Abstract
Aim
Since previous functional studies of short haplotypes and polymorphic sites of SLC6A3 have shown variant-dependent and drug-sensitive promoter activity, this study aimed to understand whether a large SLC6A3 regulatory region, containing these small haplotypes and polymorphic sites, can display haplotype-dependent promoter activity in a drug-sensitive and pathway-related manner.
Materials & methods
By creating and using a single copy number luciferase-reporter vector, we examined regulation of two different SLC6A3 haplotypes (A and B) of the 5′ 18-kb promoter and two known downstream regulatory variable number tandem repeats by 17 drugs in four different cellular models.
Results
The two regulatory haplotypes displayed up to 3.2-fold difference in promoter activity. The regulations were drug selective (37.5% of the drugs showed effects), and both haplotype and cell type dependent. Pathway analysis revealed at least 13 main signaling hubs targeting SLC6A3, including histone deacetylation, AKT, PKC and CK2 α-chains.
Conclusion
SLC6A3 may be regulated via either its promoter or the variable number tandem repeats independently by specific signaling pathways and in a haplotype-dependent manner. Furthermore, we have developed the first pathway map for SLC6A3 regulation. These findings provide a framework for understanding complex and variant-dependent regulations of SLC6A3.
Keywords: DAT, epigenetics, haplotypic activity, promoter, signaling regulation, single copy number reporter vector
The dopamine (DA) transporter (DAT) is the key regulator of DA transmission [1] for many physiologic processes including reward, voluntary movement, motivation, cognition, attention, working memory and behavioral organization. Consistently, the human DAT gene (hDAT or SLC6A3) is associated with neuropsychiatric disorders such as substance abuse, depression, bipolar disorder, schizophrenia, attention deficit hyperactivity disorder (ADHD) and Parkinson's disease [2,3]. In knockout or transgenic mice, altered DAT expression levels may cause altered behaviors mimicking various brain disorders [4–8]. These genetic findings all imply the contribution of altered SLC6A3 activity to related brain disorders.
DAT gene activity is known to be regulated by various internal as well as external factors. For example, substance abuse (such as cocaine abuse) may downregulate SLC6A3 expression [9–13]. In laboratory animals, DAT mRNA levels can be altered by many small molecules including the DAT inhibitor bupropion, the norepinephrine transporter inhibitor desipramine as well as insulin, estrogen, β-cytotoxics and diabetes-inducing drugs [14–19]. These findings suggest that there could be different regulatory pathways for DAT regulations.
Affected by aging [20,21], SLC6A3 expression levels differ significantly among individuals [22,23], this is attributable to the presence of several functionally polymorphic regulatory regions located throughout the 70-kb SLC6A3 gene. Known regulatory regions include an 18-kb promoter region [23], two functional variable number tandem repeats (VNTRs) – one located in intron 8 (Int8VNTR) and another located in the 3′-UTR (3′-VNTR) [24–33]. As risk alleles, the longer 6-repeat and 10-repeat alleles of Int8VNTR and 3′-VNTR both conferred lower gene activity than the shorter (5-/9-repeat) alleles [24,25,31–34].
However, these multiple known regulatory regions have not been studied together as a whole. It remains elusive whether and how these polymorphic regulatory regions (entire promoter, Int8VNTR and 3′-VNTR) together affect the regulated promoter activity and which signaling pathways regulate SLC6A3 via these cis-acting elements. This study is designed to address these questions, for the first time, by examining the 18-kb regulatory haplotypes together with the two VNTRs and the haplotype-dependent regulation of the promoter activity by 17 drugs in various model systems.
Materials & methods
Construction of reporter vector pGL3eBelo1
pGL3eBelo1, a single copy number reporter vector that is able to carry large genomic DNA of up to 300 kb and report promoter activity by firefly luciferase (Luc) activity, was generated in our laboratory by shuttling two fragments of Promega's (WI, USA) pGL3-enhancer, one was a 2421 bp fragment containing the synthetic poly(A), luc+, Simian vacuolating virus 40 (SV40) late poly(A) and enhancer, the other was a 222 bp fragment containing the SV40 late poly(A) signal, into pBeloBAC11 (NEB, MA, USA). pGL3eBelo1 has seven unique restriction sites including NotI, ApaLI, BamHI, HindIII, MluI, NheI and SrfI for cloning regulatory regions. NotI or HindIII digestion inactivates the LacZα for recombination selection. The lambda cos site may facilitate cloning of large DNAs by packaging into phage lambda particles. The vector displayed very little background Luc activity in the cultured cells and was used to construct all the SLC6A3 expression plasmids in this study.
Haplotype–reporter hybrid (18-kb luc) construction
An 18,909 bp fragment, covering from -16,672 (679 bp upstream of the 3′ end of the upstream gene LPCAT1, lysophosphatidylcholine acyltransferase 1) to +2236 (5′ to the ATG codon), assuming the transcription start site as +1 according to Genbank accession number NM_001044, was cloned into pGL3eBelo1. To generate SLC6A3 promoter haplotype A and B reporter constructs, we cloned the 18.9-kb fragments from two bacterial artificial chromosome (BAC) clones (AC091933.2 for A and AC026748.7 for B) into the 5′ side of luc+ in pGL3eBelo1, generating 18A and 18B. The construction is described in Supplementary Figure 1 (see www.futuremedicine.com/doi/suppl/10.2217/pgs.13.141).
18-kb luc-VNTRs construction
Cloning of Int8VNTR and 3′-VNTR into the 3′ end of luc+, at an FseI site located between luc+ and SV40 late poly(A) in 18A and 18B, generating 18A/B-luc-VNTRs. We termed the Int8VNTR 6 repeat as long (L), 5 repeat as short (S), the 3′-VNTR 10 repeat as L and 9 repeat as a S allele as well. We thus prepared four 18-kb luc-VNTRs plasmids termed as 18ALL, 18ASS, 18BLL and 18BSS.
Transformation with DNAs of large plasmids
A total of 1 μl of ligation reaction solution was mixed with 10 μl distilled deionized H2O and 10 μl of ice-thawed One Shot® TOP10 Electrocomp Escherichia coli (Invitrogen, now Life Technologies, NY, USA) in an ice-cold micro-tube, and this mixture was transferred into an ice-cold 2-mm cuvette (Eppendorf, Hamburg, Germany). Electroporation was carried out in an Gibco BRL Cell-Porator® Electroporation System (Gibco BRL, now Invitrogen, NY, USA) at 2.5 kV fast charge rate, followed immediately by addition of 0.5 ml of room temperature SOC medium (Invitrogen), and incubation at 225 rpm shaking and 37°C for 1 h before colony selection on LB agar plates.
Cell lines & culture
Four human neuroblastoma cell lines, SK-N-AS, BE(2)-M17, IMR-32 and SHSY5Y, were purchased from ATCC (VA, USA) and maintained at 37°C, 5% CO2 humidified atmosphere in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 U/ml; Invitrogen). SN4741 cells, an immortalized mouse embryonic substantia nigra-derived cell line (a kind gift from MJ Bannon of Wayne State University, MI, USA), were grown at 33°C in DMEM, supplemented with 10% (v/v) FBS, penicillin (100 U/ml), streptomycin (100 U/ml) and 0.6% D-glucose high glucose (Sigma, MO, USA).
Transfection with SLC6A3 promoter expression plasmids & Luc assay
Expression of the reporter (Luc) activity varies from cell line to cell line, from well to well, and from cell passage to cell passage, as we have observed during the last 10 years. To control these variations and identify genotype- or drug-specific activity, we used at least three independent preparations of plasmid DNA each with same quality (optical density260/280 ratio: ∼1.8) for each haplotype to rule out DNA quality-related artefact; we used three wells in each experiment and repeated the experiment four to eight independent times in different plates and cell passages for the same treatment/condition, in order to rule out well/plate-related artefact; we used relative Luc activity within a cell line, for comparison of genotype or drug effects among different cell lines. Since small control plasmids such as pRL-TK cannot be used for large plasmids, transfection efficiency of large plasmid DNAs of different genotypes was controlled by both high quality of multiple plasmid DNA preparations and multiple independent transfections of cells with different passages. Data are all presented as mean ± standard error of the mean in this study.
