagram represent individual sequenced clones from experiments. The overall percentage of observed methylation at each position is noted.
Figure 3. Lhx3b promoter and its methylation pattern.
(A) The upper diagram represents the mouse Lhx3 gene, with black rectangles signifying protein-coding exons and white areas denoting untranslated exonic regions. Two upstream promoters (aP and bP) produce the Lhx3a and Lhx3b mRNAs. The diagram below shows sites of potential methylation in the Lhx3b promoter, spanning from position −235 to +85 with the transcription start site (TSS) set at position 0. The positions of SP1 binding sites are shown. (B) The methylation pattern of the Lhx3b promoter in mouse pituitary Pit-1/0 cells was determined by bisulfite sequencing (materials and methods). Black boxes indicate methylated CpGs and open boxes indicate non methylated CpGs.
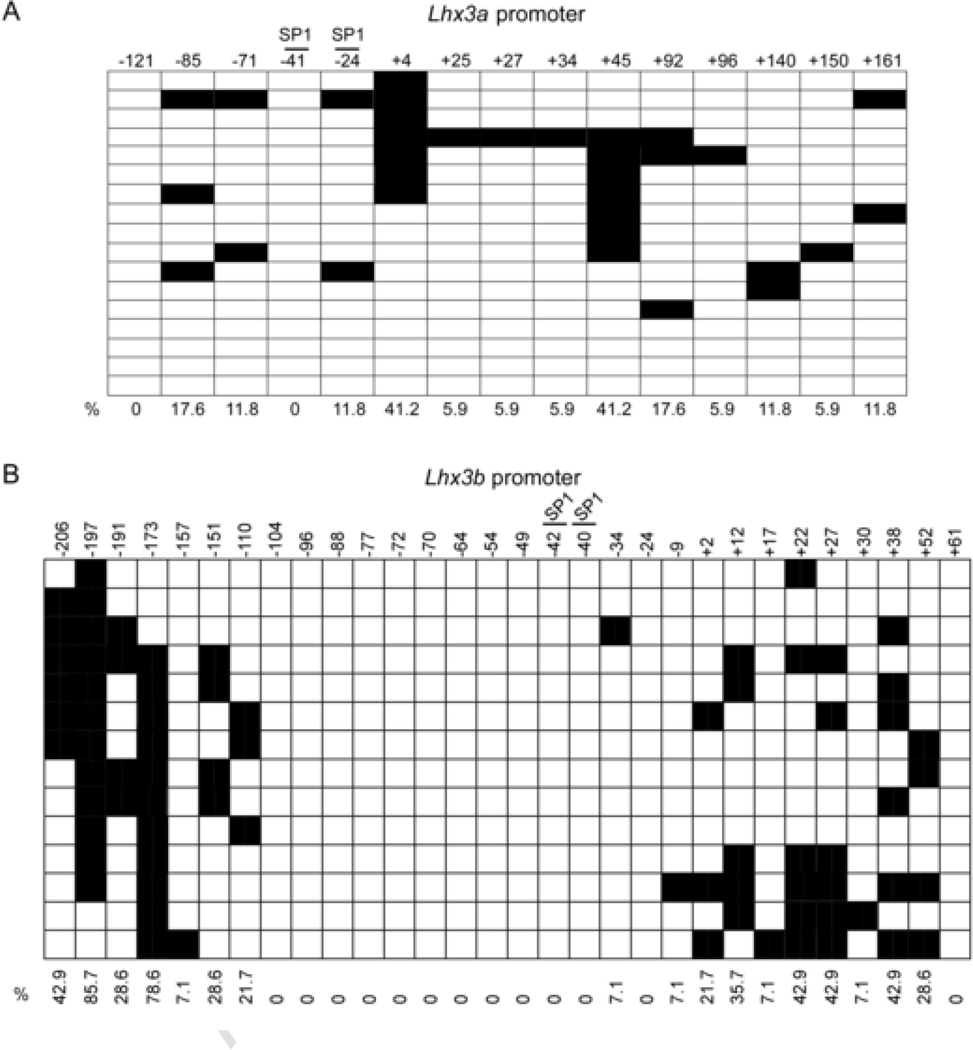
In order to characterize the methylation status of the Lhx3 promoters in the Pit-1/0 and Pit-1/Triple cell lines, genomic DNA was subjected to bisulfite conversion and sequencing. During bisulfite conversion, cytosines (C) are converted to thymines (T), but 5-methylcytosines remain unaltered. Bisulfite sequencing data from a series of individual DNA clones extracted from Pit-1/0 cells demonstrated that the Lhx3b promoter is highly methylated at specific sites, while the Lhx3a promoter is less methylated (Figures 3B and 2B, respectively). The transcription start site region of the Lhx3b promoter has low DNA methylation (Figure 3B). Pit-1/Triple cells were also subjected to bisulfite sequencing analysis (Figure 4). The Lhx3b promoter of the Pit-1/Triple cells is somewhat less methylated than the Lhx3b promoter in the Pit-1/0 cells, especially in the central section of the assayed region.
