Abstract
Development of multicellular organisms requires specification of diverse cell types. In plants, development is continuous and because plant cells are surrounded by rigid cell walls, cell division and specification of daughter cell fate must be carefully orchestrated. During embryonic and postembryonic plant development, the specification of cell types is determined both by positional cues and cell lineage. The establishment of distinct transcriptional domains is a fundamental mechanism for determining different cell fates. In this review, we focus on four examples from recent literature of switches operating in cell fate decisions that are regulated by transcriptional mechanisms. First, we highlight a transcriptional mechanism involving a mobile transcription factor in formation of the two ground tissue cell types in roots. Specification of vascular cell types is then discussed, including new details about xylem cell-type specification via a mobile microRNA. Next, transcriptional regulation of two key embryonic developmental events is considered: establishment of apical–basal polarity in the single-celled zygote and specification of distinct root and shoot stem cell populations in the plant embryo. Finally, a dynamic transcriptional mechanism for lateral organ positioning that integrates spatial and temporal information into a repeating pattern is summarized.
1. Introduction
Plant growth and development constitute a continuous process. The plant embryo does not contain most of the organs found in adult plants; instead, they have a simple structure composed of an embryonic root or radicle, one or two embryonic leaves or cotyledons, and a connecting stem or hypocotyl (Esau, 1977). Importantly, the two primary stem cell populations (meristems) are formed during embryogenesis, which will give rise to all adult organs. Thus, growth and development are largely postembryonic with new organs being forming throughout the plant’s entire life. In addition, plants do not have a fixed body plan so individuals of the same species can have a variable number of organs. In contrast, development in most animals is more finite; the number of organs is strictly defined and organ formation is generally limited to embryogenesis. Plants are also exposed to a vast range of environmental conditions during development. As immobile organisms, plants must integrate endogenous and exogenous cues and respond in an accurate and timely manner to form and pattern organs.
Organ patterning relies on specification of different cell types and tissues with each cell type having specialized features. Plant cells are constrained by interconnected cell walls that prevent cellular movement. Therefore, a plant cell must integrate information about its relative position from neighboring cells and the lineage from which it is derived to make cell fate decisions. Thus, cues required for cell fate specification can be positional, inherited, or rely on both the ancestry and the position of the cell. For instance, positional cues in plants include hormones, short peptides, mobile transcriptional regulators, and, as recently reported, microRNAs. In addition, some transcription factors (TFs) are differentially inherited and/or expressed after cell division, which can also establish distinct transcriptional domains that determine new cell fates. Here, we discuss several recent examples of transcriptional regulators that act as switches for cell fate specification during development.
2. Cell Fate Specification in the Cortex–Endodermal Cell Lineage
The outer tissues of the Arabidopsis thaliana root are organized in concentric cell layers around the stele. From the stele outward there are two ground tissue layers, with the endodermis immediately adjacent to the stele followed by the cortex and the exterior epidermal layer (Fig. 9.1A). The two ground tissue cell types are generated through asymmetric division of a single stem cell lineage, the cortex/endodermal initial (CEI). The CEI undergoes a transverse asymmetric division to renew itself and generate a CEI daughter (CEID). The CEID then undergoes another asymmetric cell division, this time in a longitudinal orientation, to produce one cell each in the endodermal and cortical cell layers (Fig. 9.1B; Benfey et al., 1993; Di Laurenzio et al., 1996; Dolan et al., 1993; Scheres et al., 1994). The asymmetric division of the CEID and the switch to endodermal or cortical fate in the daughter cells are regulated by a transcriptional mechanism that links patterning, development, and the cell cycle.
Figure 9.1.
Ground tissue formation in the root. (A) Schematic of a longitudinal section of the Arabidopsis root tip. Individual or groups of cell types are depicted in different colors. (B) Cut away of the ground tissue cell types from (A) with an emphasis on the asymmetric divisions and cellular defects in short root (shr) and scarecrow (scr) mutants. (B, upper panel) Ground tissue formation in wild type. (B upper panel, left to right) The CEI (dark green) divides transversely (white arrowheads) to regenerate itself and produce the CEID (light green). The CEID then divides longitudinally (black arrowhead) to generate the cells of the endodermis (blue) and cortex (yellow). (B, center panel) Ground tissue formation in short root. In the absence of the longitudinal asymmetric cell division, a single layer of ground tissue with cortical cell features (yellow with white stripes) forms. (B, lower panel) Ground tissue formation in scarecrow. The longitudinal asymmetric cell division also does not occur; however, the single layer of ground tissue exhibits both endodermal and cortical cell features (yellow with blue stripes). (C) Schematic of a portion of the Arabidopsis root tip, focusing on the localization of SHR mRNA, SHR protein and SCR mRNA and protein. Yellow arrows depict SHR protein movement from the stele into the adjacent cell layer. Note that SHR and SCR proteins are colocalized in the nuclei (small circles within the cells) of the adjacent cell layer. This figure was adapted from Petricka et al. (2009).