On day 1, cells were split at a ratio of 1:2 into 24-well plates with 30–40% confluence. On day 2, transfection medium was prepared using 0.4 μg plasmid DNA in 19 μl of FBS-free medium, mixed briefly with 2.4 μl of Superfect Transfection Reagent (Qiagen, CA, USA), incubated at room temperature for 20 min, followed by mixing with 0.12 ml of the normal growth medium. The transfection mixture/cells were incubated for 24 h, then added with 0.5 ml of normal growth medium, incubated for another 24 h. Cells in each well were washed with PBS and harvested for Luc activity measurements using the Luciferase Assay System (Promega) in a 96-well format and in Bio-Tek (VT, USA) Synergy™ HT/KC4, according to manufacturer's instructions. Cell number in each well was estimated by protein amount based on Bio-Rad's (CA, USA) Protein Assay Reagent. Arbitrary unit of SLC6A3 promoter activity was calculated as Luc activity: (readout/protein)(SLC6A3–MOCK) (MOCK, transfection without DNA). 18ALL was used to normalize haplotypic promoter activities.
Drug treatments
On day 1, cells were split at a ratio of 1:2 in 75 cm2 flasks with 30–40% confluence. On day 2, transfection mixture was prepared using 20 μg plasmid DNA of SLC6A3 promoter expression plasmids in 0.74 ml of FBS-free medium, mixed briefly with 94 μl of Superfect, incubated at room temperature for 20 min, followed by mixing with 4.7 ml of the normal growth medium. Transfection began by replacing media in each flask with this mixture, gently covering all the cells, and the transfection mixture/cells were incubated for another day. On day 3 (24 h after transfection), cells in each flask were harvested by brief Trypsin/EDTA treatment, resuspended in fresh medium and distributed evenly into 24-well plates. Drugs to be tested were added into the designated wells to a final desired concentration of each of 17 drugs, as listed in Supplementary Table 1 [35]. On day 4 (18 h later), cells in each well were washed with PBS and harvested for Luc activity measurements. We used 10 μM final concentration for DA, 6-hydroxydopamine (6-OHDA), d-amphetamine (d-AMPH), cocaine, metham-phetamine, U0126, SB202190 and forskolin; 1 μM for MK801 and phorbol-12-myristate-13-acetate (PMA); 10 nM for PACAP38, 0.25 μM for Dynorphin A porcine, 50 ng/ml for IGF-I, 100 ng/ml for TNF-α, 5 mM for valproate, 20 nM for okadaic acid and 3 μM for LY294002. The treatments were all at 37°C for 18 h unless indicated elsewhere.
Quantitative real-time PCR analysis of endogenous SLC6A3 mRNA in SK-N-AS
Quantitative real-time PCR analysis of endogenous SLC6A3 mRNA was carried out as previously described [23]. Quantitative real-time PCR reactions used the SYBR® Green PCR Master Mix (Fermentas, MD, USA). Data were normalized with GAPDH and the primers used in this study were: SLC6A3 5′-caaaagct-gctttccatggcacact-3′ and 5′-cggctcccaccgagcat-tacact-3′; hGAPDH 5′-tgccctcaacgaccactttg-3′ and 5′-tctctcttcctcttgtgctcttgc-3′.
BStop-PCR quantification of relative mRNA abundance of two rs6347 alleles (G/A)
To quantify allelic cDNA levels, BStop-PCR was used as previously described [23,36]. First, a 323 bp amplicon was prepared by using forward primer 5′-gggtacatggcacagaagca-3′, and reverse primer 5′-tccaggagcgtgaagacgta-3′ and a PCR cycle of 95°C for 5 min, 28 cycles of 95°C for 1 s and 57°C for 10 s and 72°C for 30 s, followed by extension at 72°C for 3 min. Biotin labeled primer 5′-biotin-tccaggagcgtgaagacgta-3′ was then added to the PCR reaction, followed by incubation at 94°C for 30 s, 62°C for 30 s and 72°C for 1 min. After digestion by BbvcI, which was ‘CCTCAGC’ sensitive, of PCR products with the A allele into biotin-labeled 215 bp and 108 bp (the resistant G allele retained the biotin-labeled 323 bp), the digests were subjected to 1.5% agarose gel electrophoresis for resolution. After electrophoresis, DNA was capillarily transferred to positively charged nylon membrane (Ambion, now Life Technologies) and biotin on the membrane was detected by using Ambion's BrightStar® BioDetect™ kit. Band intensity was analyzed by FOTO/analyst Investigator (FOTODYNE, CA, USA) and TL100 (Totallab, Newcastle upon Tyne, UK).
Pathway analysis
Drugs or kinase effectors that regulated SLC6A3 promoter activity in vitro or endogenously were used to map out the signaling pathways. The software/database that was used was the frequently updated MetaCore [37].
Expression levels of pathway members in DA (A10) neurons were consulted with a published gene-expression profiling analysis [38]. DRD2 is the auto receptor and is used as a standard active protein of DA neurons [39]. We arbitrarily used 0.25-fold as a cutoff, in order not to miss any informative pathways. At the same time, the Supplementary Material provides the details on all related pathways and relative gene expression in DA neurons. ‘Hub genes’ highlight the top members that can regulate SLC6A3 with the most (eight or more) paths.
Results
SLC6A3 18-kb promoter haplotypes A & B
This study included two different 18-kb haplotypes carried by two BAC clones (AC091933.2 and AC026748.7). Sequence alignments showed that these two BAC clones carried 34 informative polymorphisms including 32 SNPs and two VNTRs, one of them was the 5′-VNTR at -11 kb (Figure 1A). Of the 32 SNPs, rs2975226, rs2652511 and rs2975223 are already implicated in ADHD, schizophrenia, treatment efficacy and functional activity [28,40–42]. The 5′-VNTR was associated with mRNA levels in the ventral midbrain tissue of postmortem human brains [23]. We termed AC091933.2 as haplotype A and AC026748.7 as haplotype B (Figure 1A). We cloned these 18-kb haplotypes, as well as VNTRs into pGL3eBelo1 (Figure 1B), as described in Supplementary Figure 1.
Figure 1. Two 18-kb SLC6A3 promoter region haplotypes and the expression vector used in this study.
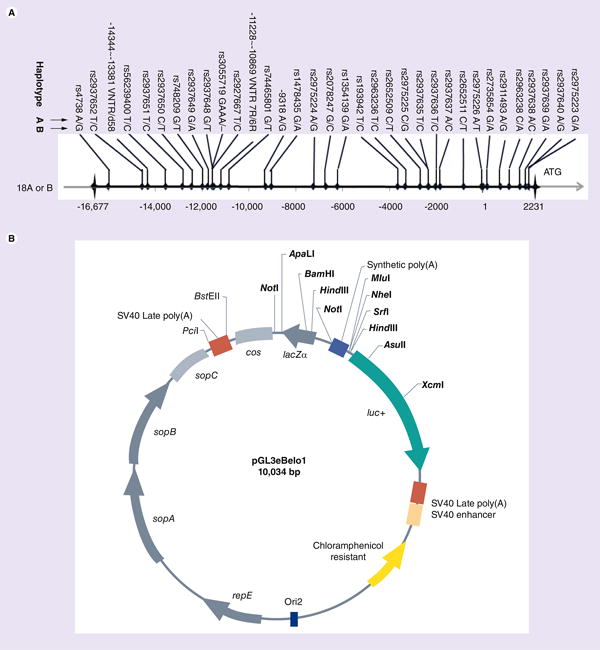
(A) Haplotypes A and B of SLC6A3 18-kb promoter region: 34 polymorphisms (32 SNPs and two variable number tandem repeats [VNTRs]) revealed by BAC alignment (AC091933 vs AC026748), each polymorphism indicated by two alleles (separated by '/') of two haplotypes; horizontal gray arrow: chromosome DNA, with coordinates for 18 kb upstream of ATG codon (black horizontal line) using the transcription start site as +1 (-14344∼-13381VNTR missed 58 bp in haplotype B; rs3055719 missed ‘GAAA’ in haplotype B). (B) A single copy number reporter vector pGL3eBelo1 developed to harbor SLC6A3 regulatory regions. Aquamarine, firefly luciferase gene (luc+); orange box, SV40 enhancer for luc+ expression; cos, the lambda cos site for packaging into phage lambda particles and facilitating cloning of large fragments; yellow arrow, the chloramphenicol-resistant gene. Cloning sites are labeled in bold; lacZα for recombination selection (see ‘Materials & methods’ section for description of construction). A unique FseI site located between luc+ and SV40 late poly(A) used for VNTR insertion is not shown here.