Figure 4. Lhx3 promoter methylation patterns in Pit-1/Triple cells.
(A) The methylation pattern of the Lhx3a promoter in mouse pituitary Pit-1/Triple cells was determined by bisulfite sequencing. Black boxes indicate methylated CpGs and open boxes indicate non methylated CpGs. Horizontal rows in each diagram represent individual sequenced clones from experiments. The overall percentage of observed methylation at each position is noted. (B) The methylation pattern of the Lhx3b promoter in mouse pituitary Pit-1/Triple cells.
3.3. 5-Aza-2’-deoxycytidine induces Lhx3 expression
To determine whether Lhx3 promoter methylation could be linked to the lack of detectable expression in Pit-1/0 cells, the cells were treated with the 5-Aza-2’-deoxycytidine (5-aza-dc) demethylating reagent. Lhx3a expression was induced 24 hours after 2.5 µM 5-aza-dc treatment, and both transcripts were expressed 48 hours post treatment (Figure 5A, B), suggesting that methylation plays a role in Lhx3 expression. Bisulfite sequencing of individual DNA clones from Pit-1/0 cells treated with 5-aza-dc for 48 hours (higher level of Lhx3 transcripts) showed modest decreases in methylation of both the Lhx3a and Lhx3b promoters (Figure 5C). The CpG sites at +12 and +17 of the Lhx3b promoter were not as affected by the treatment, with 57% and 50% of the clones methylated at the sites, respectively.
Figure 5. Lhx3 expression is activated in mouse pituitary Pit-1/0 cells after treatment with 5’-aza-dc.
Pit-1/0 cells were treated with 2.5 µM 5-aza-dc and RNA was harvested 24 hours (A) or 48 hours (B) after treatment and RT-PCR was performed. (C) Pit-1/0 cells were treated with 2.5 µM 5-aza-dc and genomic DNA was harvested for bisulfite conversion 48 hours post treatment. The methylation patterns of the Lhx3a (top) and Lhx3b (bottom) promoters were determined by bisulfite sequencing as described in the materials and methods. Black boxes indicate methylated CpGs and open boxes indicate non methylated CpGs.
3.4. Trichostatin A and 5-aza-dc treatment induce Lhx3 expression
DNA methylation has been shown to recruit chromatin modifying proteins to modulate gene expression (reviewed in Berger, 2002; Cosgrove and Wolberger, 2005; Vaissiere et al., 2008; Bogdanovic and Veenstra, 2009; Zhang and Ho, 2011; Buck-Koehntop and Defossez, 2013). To investigate whether the interaction between DNA methylation and chromatin modification can alter Lhx3 expression, Pit-1/0 cells were treated with both 5-aza-dc and the HDAC inhibitor, trichostatin A (TSA). The optimal dosage of the dual treatment that did not lead to cell death was determined to be 2.0 µM 5-aza-dc plus 100 nM TSA for 24 hours. The combined treatment induced expression of both Lhx3 transcripts (Figure 6A). Bisulfite sequencing of the Pit-1/0 cells treated with 5-aza-dc plus TSA indicated some decreased methylation of both Lhx3 promoters (Figure 6B, C). Treatment with TSA plus 5-aza-dc decreased methylation at the CpG sites +12, +17, +22, +27, +38 and +52 of the Lhx3b promoter compared to the bisulfite sequencing of untreated Pit-1/0 cells. In dual treated cells, bisulfite sequencing revealed that although methylation decreased at CpG sites −206 and −197 of the Lhx3b promoter, 50% or higher of clones still remained methylated after treatment.
Figure 6. Lhx3 expression after treatment with 5’-aza-dc plus TSA in Pit-1/0 cells.