The asymmetric division of the CEID is regulated by the activity of two TFs, SHORT ROOT (SHR) and SCARECROW (SCR). These proteins are both members of the GRAS family of transcriptional regulators (Benfey et al., 1993; Di Laurenzio et al., 1996; Helariutta et al., 2000; Pysh et al., 1999). shr and scr mutant plants each have only a single layer of ground tissue because the CEID fails to undergo the longitudinal asymmetric cell division. In shr mutants, the single ground tissue layer has some cortical cell features but no endodermal features. Whereas in scr, the mutant layer has both endodermal and cortical cell features (Fig. 9.1B; Benfey et al., 1993; Di Laurenzio et al., 1996; Helariutta et al., 2000). Because these two genes are both required for CEID division but only SHR appears to be necessary for specification of endodermal fate, SHR was predicted to be upstream of SCR in the ground tissue developmental pathway. This hypothesis was confirmed by epistasis of shr to scr and the decrease in SCR expression in shr plants (Helariutta et al., 2000). These data indicate that SHR and SCR act together to regulate the asymmetric CEID division and specification of endodermal and cortical fates in the daughter cells.
2.1. SHR reveals a twist on transcriptional regulation of cell fate specification
Differences in SHR mRNA and SHR protein localization suggested that a nonautonomous transcriptional mechanism functioned to specify the endodermis. SHR transcripts are restricted to the stele, whereas SHR protein is found both in the stele and the immediately adjacent cell layer, which includes the CEI, CEID, and endodermis (Fig. 9.1C). The subcellular localization of SHR changes between different root tissues: in the stele SHR is nuclear and cytoplasmic, whereas in the adjacent layer SHR is nuclear. Ectopic expression of SHR in other root cell types results in formation of endodermal features (Helariutta et al., 2000; Nakajima et al., 2001). These results revealed that TFs were not strictly cell autonomous but could function as intercellular signaling molecules with the ability to directly activate a new transcriptional program in neighboring cells.
The capacity for SHR to function as a positional cue as well as a transcriptional switch in root patterning is evident when SHR is ectopically expressed (Helariutta et al., 2000; Nakajima et al., 2001; Sena et al., 2004). Ectopic expression of SHR in the adjacent cell layer increased the number of cell layers between the epidermis and stele; these layers exhibited cellular markers of endodermal identity, including expression of SCR (Nakajima et al., 2001). Additionally, ectopic SHR expression in cell types outside the stele, such as the epidermis, can induce these cells to exhibit endodermal features (Sena et al., 2004). This suggests that SHR movement from the stele is not a prerequisite for its activity in specifying endodermal cell fate. However, SHR movement across only one cell layer is highly regulated appearing to require both cytoplasmic and nuclear localization prior to trafficking out of the stele, suggesting that regulation of SHR movement is important for its function (Gallagher and Benfey, 2009; Gallagher et al., 2004). SCR was implicated in limiting SHR movement because ectopic SHR movement and decreased nuclear localization were observed in the scr mutant background (Cui et al., 2007; Heidstra et al., 2004). Additionally, SHR expressed in the epidermis was more cytoplasmic and showed movement into the adjacent (inner) mutant cell layer in scr mutants (Sena et al., 2004). These observations reveal the interdependent nature of SHR and SCR in root radial patterning, including an additional role for SCR in endodermal specification via modulation of SHR intercellular movement and subcellular localization.
The observation that cell-specific factors could limit SHR movement provided a straightforward mechanism for forming a single endodermal tissue layer. In vivo molecular evidence supporting this hypothesis, with SCR as a key component in this process, has been obtained. SHR and SCR proteins physically interact and have many common transcriptional targets suggesting that they form a transcriptional regulatory complex (Cui et al., 2007; Levesque et al., 2006; Sozzani et al., 2010). Additionally, binding of SHR and SCR to the SCR promoter was detected by chromatin immunoprecipitation experiments. SCR expression is reduced in both shr and scr mutants suggesting that SCR expression is controlled by a SHR–SCR-dependent positive feedback loop (Cui et al., 2007; Levesque et al., 2006). This predicts that relatively high levels of SCR would be necessary to interact with SHR to sequester it into the nucleus. This prediction was examined using RNA interference lines with variable SCR expression levels. Plants with reduced SCR mRNA levels revealed SHR movement into adjacent cell layers, which led to ectopic endodermal cell fate specification (Cui et al., 2007). These data indicate that SCR functions to limit SHR movement via nuclear sequestration to one cell layer outside the stele, therefore preventing excess endodermis formation.
Together, these observations have resulted in the following model for SHR–SCR function in asymmetric cell division and cell fate specification in the ground tissue. First, both nuclear and cytoplasmic localization of SHR in the stele promotes SHR movement to the adjacent cell layer (Gallagher and Benfey, 2009). In the adjacent cell layer, SHR interacts with SCR and is nuclear-localized. The SHR–SCR complex activates SCR transcription forming a positive feedback loop that sequesters all the SHR protein in the nuclei, thereby restricting endodermal cell fate specification to a single layer (Cui et al., 2007). SHR and SCR then activate transcription of downstream targets, which leads to the asymmetric division in the ground tissue initials and specification of the endodermal cell layer (Cui et al., 2007; Levesque et al., 2006). In addition, other players, such as the zinc finger proteins JACKDAW, MAGPIE, and NUTCRACKER, have been tied to the SHR/SCR model for regulation of asymmetric division of the CEID cells (Levesque et al., 2006; Welch et al., 2007).