Please see colour figure at http://www.futuremedicine.com/doi/pdf/10.2217/pgs.13.141.
In vitro expression systems
We selected cell lines for SLC6A3 promoter expression by Luc-based screening of the 18-kb haplotypes A and B in five cell lines including four neuroblastoma cell lines (SK-N-AS, BE(2)-M17, IMR-32 and SH-SY5Y) of human origin and one nigral neuron-derived cell line (SN4741) of mouse origin. Luc activity showed that SK-N-AS and SN4741 both supported the expression of the SLC6A3 promoter while the three other cell lines expressed the promoter at relatively low levels (7.4–8.6% of the SK-N-AS values). In particular, BE(2)-M17 can also express endogenous SLC6A3. The high SLC6A3 promoter activity in SK-N-AS was consistent with the robust endogenous SLC6A3 expression in this cell line [43]. The endogenous DAT expression in SN4741 has also been con-firmed by different studies [44,45]. Therefore, expression analyses of the four haplotypes in this study were carried out mainly in SK-N-AS and SN4741.
Haplotype-dependent expression
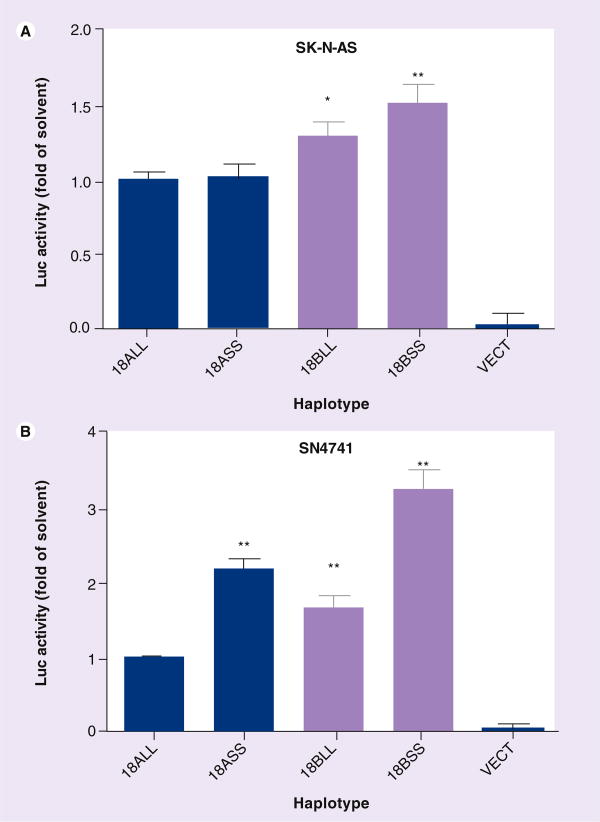
In both SK-N-AS and SN4741, the vector pGL3eBelo1 displayed very low, residual background activity (1.8 and 2.4% of 18ALL on average). Int8VNTR was a cis-acting element. Its inhibitory effects were independent of insertion location or orientation, and no matter whether it was or it was not a part of the luc RNA (Supplementary Figure 2). Therefore we combined the two VNTRs together before insertion into the end of luc+. We combined the short alleles into SS and the longer alleles into LL because the longer alleles, but not the shorter alleles, were risk factors for ADHD or cocaine addiction. In this sense, LL was the risk factor and SS, the control. Owing to low linkage disequilibrium in between, the four regulatory haplotypes of 18-kb promoter – SS/LL in this study all represent natural regulatory haplotypes in humans. The 18-kb B-shorter alleles of VNTRs (18BSS) haplotype displayed an activity 1.51-fold higher than the 18-kb A-longer alleles (18ALL) in SK-N-AS (Figure 2A). In particular, the 18BSS activity was 3.24-fold higher than the 18ALL's in SN4741 (Figure 2B). The higher activity of 18BSS, was also observed in three other cell lines BE(2)-M17, IMR-32 and SH-SY5Y (data not shown). These haplotypic activities were consistent with the previous findings on higher activity with shorter haplotypes [29,30]. In addition, our data show that even in the presence of the 18-kb promoter region, the long alleles are still associated with lower activity and the short alleles, associated with higher activity.
Figure 2. Haplotype-dependent expression of SLC6A3 in SK-N-AS and SN4741.
(A) SK-N-AS and (B) SN4741. Average residual activity of VECT was 1.8% and 2.4% of 18ALL in SK-N-AS and SN4741, respectively. One-way ANOVA Tukey, comparing with 18ALL: *p < 0.05, **p < 0.01 (n = 4–6). Darker colored bars = 18A, lighter colored bars = 18B.
LL: 6-repeat of Int8 variable number tandem repeat (VNTR) combined with 10-repeat of 3′-VNTR; SS: 5-repeat of Int8VNTR with 9-repeat of 3′-VNTR; VECT: Expression vector pGL3eBelo1.
Drug regulation
To understand whether pathways are involved in haplotype-dependent regulations, 17 different drugs were examined for their effects on SLC6A3 activity in the four in vitro model expression systems (see the drug list in Supplementary Table 1). These four in vitro model expression systems were used because of their human origins for this human gene. The listed drugs were common effectors of DA signaling among many other signaling pathways. In particular, these drugs were used to examine the regulation of another human monoamine transporter gene, SLC18A2 [35]. Results from the regulation studies would not only reveal regulatory pathways for SLC6A3 but also be comparable between the two DAT genes, helping to identify shared or unique pathways. Among many DAT protein inhibitors, we were focused on only three drugs of abuse, cocaine, d-AMPH and methamphetamine. Therapeutics such as methylphenidate and mazindol were not included. Among the 17 drugs, only valproate has been used to examine SLC6A3 regulation before [46] but this drug has never been used for haplotype-dependent regulation; forskolin was studied for allele-dependent regulation of Int8VNTR before [34] but has not yet been used to study the SLC6A3 promoter. Therefore, none of these drugs have been studied for haplotype-dependent regulation of SLC6A3 before.
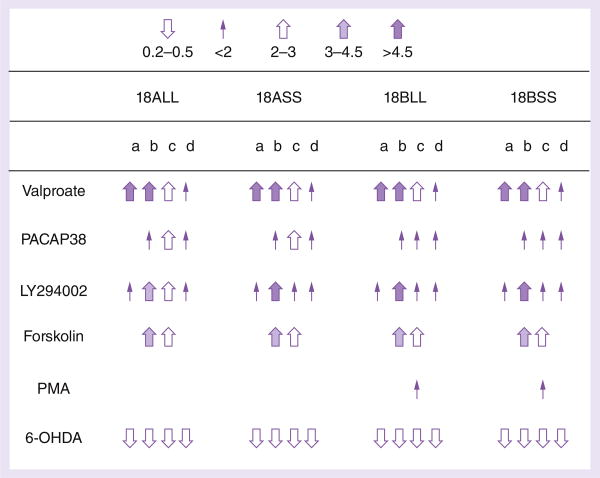
Eleven of these drugs (DA, d-AMPH, dynorphin A, IGF-I, (+)MK801, okadaic acid, SB202190, U0126, cocaine, methamphetamine and TNF-α) did not exert any significant regulation and their data are not shown here. We made sure of their activity by using fresh orders of different batches whose qualities were validated by the manufacturers. We present the statistically significant data based upon 18 h treatments of four human cell lines transiently expressing the four regulatory haplotypes with the six other agents: valproate, PACAP38, LY294002, forskolin, PMA and 6-OHDA, as summarized in Figure 3. Overall, the first four drugs upregulated the SLC6A3 promoter activity on all four haplo types. Uniformly, the neurotoxin 6-OHDA downregulated the activity to 0.2–0.5-fold independently of haplo type or cell type, suggesting that SLC6A3 be ubiquitously sensitive to oxidative stress. Among the top three upregulators including valproate, PACAP38 and LY294002, dose and time dependences of the endogenous SLC6A3 upregulation have been reported consistently for valproate before [46] and observed for both PACAP38 and LY294002 as well in SK-N-AS (Supplementary Figure 3).