(A) Mouse pituitary Pit-1/0 cells were treated with 2.0 µM 5-aza-dc plus 100 nM TSA and RNA was harvested 24 hours after treatment and RT-PCR was performed. (B), (C) Pit-1/0 cells were treated with 2.0 µM 5-aza-dc plus 100 nM TSA and genomic DNA was harvested for bisulfite conversion analysis at 24 hours post treatment. The methylation pattern of the Lhx3a (B) and Lhx3b (C) promoters were determined by bisulfite sequencing as described in the materials and methods. Black boxes indicate methylated CpGs and open boxes indicate non methylated CpGs.
3.5. The MeCP2 methyl-binding protein occupies the Lhx3 promoter
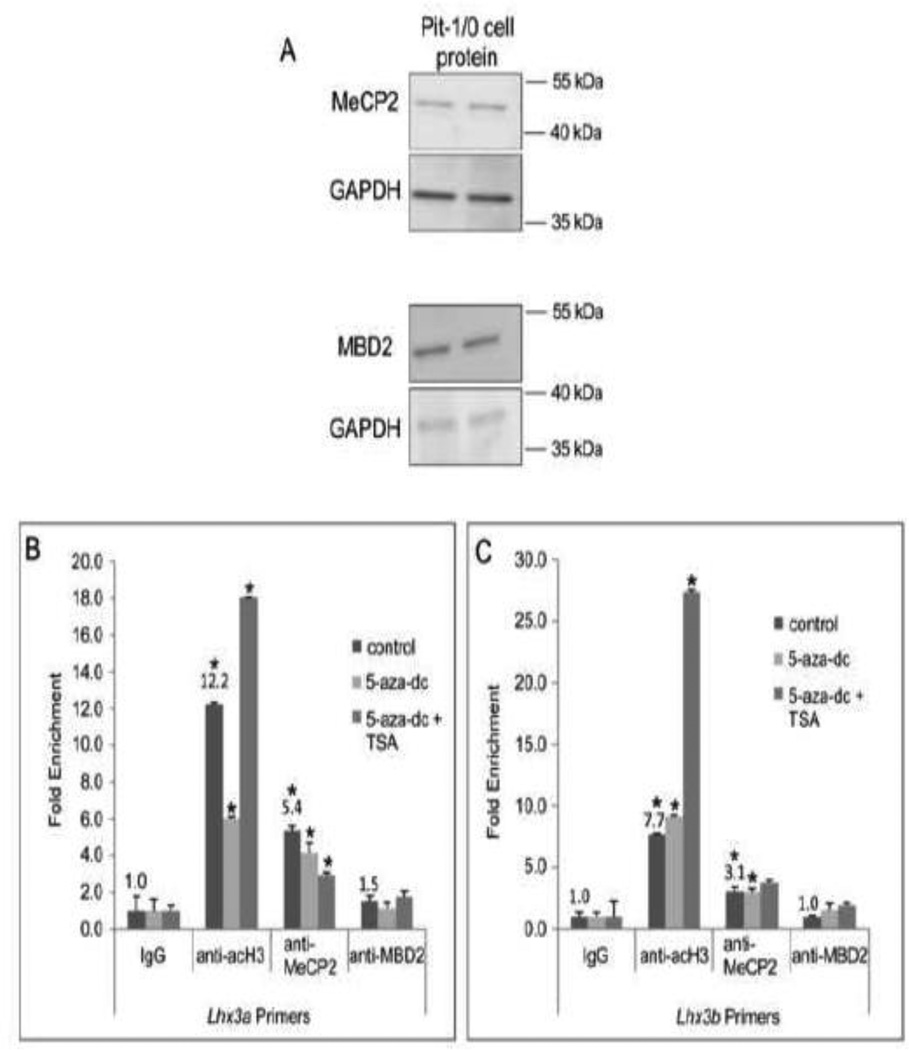
Methyl-CpG-binding proteins bind methylated DNA to regulate gene transcription (Robertson and Jones, 2000; Bogdanovic and Veenstra, 2009). MeCP2, a member of the methyl-binding domain (MBD) family of proteins is a regulator of global de novo methylation and has been shown to bind the pituitary pro-opiomelanocortin (Pomc) gene promoter (Tao et al., 2009). We determined if MeCP2 and its related protein family member, MBD2 were expressed in the Pit-1/0 pituitary cell line. Whole cell protein extracts from Pit-1/0 cells were probed by western analyses using antibodies recognizing MeCP2, MBD2 and GAPDH (positive control). Both MeCP2 and MBD2 protein were detected in the Pit-1/0 cells (Figure 7A). Because treatment with the demethylating reagent, 5-aza-dc and the HDAC inhibitor, TSA induced Lhx3 expression, we further investigated how treatment with 5-aza-dc and 5-aza-dc plus TSA might alter Lhx3 promoter occupation by MeCP2 and acetylated histone 3 (acH3). Acetylation of histones relaxes the chromatin structure and histone acetylation is often used as a marker of gene activation (Berger, 2002). To examine the effects of the inhibitors, chromatin immunoprecipitation (ChIP) assays were performed in treated and untreated Pit-1/0 cells with antibodies to MeCP2, MBD2, acH3 and IgG (negative control) and primers to the Lhx3 promoters. The 5-aza-dc plus TSA and control plates were harvested 24 hours post treatment and the 5-aza-dc treated cells were harvested for chromatin 48 hours post treatment to capture maximized Lhx3 expression in the cells. Analysis of the ChIP assay with Pit-1/0 cells indicated detection of acH3 and MeCP2 but not notable MBD2 at the Lhx3 promoters (Figure 7B, C). Pit-1/0 cells treated with 5-aza-dc showed an approximately two-fold decrease in acH3 enrichment and a modest decrease in MeCP2 enrichment at the Lhx3a promoter (Figure 7B). For the Lhx3b promoter, 5-aza-dc treatment led to a modest increase in acH3 enrichment and no change in MeCP2 enrichment (Figure 7C). Analysis of ChIP assays with Pit-1/0 cells treated with 5-aza-dc plus TSA indicated an increase in acH3 enrichment and a decrease in MeCP2 enrichment at the Lhx3a promoter region (Figure 7B). Enrichment of acH3 at the Lhx3b promoter of the dual treated cells notably increased, while there was no major change in MeCP2 enrichment of the Lhx3b promoter (Figure 7C).
Figure 7. MeCP2 is associated with the Lhx3 gene in Pit-1/0 cells.
(A) MeCP2 and MBD2 were detected in mouse pituitary Pit-1/0 cells by western blotting. GAPDH was used as a positive control. Two independent cell extracts are shown. (B, C) ChIP assays were carried out with chromatin from Pit-1/0 cells treated with 5-aza-dc, 5-aza-dc plus TSA or DMSO (solvent control), using anti-MeCP2, anti-MBD2 (negative control), anti-acetylated histone H3 (acH3) and IgG (negative control) antibodies and primers to the Lhx3a and Lhx3b promoters. A representative experiment of three independent, biological replicates is shown. Numbers indicate fold changes for untreated samples compared to IgG negative controls. Asterisks (*) indicate significant differences to IgG negative controls at P<0.05.
4. Discussion
DNA methylation has roles in a variety of cellular processes including transcriptional regulation (Bird, 1992). Methylation occurs at CpG sites so GC-rich DNA regions can be modified by DNA methylation (Smith and Meissner, 2013). The Lhx3 gene has two TATA-less, GC-rich promoters (Yaden et al., 2006) and recent studies have demonstrated methylated CpG islands in the promoters of developmental genes such as the Hox genes (Auclair and Weber, 2012) suggesting the hypothesis that DNA methylation patterns may affect Lhx3 gene expression. In this study, we describe the first investigation of the role of DNA methylation in Lhx3 gene transcription.
The recently established murine Pit-1/0 and Pit-1/Triple cell lines represent different phases of Pit-1-dependent cell differentiation in mouse anterior pituitary development (Sizova et al., 2010). The Pit-1 transcription factor is critical for differentiation of the somatotropes, lactotropes and thyrotrope cells of the anterior pituitary, and it synergistically interacts with LHX3 to regulate pituitary genes, such as Prl (Savage et al., 2003). In the Pit-1/0 cells, the Pit-1 gene is expressed, but the cells fail to express Pit-1-dependent hormones. The Pit-1/Triple cells represent a later phase of differentiation and express the Pit-1 gene and GH, PRL and TSHβ. The expression of the Lhx3 gene in these “Pit-1 lineage” cell lines had yet to be determined. In this study, we found that whereas the Pit-1/0 cells do not express either the Lhx3 gene or the LHX3 protein, the Pit-1/Triple cells produce LHX3. Therefore, the Pit-1/0 cells represent a valuable model investigate the role of DNA methylation in Lhx3 gene expression.