Figure 3. Regulation of hDAT promoter activity by 6-OHDA, PACAP38, valproate, LY294002, forskolin and PMA in four cell lines, based on Luc activity analysis.
All arrows represent statistical significances by one-way ANOVA Tukey (n = 4–8), compared with solvent controls; no arrow, no statistical significance.
a: SK-N-AS (1.8%)†; b: BE(2)-M17 (2.9%); c: IMR-32 (6.2%);
d: SH-SY5Y (0.5%).
†Average residual activity of the vector as a percentage of 18ALL activity.
Haplotype-dependent drug regulation
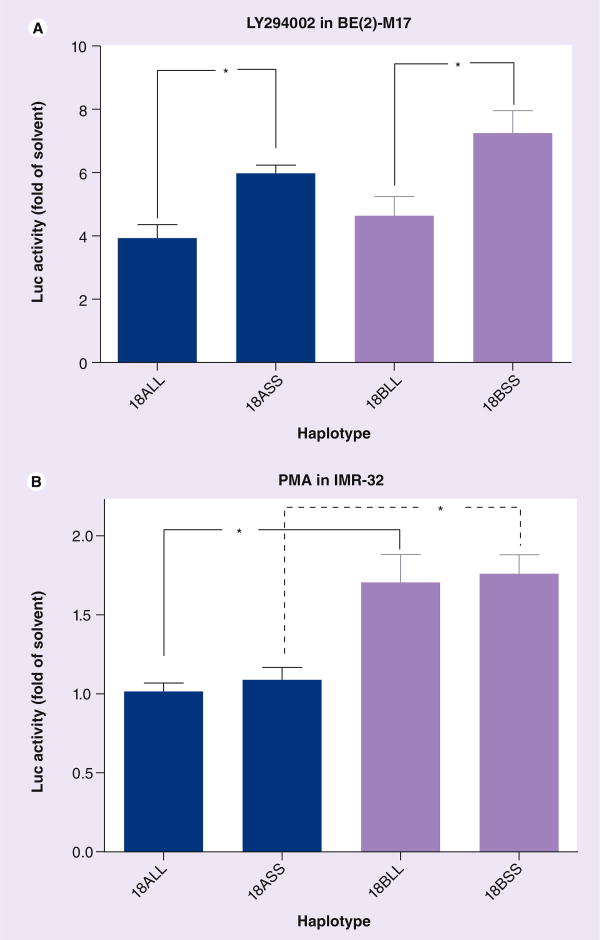
Among the drug regulations in Figure 3, two drugs displayed statistically significant haplotype dependence, which is detailed in Figure 4. The PI3K inhibitor LY294002 upregulated the shorter alleles more significantly (with a greater magnitude) than the longer ones, without affecting the 18-kb promoter. LY294002 upregulated the SS more than the LL activities, with the largest extents (1.51-fold on haplotype A and 1.56-fold on haplotype B) in the background of 18-kb promoter region (Figure 4A). The PKC activator PM A upregulated only t he promoter region of haplotype B in IMR-32, without affecting the VNTRs (Figure 4B).
Figure 4. Haplotype-dependent regulation of SLC6A3 promoter by drugs in BE(2)-M17 and IMR-32, based on Luc activity analysis.
(A) BE(2)-M17 and (B) IMR-32. One-way ANOVA Tukey: *p < 0.05 (n = 4–8). Darker colored bars = 18A, lighter colored bars = 18B.
Cell type-dependent drug regulation
Of the six drugs, valproate upregulated the promoter activity in all constructs and in all expression systems. The extent of regulation was smaller in SH-SY5Y compared to other cell lines including SK-N-AS, BE(2)-M17 and IMR-32. The PACAP38 regulation depended on cell type, without any effects in SK-N-AS. Forskolin's upregulations had larger extents in BE(2)-M17 and IMR-32 than in SK-N-AS and SH-SY5Y. 6-OHDA downregulated the promoter activity in all constructs and in all cell lines (Figure 3).
Allele-dependent drug regulation of endogenous SLC6A3 expression
To support the overall haplotype-dependent regulation of SLC6A3 observed in the transient expression systems, we further investigated whether these drugs regulated endogenous SLC6A3 expression from chromosome 5 in an allele-dependent manner by using a modification of hot-stop PCR, BStop-PCR. We used the exonic SNP rs6347 to monitor allele-dependence in BE(2)-M17 because this cell line carried informative (both alleles) rs6347. SK-N-AS was included as a homo-zygote A/A carrier for allele and digestion controls (lanes 1 and 2 in Figure 5A where 323 bp represented allele G and the digestion product 215 bp, allele A). Imbalanced intensities between the two bands indicated allele-dependent regulations (Figure 5A). Quantification of the regulations by the six drugs in Figure 5A is summarized in Figure 5B. Valproate, PACAP38, LY294002 and forskolin upregulated G- more than A-associated SLC6A3 expression (1.52-, 1.46-, 1.36- and 1.39-fold), all reaching statistical significance (Figure 5B). Cocaine did not regulate SLC6A3 in this cell line, as opposed to previously observed reduced expression in postmortem brain tissue [10–12], probably because BE(2)-M17 lacked the cocaine-related pathways. 6-OHDA did not display any allele dependence but appeared to downregulate SLC6A3 on both chromosomes, consistent with the findings from the transient expression systems (Figure 3).
Figure 5. Allele-dependent regulation of endogenous SLC6A3 activity by drugs in rs6347-carrying BE(2)-M17, based on BStop-PCR analysis of mRNA levels.
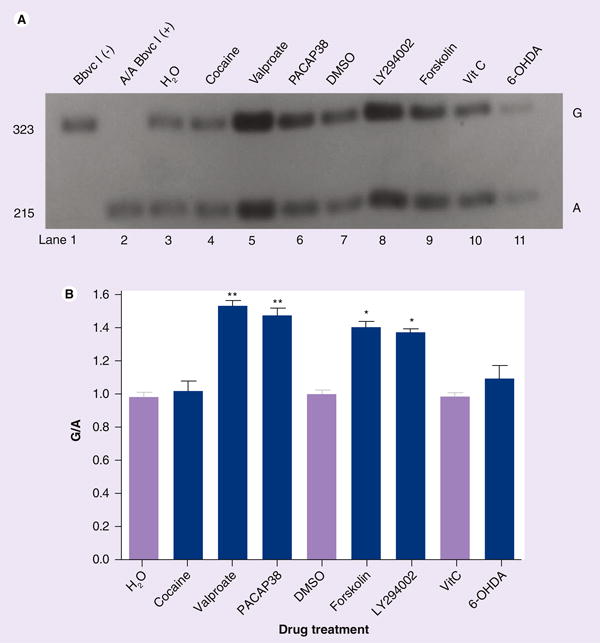
(A) Characteristic data of BStop-PCR analysis. Lane 1: PCR production without BbvcI-digestion; lane 2: rs6347 homozygote A/A (from SK-N-AS) digested completely by BbvcI; lanes 3, 7 and 10: solvents (lighter color) for drugs in following lanes (lanes 4, 5, 6, 8, 9 and 11: drug treatments; darker color). (B) Quantification of regulation data in (A). One-way ANOVA Tukey: *p < 0.05; **p < 0.01, compared with solvent control (n = 3).