Bioinformatics was used to identify CpG-rich regions in the mouse and human Lhx3 promoters. Bisulfite sequencing was performed in both the Pit-1/0 and Pit-1/Triple cell line. The Pit-1/Triple cells are less methylated in the Lhx3b promoter region compared to the Pit-1/0 cells. Although the transcription start site (TSS) of each promoter in the Pit-1/0 cells is nearly devoid of methylation, surrounding sites within each promoter region are methylated. Since promoters are regions of regulatory control, it is possible that the surrounding methylated sites help coordinate protein-DNA interactions, and subsequent activities to regulate gene expression. Also, the DNA methylation pattern at or near the TSS may be determined by the local chromatin conformation (or conversely, such that DNA methylation dictates chromatin conformation), contributing to a pattern conducive for gene control (Cedar and Bergman, 2009).
CpG-rich regions in the Lhx3a and Lhx3b promoters contained previously identified SP1 binding sites (Yaden et al., 2006). SP1 has established roles in basal transcriptional activity of genes with GC-rich promoters, and SP1 binding sites have been noted as sites of DNA methylation control in some cases (Li et al., 2004). Although there are multiple studies examining the methylation status of SP1 binding sites, the findings are conflicting. Some studies have found that methylation of SP1 binding sites prevents its binding and subsequent gene activation, while others have shown that methylation of SP1 binding sites does not alter its binding (e.g. Harrington et al., 1988; Holler et al., 1988; Mancini et al., 1999; Butta et al., 2006; Douet et al., 2007). We found that there were no obvious differences in the DNA methylation patterns associated with the SP1 binding sites between the Pit-1/0 cell lines, which do not express the Lhx3 gene and the Pit-1/Triple cell line that does express the Lhx3 gene. These findings suggest that the methylation status of the promoter SP1 sites is not critical for the transcriptional status of the Lhx3 gene.
To further clarify how DNA methylation may impact the expression of the Lhx3 gene, we treated the cells with the demethylating reagent, 5-aza-dc. This reagent inhibits DNA methyltransferase activity causing hypomethylation. Treatment with 5-aza-dc induced expression of both Lhx3 mRNA transcripts in the Pit-1/0 cells, with increased expression correlating with longer exposure to the reagent. DNA samples from treated cells were subjected to bisulfite sequencing, and the results showed some decreases in methylation at both the Lhx3a and the Lhx3b promoter CpG-rich regions. Together, this data suggests that methylation may be part of Lhx3 gene inactivity in the Pit-1/0 cells. To our knowledge, this is the first report of such an activation of Lhx3 gene expression in a model system.
Acetylation is a well characterized post translational histone modification that is often a marker of gene activation (reviewed in Berger, 2002). The addition of an acetyl group is catalyzed by histone acetyl transferases (HATs), whereas the removal of acetyl groups is catalyzed by histone deacetylases (HDACs). Interestingly, treatment of Pit-1/0 cells with both 5-aza-dc and the HDAC inhibitor TSA led to a rapid induction of Lhx3 mRNA expression, suggesting possible cross-talk between the DNA methylation and chromatin microenvironment. Similar results have been seen in other studies, involving 5-aza-dc and TSA treatment to induce gene activity. For example, 5-aza-dc weakly activated expression of the retinoic acid receptor and estrogen receptor α; genes in MDA-MB-231 breast cancer carcinoma cells, while the combination treatment of 5-aza-dc plus TSA resulted in greater activation of both genes (Bovenzi and Momparler, 2001).
Methyl-CpG binding proteins have been shown to recruit histone modifiers to modulate transcription (Nan et al., 1998; Jones and Liang, 2009), so communication between methyl-CpG binding proteins and chromatin modifiers may influence Lhx3 mRNA expression. The methyl-binding protein, MeCP2 has been shown to bind the pituitary proopiomelanocortin (Pomc) gene promoter (Tao et al., 2009) and we therefore hypothesized that it may be similarly associated with the methylated Lhx3 promoters. ChIP assay analysis indicated that MeCP2 is associated with both Lhx3 promoters in Pit-1/0 cells. Studies in Xenopus and mice have demonstrated that MeCP2 interacts with Sin3 and histone deacetylase to trigger deacetylation of histones (Jones et al., 1998; Nan et al., 1998). Another study showed that MeCP2 recruited the mSin3a-HDAC1 complex to repress transcription of the brain-derived neurotrophic factor promoter in cortical cells (Chen et al., 2003; Martinowich et al., 2003; Klose and Bird, 2004). Therefore, we performed ChIP assays to investigate how inhibiting HDACs and/or blocking methylation may alter MeCP2 and acetylated histone H3 (acH3) occupation of the Lhx3 promoters. Interestingly, the treatments resulted in different effects on each Lhx3 promoter. Analysis of the ChIP assay with Pit-1/0 cells treated with 5-aza-dc showed only a very slight decrease in both acH3 occupation and MeCP2 occupation of the Lhx3a promoter. A decrease is MeCP2 occupation was expected, since the reagent blocks methylation and thereby potentially prevents MeCP2 from binding the methylated DNA. The decrease in acH3 occupation of the Lhx3a promoter with 5-aza-dc treatment may suggest potential cross-talk between chromatin status and DNA methylation. One possible explanation for the decrease in acH3 occupation of the promoter is that MeCP2 also interacts with chromatin modifying proteins to activate genes (Jones et al., 1998; Nan et al., 1998; Lunyak et al., 2002; Chen et al., 2013). For example, a study showed the MeCP2 recruits CREB1 to activate the murine neuronal glucose transporter isoform 3 (glut3) gene in murine neurons (Chen et al., 2013). Therefore, blocking methylation with 5-aza-dc may prevent MeCP2 from binding and recruiting other proteins that could potentially increase or maintain basal histone acetylation at the promoter.
At the Lhx3b promoter, 5-aza-dc treatment led to a slight increase in acH3 occupation and no change in MeCP2 occupation compared to untreated cells. Because MeCP2 recruits HDACs, blocking methylation may prevent binding of MeCP2 to methylated DNA and subsequent recruitment of HDACs to histones. The absence of recruited HDACs to histones may translate to a decrease in histone deacetylation and possibly an increase in acetylation of histone H3. The lack of change in Lhx3b promoter occupation by MeCP2 may be due to the fact that some sites still remain methylated in the Lhx3b promoter after treatment and therefore facilitate the binding of MeCP2 binding to the promoter. Additionally, the resolution of the ChIP assay is limited by sonication of DNA fragments less than 1 kB so precise resolution of changes in MeCP2 occupation may be challenging; nonetheless, the examined sequence is within a key regulatory region of the Lhx3 gene.
Unlike ChIP assays performed with the 5-aza-dc treatment, analysis of ChIP assays performed with Pit-1/0 cells treated with 5-aza-dc plus TSA indicated an increase in acH3 occupation and a decrease in MeCP2 occupation at the Lhx3a promoter region. These results suggest cross-talk and possible synergy between methyl-binding proteins and chromatin modifying proteins in regulating gene transcription, which has been seen in other studies (Jones et al., 1998; Nan et al., 1998; Klose and Bird, 2004). An increase in occupation of acH3 at the Lhx3b promoter resulted from the 5-aza-dc plus TSA treatment, while there was no change in MeCP2 occupation of the Lhx3b promoter, compared to untreated cells. Again, the lack of change in MeCP2 occupation may be due to the remaining methylated sites after treatment.
Transcription is a dynamic process. Modifying epigenetic marks of the genes in a cell by treatment with HDAC inhibitors and/or demethylating reagents may alter the temporal expression of some genes, which in turn may result in downstream changes to both protein-protein interactions and protein-DNA interactions. Therefore, expression of the Lhx3 gene induced by treatment with the HDAC inhibitor and/or the demethylating reagent is most likely the result of both direct and indirect effects. The change in methylation patterns after treatment (demonstrated by the Lhx3 bisulfite sequencing) and the subsequent induction of the Lhx3 gene suggest some roles for direct effects, but it should be noted that other unassayed regions of the gene also likely contribute to its expression as well. Furthermore, the transcription of upstream genes that regulate Lhx3, as well as chromatin conformation may have been altered by the inhibitor treatment, contributing to the indirect induction of the Lhx3 gene.
In future studies it would be interesting to examine the role of methylation in the human LHX3 gene, including testing whether epigenetic mechanisms play a role in the actions of the distal enhancer that is located downstream of the gene and has roles in pituitary and nervous system expression of LHX3 (Mullen et al., 2012). However, the 3′ pituitary/neural enhancer region has very few CpG sites in the “R3” region that is the critical cis element for pituitary expression making such a mechanism perhaps less likely at least in cells such as those tested here. Similarly, it will be interesting to examine whether there is interplay between LHX3 gene regulation and the actions of the Inhibitor of Histone Acetyltransferase complex (INHAT) proteins that were recently suggested to be part of the mechanism of action of LHX3 (Hunter et al., 2013).