Haplotype-dependent signaling pathways
The purpose of this pathway analysis was to develop an initial regulatory map for DA neurons based on findings from in vitro studies and on active genes in DA neurons. Based on the significant regulation findings mentioned above, we used the ‘build network’ function with the building algorithm of ‘shortest paths’ in Meta-Core to generate a preliminary pathway map that collected information from all five types of cells (see Supplementary Figure 4). Manually, we screened all the proteins in this preliminary map for expression levels in DA neurons, based on DA neuron-specific gene-expression profiling [38]. Any expression levels below a quarter of the DRD2 receptor level were considered nonexpression and the related pathways were removed from the map, resulting in a DA neuron-related pathway map as in Figure 6. Among the six drugs that endogenously regulated SLC6A3 haplotypes, 6-OHDA was not yet included in the MetaCore database. The pathways mediating the valproate, LY294002, PMA, forskolin and PACAP38 regulation were mapped out but only approximately two-thirds of the members were expressed at levels of atleast 25% of the DRD2 receptor (the DA neuron autoreceptor) expression levels (Supplementary Figure 4). After removing those with expression <25% of the DRD2 levels, a map that likely works in DA neurons was described (Figure 6). In the valproate pathway, histone deacetylation was a major hub because HDAC1, for example, had more than ten interactions with other targets. LY294002 regulated SLC6A3 mainly through the CK2 α chains–SP1/SP3 complex as well as the DNA–PK–PKC–SP1/SP3 complex pathways. PMA regulated SLC6A3 through the PKC–SP1/SP3 complex pathways. Forskolin regulated SLC6A3 through FXR (also known as NR1H4, a bile acid-binding transcription factor), a pathway that might work only in our in vitro assay systems because of very low FXR expression levels in DA neurons. PACAP38 regulated SLC6A3 through cAMP but how cAMP regulated SLC6A3 remained unclear. cAMP was supposed to regulate ERK1/2 via MEK1 but in the different assay systems the ERK1/2 inhibitor did not affect SLC6A3 in our assays. Instead, PACAP38, perhaps through the cAMP, regulated SLC6A3 indirectly through unknown transcription factors. These known pathways all targeted SLC6A3 through the three known transcription factors HEY1, NURR1, GCR-α and the SP1/SP3 complex. All of these five drugs differentially regulated SLC6A3 between the two haplotypes or the two rs6347 alleles under this study.
Figure 6. Signaling cascades that regulate SLC6A3 (located at bottom) in a haplotype-dependent manner.
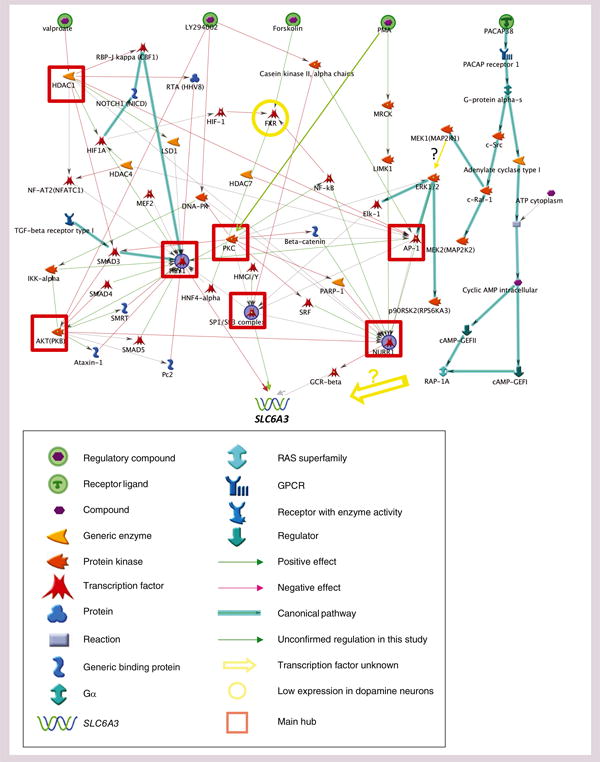
Insert: symbol description. Included are all genes expressed at levels over a quarter of DRD2 expression levels in the dopamine neurons, except the open circle, for over a quarter of DRD2 expression levels (other genes with low expression levels are not shown). Oxidative stress-related (6-hydroxydopamine) pathways are not included.
Discussion
For the first time, we have examined drug regulation of the large regulatory haplotypes, especially the promoter regions, of SLC6A3. To analyze these large regulatory haplotypes, we have developed the first single copy number plasmid Luc-reporter constructs facilitating the mapping of the first SLC6A3 signaling pathways. These comprehensive SLC6A3 haplotypes covering from the 18-kb promoter region to the 3′-VNTR display promoter activity of more than threefold difference, the largest haplotypic difference reported to date for this gene (Figure 2). Our data, especially the drug regulation data, suggest that genetic evaluation of SLC6A3 activity needs to consider the contribution from both VNTRs and the 5′ promoter region (Figure 4).
It has been known for the last two decades that F-plasmid-based single copy number vectors may allow for cloning and maintainence of chromosome DNA fragments of up to 300 kb in length stably in E. coli [47]. In this study, we have adopted this single copy number plasmid approach and developed pGL3eBelo1 for investigation of regulatory activity of large chromosomal fragments (Figure 1). As more and more information is revealed by genome-wide association studies and gene–gene interaction studies, it is critical to have the ability to assess functional relevance of the statistical findings in human genetics across different fields [48–51]. It is therefore likely that the combination of single copy number vectors and gene activity reporters will find wide applications in the near future.
Our study in these large regulatory haplotypes confirms the previous findings on allelic activity of the individual VNTRs. As mentioned above, the 5-repeat or 9-repeat alleles were found to be associated with higher promoter activity whereas the longer alleles, were found to be associated with lower activity, especially in healthy subjects. These reported results were obtained when each VNTR was studied independently. In the 18-kb promoter region background, these results remain valid. Conceivably, these VNTRs responded to different signaling pathways for SLC6A3 regulation. This explains why the 9-repeat allele is also associated with the higher DAT protein density in human brains carrying this allele, based upon imaging analysis [33].
The signaling pathways turned out to be complicated already, based on only a limited number of transcription factors for SLC6A3, making three implications. Foremost, the new finding on the involvement of histone deacetylation, together with recently reported promoter DNA methylation, suggests that the DAT gene is also subject directly or indirectly to extensive epigenetic regulation [21,52,53]. Therefore, when a polymorphism in the promoter is associated with a disease but does not confer any functional variation in transient expression systems, it is worth considering examining the epigenetic status including DNA methylation and histone modifications around the polymorphic site in endogenous systems. Second, signaling regulation can be achieved through different regulatory regions. AP-1, GCR-α and the SP1/SP3 complex act through the promoter region while HEY1 regulates SLC6A3 through 3′-VNTR [54,55]. Specifically, forskolin may upregulate SLC6A3 through NURR1–SP1/SP3 complex, but downregulate SLC6A3 through HEY1. Since HEY1 did not differentiate between the 9-repeat and 10-repeat alleles of 3′-VNTR in transcriptional regulation in vitro, it could be various expression levels of HEY1 that underlie the reported 3′-VNTR associations with ADHD [56–58]. Third, SLC6A3 could be regulated by complicated signaling cascades in various DA neurons. Only five SLC6A3-selective drugs already imply relatively complex signaling cascades here. In fact, the DAT gene is regulated by many external and internal factors, suggesting that additional related signaling cascades wait for future delineation. Furthermore, balance or imbalance between different pathways may partly explain the cell-type dependence in SLC6A3 promoter regulation (Figure 3). Importantly, the regulation by these signaling cascades can be affected by genetic variation in their genomic target (Figures 4 & 5), providing mechanistic insight into individual variation in SLC6A3 activity and in response to environment. This initial pathway map merits experimental validation in future studies.
SLC6A3 variant-dependent regulation by valproate could imply individual dependent efficacy of this medicine for mood stabilization. Valproate has been used as a mood stabilizer for patients with bipolar disorder and schizophrenia [59–63]. Mice with reduced expression of DAT display hyperactive activity and perseverative motor behavior, mimicking certain symptoms observed in these patients. Treatment of these transgenic mice with valproate was able to attenuate these symptoms significantly [6]. Indeed, it was shown that valproate could upregulate SLC6A3 activity [46]. We have shown that valproate upregulates the G more than the A allele of rs6347 endogenously in BE(2)-M17. This data is consistent with the clinical observations that valproate treatment can be effective in some patients but not in others [64–68]. Future study will need to verify these pathways in DA neurons as well as the underlying polymorphisms in order to provide evidence-based guidelines for individualized treatment [69].
Finally, we have shown that 6-OHDA down-regulated SLC6A3 in all haplotypes and all expression systems used (Figure 3). By contrast, 6-OHDA was able to upregulate SLC18A2 that encodes VMAT2 [35]. As a result, exposure to this neurotoxin reduces accumulation of cytosolic DA molecules by decreasing the DAT-mediated entry of extracellular DA and increasing the VMAT2-mediated sequestering of cytosolic DA into intra-cellular vesicles. Although we do not yet understand the pathways, there might be a feedback mechanism to respond to oxidative environment for self-defense.
Conclusion
To date, most studies of functional variation of SLC6A3 have focused on pharmacologic regulation of single polymorphisms or promoter activity of short haplotypes (<7.4 kb). No reported study has looked at activity of large haplotypes containing multiple functional or potentially functional alleles, mainly owing to the lack of appropriate methodologies for large haplotypes. In this study, we have developed a single copy number reporter vector, pGL3eBelo1, which can potentially harbor up to 300 kb of haplotypes. By using pGL3eBelo1, we are able to analyze the 18-kb promoter region haplotypes together with the two well-studied downstream VNTRs in SLC6A3. Findings from these analyses show that 18-kb haplotypes may display large differences in SLC6A3 promoter activity. Specific signaling pathways that regulate SLC6A3 may target various regulatory regions throughout the 70-kb gene and be affected by both haplotype and cell type. In conclusion, our data help build the first pathway map for SLC6A3 regulation, suggesting that both transcription factors and epigenetic events, and perhaps miRNAs and other ncRNAs, may contribute to varying SLC6A3 activity. It is anticipated that a comprehensive map may play an instructive role in the development of individualized medicine for SLC6A3 -related brain disorders.
Future perspective
With a single copy number reporter vector available, we are for the first time able to measure overall promoter activity of 18-kb haplotypes in SLC6A3. Additionally, by investigating 17 drugs' effects, this study provides the most comprehensive regulatory information to date on this important brain gene. The pharmacological data have suggested that there are more transcription factors for this gene but there is no clue about what transcription factors are mediating the regulatory pathways (e.g., PACAP38) and where these transcription factors bind in the regulatory regions. By progressive deletions of the 18-kb reporter constructs, we are currently mapping novel cis-acting elements in the promoter regions and will further clone transcription factors that bind to polymorphic sites in these elements, delineating individual cis-acting elements. We will, therefore, better understand how SLC6A3 variants are differentially regulated through its promoter regions. This large-construct-guided approach can also be applied to the transcribed and downstream regions, generating a comprehensive regulatory map for individualized treatment of SLC6A3-related diseases.
Supplementary Material
Executive summary.
Background
SLC6A3 encodes the plasma membrane dopamine transporter that regulates dopamine transmission in the brain and is associated with more than ten different brain disorders.
Previous analyses have focused on regulation of downstream variable number tandem repeats (VNTRs) by a few chemicals or on haplotypic activity of the core promoter regions of SLC6A3.
pGL3eBelo1-based measurement of comprehensive haplotypic activity in SLC6A3
In order to analyze transcriptional activity of 18-kb chromosomal fragments in SLC6A3, a single copy number luciferase reporter vector, pGL3eBelo1, was created.
By using pGL3eBelo1, we demonstrated that two 18-kb regulatory haplotypes containing the entire 18-kb promoter regions and two known downstream VNTRs display up to 3.2-fold difference in SLC6A3 promoter activity.
Haplotypic difference in promoter activity varies in different expression systems.
SLC6A3 promoter regulation by six (35.3%) of the 17 drugs examined
Valproate, PACAP38, the PI3K inhibitor LY294002, the adenylyl cyclase activator forskolin and the protein kinase activator PMA all upregulated the promoter activity of the 18-kb regulatory haplotypes; the neurotoxin 6-OHDA downregulated the promoter activity.
The upregulations were both haplotype and cell type dependent. LY294002 and PMA could differentially regulate the two haplotypes, in a cell type-dependent manner.
First pathway map for SLC6A3 regulation
SLC6A3 was regulated by selected pathways, which involved 13 hubs.
Histone-deacetylase-related epigenetic regulation is a major pathway.
Conclusion & future perspective
Our pGL3eBelo1-based study with 17 drugs provided the most comprehensive data to date on regulation of SLC6A3 promoter activity and haplotype or cell-type dependence.
The 18-kb haplotypes showed a large difference in promoter activity, attributable to independent effect of promoter versus VNTRs; most pathways upregulated the promoters.
The pGL3eBelo1-based approach may help map novel cis-acting elements and build a signaling map of SLC6A3 regulation, guiding the development of individualized treatment of SLC6A3-related brain disorders.
Acknowledgments
This work was supported by US NIH grant R01DA021409 (to Z Lin). Y Zhao has received National Nature Science Foundation of China 81100956.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Coyle JT, Snyder SH. Antiparkinsonian drugs: inhibition of dopamine uptake in the corpus striatum as a possible mechanism of action. Science. 1969;166(3907):899–901. doi: 10.1126/science.166.3907.899. [DOI] [PubMed] [Google Scholar]
- 2.Lin Z, Canales JJ, Bjorgvinsson T, et al. Monoamine transporters: vulnerable and vital doorkeepers. Prog Mol Biol Transl Sci. 2011;98:1–46. doi: 10.1016/B978-0-12-385506-0.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannon MJ, Michelhaugh SK, Wang J, Sacchetti P. The human dopamine transporter gene: gene organization, transcriptional regulation, and potential involvement in neuropsychiatric disorders. Eur Neuropsychopharmacol. 2001;11(6):449–455. doi: 10.1016/s0924-977x(01)00122-5. [DOI] [PubMed] [Google Scholar]
- 4.Berridge KC, Aldridge JW, Houchard KR, Zhuang X. Sequential super-stereotypy of an instinctive fixed action pattern in hyper-dopaminergic mutant mice: a model of obsessive compulsive disorder and Tourette's. BMC Biol. 2005;3:4. doi: 10.1186/1741-7007-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pecina S, Cagniard B, Berridge KC, Aldridge JW, Zhuang X. Hyperdopaminergic mutant mice have higher “wanting” but not “liking” for sweet rewards. J Neurosci. 2003;23(28):9395–9402. doi: 10.1523/JNEUROSCI.23-28-09395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralph-Williams RJ, Paulus MP, Zhuang X, Hen R, Geyer MA. Valproate attenuates hyperactive and perseverative behaviors in mutant mice with a dysregulated dopamine system. Biol Psychiatry. 2003;53(4):352–359. doi: 10.1016/s0006-3223(02)01489-0. [DOI] [PubMed] [Google Scholar]