Collectively, these findings implicate DNA methylation in the regulation of mouse Lhx3 gene transcription. We have shown that the demethylating reagent 5-aza-dc can induce Lhx3 gene activation both independently, and in combination with the HDAC inhibitor, TSA, in a pituitary precursor cell suggesting possible crosstalk between DNA methylation and chromatin modifiers. The methyl-binding protein, MeCP2 has been shown to occupy the Lhx3 promoter in Pit-1/0 cells and it may function as either a transcriptional repressor or activator, depending on the specific chromatin architecture. LHX3 plays a critical role in pituitary development, and mutations in LHX3 lead to severe pediatric combined pituitary hormone deficiency (CPHD) diseases. Characterization of the cellular processes that influence LHX3/Lhx3 expression, including mechanisms such as DNA methylation are critical to our understanding of pituitary organogenesis and disease.
Highlights.
LHX3 is a transcription factor with critical roles in pituitary and nervous system development.
The role of DNA methylation in expression of the mouse Lhx3 transcription factor gene was examined.
Treatment of cultured Pit-1/0 pituitary cells with a demethylating reagent lead to activation of the Lhx3 gene.
DNA methylation levels of Lhx3 promoter regions in untreated and treated Pit-1/0 cells were examined.
Acknowledgements
We are grateful to Drs. Richard Day, Maureen Harrington, Stephen Liebhaber, Raghu Mirmira, and David Skalnik for reagents and advice. Supported by NIH HD42024 to SJR and an NSF graduate fellowship to REM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94:2202–2211. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse RV, 2nd, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci U S A. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold-Dalla Pozza S, Hiedl S, Roeb J, Lohse P, Malik RE, Park S, Duran-Prado M, Rhodes SJ. A recessive mutation resulting in a disabling amino acid substitution (T194R) in the LHX3 homeodomain causes combined pituitary hormone deficiency. Horm Res Paediatr. 2012;77:41–51. doi: 10.1159/000335929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bhangoo AP, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, Walvoord EC, Ten S, Rhodes SJ. Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab. 2006;91:747–753. doi: 10.1210/jc.2005-2360. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfig W, Krude H, Schmidt H. A novel mutation of LHX3 is associated with combined pituitary hormone deficiency including ACTH deficiency, sensorineural hearing loss, and short neck-a case report and review of the literature. Eur J Pediatr. 2011;170:1017–1021. doi: 10.1007/s00431-011-1393-x. [DOI] [PubMed] [Google Scholar]
- Bovenzi V, Momparler RL. Antineoplastic action of 5-aza-2'-deoxycytidine and histone deacetylase inhibitor and their effect on the expression of retinoic acid receptor beta and estrogen receptor alpha genes in breast carcinoma cells. Cancer Chemother Pharmacol. 2001;48:71–76. doi: 10.1007/s002800100294. [DOI] [PubMed] [Google Scholar]
- Buck-Koehntop BA, Defossez PA. On how mammalian transcription factors recognize methylated DNA. Epigenetics. 2013;8:131–137. doi: 10.4161/epi.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butta N, Larrucea S, Alonso S, Rodriguez RB, Arias-Salgado EG, Ayuso MS, Gonzalez-Manchon C, Parrilla R. Role of transcription factor Sp1 and CpG methylation on the regulation of the human podocalyxin gene promoter. BMC Mol Biol. 2006;7:17. doi: 10.1186/1471-2199-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen Y, Shin BC, Thamotharan S, Devaskar SU. Creb1-Mecp2-mCpG Complex Transactivates Postnatal Murine Neuronal Glucose Transporter Isoform 3 Expression. Endocrinology. 2013 doi: 10.1210/en.2012-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin SC, Malik RE, Showalter AD, Sloop KW, Rhodes SJ. Model of pediatric pituitary hormone deficiency separates the endocrine and neural functions of the LHX3 transcription factor in vivo. Proc Natl Acad Sci U S A. 2011;108:173–178. doi: 10.1073/pnas.1009501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin SC, Mullen RD, Pfaeffle RW, Rhodes SJ. LHX3 and LHX4 transcription factors in pituitary development and disease. Pediatr Endocrinol Rev. 2009;6(Suppl 2):283–290. [PubMed] [Google Scholar]
- Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH, Brinkmeier ML, Mendonça BB, Brue T, Camper SA. Molecular mechanisms of pituitary organogenesis: In search of novel regulatory genes. Mol. Cell. Endocrinol. 2010;323:4–19. doi: 10.1016/j.mce.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet V, Heller MB, Le Saux O. DNA methylation and Sp1 binding determine the tissue-specific transcriptional activity of the mouse Abcc6 promoter. Biochem Biophys Res Commun. 2007;354:66–71. doi: 10.1016/j.bbrc.2006.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Harrington MA, Jones PA, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Malik RE, Witzmann FA, Rhodes SJ. LHX3 interacts with inhibitor of histone acetyltransferase complex subunits LANP and TAF-1beta to modulate pituitary gene regulation. PloS one. 2013;8:e68898. doi: 10.1371/journal.pone.0068898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30:790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- Kristrom B, Zdunek AM, Rydh A, Jonsson H, Sehlin P, Escher SA. A novel mutation in the LIM homeobox 3 gene is responsible for combined pituitary hormone deficiency, hearing impairment, and vertebral malformations. J Clin Endocrinol Metab. 2009;94:1154–1161. doi: 10.1210/jc.2008-0325. [DOI] [PubMed] [Google Scholar]
- Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Mollard P, Hodson DJ, Lafont C, Rizzoti K, Drouin J. A tridimensional view of pituitary development and function. Trends Endocrinol Metab. 2012;23:261–269. doi: 10.1016/j.tem.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Mullen RD, Park S, Rhodes SJ. A distal modular enhancer complex acts to control pituitary- and nervous system-specific expression of the LHX3 regulatory gene. Mol Endocrinol. 2012;26:308–319. doi: 10.1210/me.2011-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, Moers A, Goossens M, Gruters A, Amselem S. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–186. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, Kumar P, Bellone J, Schoenau E, Korsch E, Bramswig JH, Stobbe HM, Blum WF, Rhodes SJ. Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92:1909–1919. doi: 10.1210/jc.2006-2177. [DOI] [PubMed] [Google Scholar]
- Pfaffle R, Klammt J. Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract Res Clin Endocrinol Metab. 2011;25:43–60. doi: 10.1016/j.beem.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Prince KL, Walvoord EC, Rhodes SJ. The role of homeodomain transcription factors in heritable pituitary disease. Nat Rev Endocrinol. 2011;7:727–737. doi: 10.1038/nrendo.2011.119. [DOI] [PubMed] [Google Scholar]
- Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, Pearce K, Hall CM, Shaikh G, Gerrelli D, Grueters A, Krude H, Dattani MT. Novel mutations in LHX3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet. 2008;17:2150–2159. doi: 10.1093/hmg/ddn114. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Davies CS, Mills KI. DNA methylation: biology and significance. Blood Rev. 1996;10:249–261. doi: 10.1016/s0268-960x(96)90009-0. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Savage JJ, Yaden BC, Kiratipranon P, Rhodes SJ. Transcriptional control during mammalian anterior pituitary development. Gene. 2003;319:1–19. doi: 10.1016/s0378-1119(03)00804-7. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Zhadanov AB, Mosinger BJ, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang S-P, Mahon KA, Westphal H. Specification of Pituitary Cell Lineages by the LIM Homeobox Gene Lhx3. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- Sizova D, Ho Y, Cooke NE, Liebhaber SA. Research resource: T-antigen transformation of pituitary cells captures three novel cell lines in the Pit-1 lineage. Mol Endocrinol. 2010;24:2232–2240. doi: 10.1210/me.2010-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloop KW, Dwyer CJ, Rhodes SJ. An isoform-specific inhibitory domain regulates the LHX3 LIM homeodomain factor holoprotein and the production of a functional alternate translation form. J Biol Chem. 2001;276:36311–36319. doi: 10.1074/jbc.M103888200. [DOI] [PubMed] [Google Scholar]
- Sloop KW, Showalter AD, Von Kap-Herr C, Pettenati MJ, Rhodes SJ. Analysis of the human LHX3 neuroendocrine transcription factor gene and mapping to the subtelomeric region of chromosome 9. Gene. 2000;245:237–243. doi: 10.1016/s0378-1119(00)00025-1. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Yaden BC, Garcia M, 3rd, Smith TP, Rhodes SJ. Two promoters mediate transcription from the human LHX3 gene: involvement of nuclear factor I and specificity protein 1. Endocrinology. 2006;147:324–337. doi: 10.1210/en.2005-0970. [DOI] [PubMed] [Google Scholar]
- Zhadanov AB, Copeland NG, Gilbert DJ, Jenkins NA, Westphal H. Genomic structure and chromosomal localization of the mouse LIM/homeobox gene Lhx3. Genomics. 1995;27:27–32. doi: 10.1006/geno.1995.1004. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ho SM. Epigenetics meets endocrinology. J Mol Endocrinol. 2011;46:R11–R32. doi: 10.1677/jme-10-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]