- 7.Salahpour A, Ramsey AJ, Medvedev IO, et al. Increased amphetamine-induced hyperactivity and reward in mice overexpressing the dopamine transporter. Proc Natl Acad Sci USA. 2008;105(11):4405–4410. doi: 10.1073/pnas.0707646105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuang X, Oosting RS, Jones SR, et al. Hyperactivity and impaired response habituation in hyperdopaminergic mice. Proc Natl Acad Sci USA. 2001;98(4):1982–1987. doi: 10.1073/pnas.98.4.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath MC, Kovacs GG, Kovari V, Majtenyi K, Hurd YL, Keller E. Heroin abuse is characterized by discrete mesolimbic dopamine and opioid abnormalities and exaggerated nuclear receptor-related 1 transcriptional decline with age. J Neurosci. 2007;27(49):13371–13375. doi: 10.1523/JNEUROSCI.2398-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little KY, McLaughlin DP, Zhang L, et al. Brain dopamine transporter messenger RNA and binding sites in cocaine users: a postmortem study. Arch Gen Psychiatry. 1998;55(9):793–799. doi: 10.1001/archpsyc.55.9.793. [DOI] [PubMed] [Google Scholar]
- 11.Bannon MJ, Pruetz B, Manning-Bog AB, et al. Decreased expression of the transcription factor NURR1 in dopamine neurons of cocaine abusers. Proc Natl Acad Sci USA. 2002;99(9):6382–6385. doi: 10.1073/pnas.092654299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Segal DM, Moraes CT, Mash DC. Dopamine transporter mRNA in autopsy studies of chronic cocaine users. Brain Res Mol Brain Res. 1999;73(1–2):181–185. doi: 10.1016/s0169-328x(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Huang C, Tong J, Hong WC, Liu YJ, Xia XG. Temporal expression of mutant LRRK2 in adult rats impairs dopamine reuptake. Int J Biol Sci. 2011;7(6):753–761. doi: 10.7150/ijbs.7.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrie EC, Veith RC, Szot P. Bupropion and desipramine increase dopamine transporter mRNA expression in the ventral tegmental area/substantia nigra of rat brain. Prog Neuropsychopharmacol Biol Psychiatry. 1998;22(5):845–856. doi: 10.1016/s0278-5846(98)00044-x. [DOI] [PubMed] [Google Scholar]
- 15.Patterson TA, Brot MD, Zavosh A, Schenk JO, Szot P, Figlewicz DP. Food deprivation decreases mRNA and activity of the rat dopamine transporter. Neuroendocrinology. 1998;68(1):11–20. doi: 10.1159/000054345. [DOI] [PubMed] [Google Scholar]
- 16.Figlewicz DP, Szot P, Chavez M, Woods SC, Veith RC. Intraventricular insulin increases dopamine transporter mRNA in rat VTA/substantia nigra. Brain Res. 1994;644(2):331–334. doi: 10.1016/0006-8993(94)91698-5. [DOI] [PubMed] [Google Scholar]
- 17.Dluzen DE, McDermott JL. Neuroprotective role of estrogen upon methamphetamine and related neurotoxins within the nigrostriatal dopaminergic system. Ann NY Acad Sci. 2000;914:112–126. doi: 10.1111/j.1749-6632.2000.tb05189.x. [DOI] [PubMed] [Google Scholar]
- 18.Salkovic-Petrisic M, Lackovic Z. Intracerebroventricular administration of betacytotoxics alters expression of brain monoamine transporter genes. J Neural Transm. 2003;110(1):15–29. doi: 10.1007/s00702-002-0773-9. [DOI] [PubMed] [Google Scholar]
- 19.Figlewicz DP, Brot MD, McCall AL, Szot P. Diabetes causes differential changes in CNS noradrenergic and dopaminergic neurons in the rat: a molecular study. Brain Res. 1996;736(1–2):54–60. doi: 10.1016/0006-8993(96)00727-5. [DOI] [PubMed] [Google Scholar]
- 20.Shumay E, Chen J, Fowler JS, Volkow ND. Genotype and ancestry modulate brain's DAT availability in healthy humans. PLoS ONE. 2011;6(8):e22754. doi: 10.1371/journal.pone.0022754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪▪.Shumay E, Fowler JS, Volkow ND. Genomic features of the human dopamine transporter gene and its potential epigenetic states: implications for phenotypic diversity. PLoS ONE. 2010;5(6):e11067. doi: 10.1371/journal.pone.0011067. Discusses epigenetic regulation of SLC6A3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannon MJ, Whitty CJ. Age-related and regional differences in dopamine transporter mRNA expression in human midbrain. Neurology. 1997;48(4):969–977. doi: 10.1212/wnl.48.4.969. [DOI] [PubMed] [Google Scholar]
- 23▪▪.Zhou Y, Michelhaugh SK, Schmidt CJ, Liu JS, Bannon MJ, Lin Z. Ventral midbrain correlation between genetic variation and expression of the dopamine transporter gene in cocaine-abusing versus non-abusing subjects. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00391.x. (Epub ahead of print). Shows potential functionality of the 5′ variable number tandem repeat (VNTR) in the distal region of the SLC6A3 promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24▪.Hill M, Anney RJ, Gill M, Hawi Z. Functional analysis of intron 8 and 3′ UTR variable number of tandem repeats of SLC6A3: differential activity of intron 8 variants. Pharmacogenomics J. 2010;10(5):442–447. doi: 10.1038/tpj.2009.66. Shows functional Int8VNTR. [DOI] [PubMed] [Google Scholar]
- 25.Miller GM, Madras BK. Polymorphisms in the 3′-untranslated region of human and monkey dopamine transporter genes affect reporter gene expression. Mol Psychiatry. 2002;7(1):44–55. doi: 10.1038/sj.mp.4000921. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Zhou Y, Xiong N, Lin Z. Identification of an intronic cis-acting element in the human dopamine transporter gene. Mol Biol Rep. 2012;39(5):5393–5399. doi: 10.1007/s11033-011-1339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacchetti P, Brownschidle LA, Granneman JG, Bannon MJ. Characterization of the 5′-fanking region of the human dopamine transporter gene. Brain Res Mol Brain Res. 1999;74(1–2):167–174. doi: 10.1016/s0169-328x(99)00275-2. [DOI] [PubMed] [Google Scholar]
- 28▪▪.Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82(5):511–520. doi: 10.1016/s0888-7543(03)00142-3. Shows haplotypic difference in the core promoter activity of SLC6A3. [DOI] [PubMed] [Google Scholar]
- 29.Kelada SN, Costa-Mallen P, Checkoway H, et al. Dopamine transporter (SLC6A3) 5′ region haplotypes significantly affect transcriptional activity in vitro but are not associated with Parkinson's disease. Pharmacogenet Genomics. 2005;15(9):659–668. doi: 10.1097/01.fpc.0000170917.04275.d6. [DOI] [PubMed] [Google Scholar]
- 30.Bamne MN, Talkowski ME, Chowdari KV, Nimgaonkar VL. Functional analysis of upstream common polymorphisms of the dopamine transporter gene. Schizophr Bull. 2010;36(5):977–982. doi: 10.1093/schbul/sbp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Giessen E, de Win MM, Tanck MW, van den Brink W, Baas F, Booij J. Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med. 2009;50(1):45–52. doi: 10.2967/jnumed.108.053652. [DOI] [PubMed] [Google Scholar]
- 32.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. BMC Genet. 2005;6:55. doi: 10.1186/1471-2156-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dyck CH, Malison RT, Jacobsen LK, et al. Increased dopamine transporter availability associated with the 9-repeat allele of the SLC6A3 gene. J Nucl Med. 2005;46(5):745–751. [PubMed] [Google Scholar]
- 34▪.Guindalini C, Howard M, Haddley K, et al. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proc Natl Acad Sci USA. 2006;103(12):4552–4557. doi: 10.1073/pnas.0504789103. First evidence for functional Int8VNTR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Z, Zhao Y, Chung CY, et al. High regulatability favors genetic selection in SLC18A2, a vesicular monoamine transporter essential for life. FASEB J. 2010;24(7):2191–2200. doi: 10.1096/fj.09-140368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uejima H, Lee MP, Cui H, Feinberg AP. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet. 2000;25(4):375–376. doi: 10.1038/78040. [DOI] [PubMed] [Google Scholar]
- 37.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 38.Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14(13):1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones SR, Gainetdinov RR, Hu XT, et al. Loss of autoreceptor functions in mice lacking the dopamine transporter. Nat Neurosci. 1999;2(7):649–655. doi: 10.1038/10204. [DOI] [PubMed] [Google Scholar]
- 40.Huang SY, Chen HK, Ma KH, et al. Association of promoter variants of human dopamine transporter gene with schizophrenia in Han Chinese. Schizophr Res. 2010;116(1):68–74. doi: 10.1016/j.schres.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Genro JP, Polanczyk GV, Zeni C, et al. A common haplotype at the dopamine transporter gene 5′ region is associated with attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1568–1575. doi: 10.1002/ajmg.b.30863. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Mill J, Sun B, et al. Association study of promoter polymorphisms at the dopamine transporter gene in attention deficit hyperactivity disorder. BMC Psychiatry. 2009;9:3. doi: 10.1186/1471-244X-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Bannon MJ. Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J Neurochem. 2005;93(2):474–482. doi: 10.1111/j.1471-4159.2005.03051.x. [DOI] [PubMed] [Google Scholar]
- 44.Son JH, Chun HS, Joh TH, Cho S, Conti B, Lee JW. Neuroprotection and neuronal differentiation studies using substantia nigra dopaminergic cells derived from transgenic mouse embryos. J Neurosci. 1999;19(1):10–20. doi: 10.1523/JNEUROSCI.19-01-00010.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fishman-Jacob T, Reznichenko L, Youdim MB, Mandel SA. A sporadic Parkinson disease model via silencing of the ubiquitin-proteasome/E3 ligase component SKP1A. J Biol Chem. 2009;284(47):32835–32845. doi: 10.1074/jbc.M109.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Michelhaugh SK, Bannon MJ. Valproate robustly increases Sp transcription factor-mediated expression of the dopamine transporter gene within dopamine cells. Eur J Neurosci. 2007;25(7):1982–1986. doi: 10.1111/j.1460-9568.2007.05460.x. [DOI] [PubMed] [Google Scholar]
- 47.Shizuya H, Birren B, Kim UJ, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89(18):8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartlett CW, Flax JF, Fermano Z, et al. Gene x gene interaction in shared etiology of autism and specific language impairment. Biol Psychiatry. 2012;72(8):692–699. doi: 10.1016/j.biopsych.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yacubian J, Sommer T, Schroeder K, et al. Gene–gene interaction associated with neural reward sensitivity. Proc Natl Acad Sci USA. 2007;104(19):8125–8130. doi: 10.1073/pnas.0702029104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma L, Brautbar A, Boerwinkle E, Sing CF, Clark AG, Keinan A. Knowledge-driven analysis identifies a gene–gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 2012;8(5):e1002714. doi: 10.1371/journal.pgen.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada Y, Kubo M, Ohmiya H, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44(3):302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frieling H, Romer KD, Scholz S, et al. Epigenetic dysregulation of dopaminergic genes in eating disorders. Int J Eat Disord. 2010;43(7):577–583. doi: 10.1002/eat.20745. [DOI] [PubMed] [Google Scholar]
- 53.Vucetic Z, Carlin JL, Totoki K, Reyes TM. Epigenetic dysregulation of the dopamine system in diet-induced obesity. J Neurochem. 2012;120(6):891–898. doi: 10.1111/j.1471-4159.2012.07649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54▪.Fuke S, Sasagawa N, Ishiura S. Identification and characterization of the Hesr1/Hey1 as a candidate trans-acting factor on gene expression through the 3′ non-coding polymorphic region of the human dopamine transporter (DAT1) gene. J Biochem. 2005;137(2):205–216. doi: 10.1093/jb/mvi020. First report of a transcription factor binding to a SLC6A3 polymorphic site. [DOI] [PubMed] [Google Scholar]
- 55.Fuke S, Minami N, Kokubo H, et al. Hesr1 knockout mice exhibit behavioral alterations through the dopaminergic nervous system. J Neurosci Res. 2006;84(7):1555–1563. doi: 10.1002/jnr.21062. [DOI] [PubMed] [Google Scholar]
- 56.Yang B, Chan RC, Jing J, Li T, Sham P, Chen RY. A meta-analysis of association studies between the 10-repeat allele of a VNTR polymorphism in the 3′-UTR of dopamine transporter gene and attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(4):541–550. doi: 10.1002/ajmg.b.30453. [DOI] [PubMed] [Google Scholar]
- 57.Joober R, Grizenko N, Sengupta S, et al. Dopamine transporter 3′-UTR VNTR genotype and ADHD: a pharmaco-behavioural genetic study with methylphenidate. Neuropsychopharmacology. 2007;32(6):1370–1376. doi: 10.1038/sj.npp.1301240. [DOI] [PubMed] [Google Scholar]
- 58.Hawi Z, Kent L, Hill M, et al. ADHD and DAT1: further evidence of paternal over-transmission of risk alleles and haplotype. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):97–102. doi: 10.1002/ajmg.b.30960. [DOI] [PubMed] [Google Scholar]
- 59.Sachs GS, Ice KS, Chappell PB, et al. Efficacy and safety of adjunctive oral ziprasidone for acute treatment of depression in patients with bipolar I disorder: a randomized, double-blind, placebo-controlled trial. J Clin Psychiatry. 2011;72(10):1413–1422. doi: 10.4088/JCP.09m05934. [DOI] [PubMed] [Google Scholar]
- 60.Li SC, Aggarwal SK. Estimation of resource utilisation difference between lithium and valproate treatment groups from the VALID study. J Med Econ. 2011;14(3):350–356. doi: 10.3111/13696998.2011.581321. [DOI] [PubMed] [Google Scholar]
- 61.Casey DE, Daniel DG, Tamminga C, et al. Divalproex ER combined with olanzapine or risperidone for treatment of acute exacerbations of schizophrenia. Neuropsychopharmacology. 2009;34(5):1330–1338. doi: 10.1038/npp.2008.209. [DOI] [PubMed] [Google Scholar]
- 62.Meltzer HY, Bonaccorso S, Bobo WV, Chen Y, Jayathilake K. A 12-month randomized, open-label study of the metabolic effects of olanzapine and risperidone in psychotic patients: influence of valproic acid augmentation. J Clin Psychiatry. 2011;72(12):1602–1610. doi: 10.4088/JCP.10m05997. [DOI] [PubMed] [Google Scholar]
- 63.Leon AC, Solomon DA, Li C, et al. Antiepileptic drugs for bipolar disorder and the risk of suicidal behavior: a 30-year observational study. Am J Psychiatry. 2012;169(3):285–291. doi: 10.1176/appi.ajp.2011.11060948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vieta E, Owen R, Baudelet C, McQuade RD, Sanchez R, Marcus RN. Assessment of safety, tolerability and effectiveness of adjunctive aripiprazole to lithium/valproate in bipolar mania: a 46-week, open-label extension following a 6-week double-blind study. Curr Med Res Opin. 2010;26(6):1485–1496. doi: 10.1185/03007991003779380. [DOI] [PubMed] [Google Scholar]
- 65.Glick ID, Bosch J, Casey DE. A double-blind randomized trial of mood stabilizer augmentation using lamotrigine and valproate for patients with schizophrenia who are stabilized and partially responsive. J Clin Psychopharmacol. 2009;29(3):267–271. doi: 10.1097/JCP.0b013e3181a443d0. [DOI] [PubMed] [Google Scholar]
- 66.Houston JP, Ahl J, Meyers AL, Kaiser CJ, Tohen M, Baldessarini RJ. Reduced suicidal ideation in bipolar I disorder mixed-episode patients in a placebo-controlled trial of olanzapine combined with lithium or divalproex. J Clin Psychiatry. 2006;67(8):1246–1252. doi: 10.4088/jcp.v67n0811. [DOI] [PubMed] [Google Scholar]
- 67.Sokolski KN, Denson TF. Adjunctive quetiapine in bipolar patients partially responsive to lithium or valproate. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27(5):863–866. doi: 10.1016/S0278-5846(03)00145-3. [DOI] [PubMed] [Google Scholar]
- 68.Tohen M, Chengappa KN, Suppes T, et al. Efficacy of olanzapine in combination with valproate or lithium in the treatment of mania in patients partially nonresponsive to valproate or lithium monotherapy. Arch Gen Psychiatry. 2002;59(1):62–69. doi: 10.1001/archpsyc.59.1.62. [DOI] [PubMed] [Google Scholar]
- 69.Geiger H, Pawar SA, Kerschen EJ, et al. Pharmacological targeting of the thrombomodulin-activated protein C pathway mitigates radiation toxicity. Nat Med. 2012;18(7):1123–1129. doi: 10.1038/nm.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.