Recently, this spatial model for asymmetric cell division and endodermal specification has been expanded to include temporal components. Tissue-specific inducible versions of SHR and SCR were utilized to examine the temporal progression of asymmetric cell divisions and the changes in gene expression induced by these proteins. The timing of cell division after SHR or SCR induction coincided with induced expression of direct target genes. Remarkably, a component of the cell cycle machinery, a D-type cyclin, is directly regulated by SHR and SCR and involved in asymmetric CEID division providing an unexpected direct link between patterning and the cell cycle (Sozzani et al., 2010). Thus, the model for regulating CEI/CEID cell division likely involves more intricate transcriptional regulatory complexes and is possibly more tunable than previously understood. Another open question involves the shutdown of the SHR/SCR transcriptional switch following the asymmetric division of the CEID. Why does not SHR movement into the endodermal cells cause them to divide like the CEID, forming additional cell layers? This suggests that the transcriptional network mediated by SHR and SCR is a dynamic switch that functions to integrate ancestry and positional cues in ground tissue development.
3. Cell Fate Switches During Vascular Tissue Development
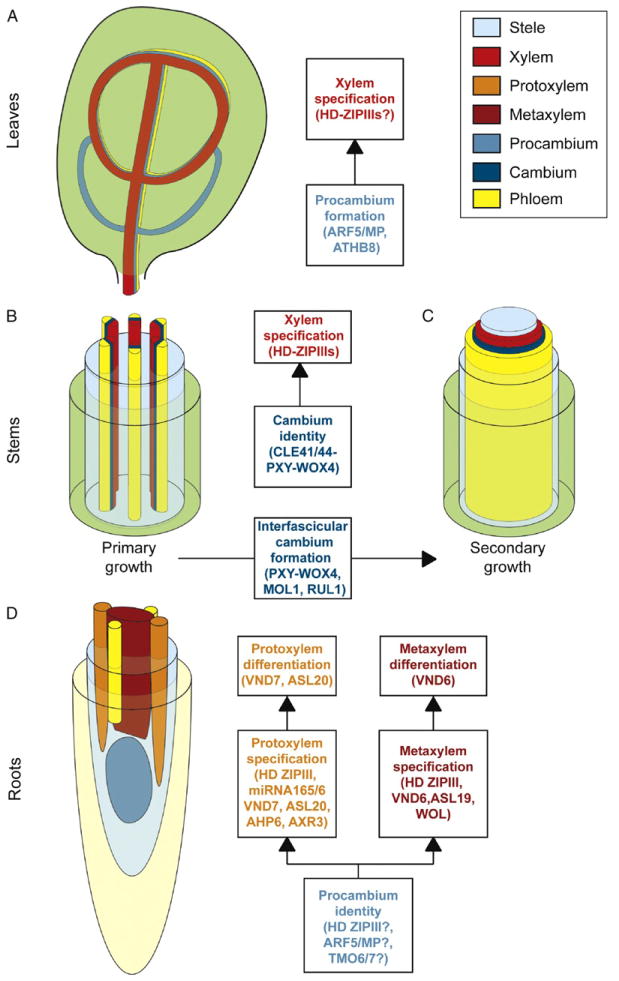
Plant vasculature comprises many different tissues with the predominant ones being the xylem and phloem (Brady et al., 2007; Esau, 1977). Xylem is the water-conducting tissue, while phloem specializes in nutrient transport. In above and below ground organs, the spatial organization of the xylem and phloem is different (Fig. 9.2). In roots, these tissues are typically arranged in a central cylinder while in the above ground organs they are arranged in bundles that are stereotypic in number and disposition. In the Arabidopsis root, primary xylem develops from vascular stem cells (procambial cells; Ohashi-Ito and Fukuda, 2010) and is made up of a single row of cells that extends across the central vascular cylinder (Figs. 9.2D and 9.3A). In addition, the two most peripheral xylem cells, which are in contact with the pericycle, are subsequently specified into protoxylem with spiral thickenings of the secondary cell walls. The remaining cells, in the central part of the row, form metaxylem with pitted secondary cell walls. Recent findings in Arabidopsis have identified novel TFs (Cano-Delgado et al., 2010; Zhang et al., 2011) that confer different xylem cell identities and appear to act as a multistep transcriptional switch that integrates signals and positional information from surrounding tissues (Fig. 9.2).
Figure 9.2.
Schematic of vascular patterning in Arabidopsis. (A) Patterning of leaf vasculature occurs through establishment of procambial cells and subsequent specification of vascular tissues from these cells. Regulators of procambium formation during venation are indicated. (B) Vascular tissues in stems are organized into vascular bundles. These bundles are comprised of xylem toward the inside and phloem toward the outside separated by cambial cells. Xylem is specified from cambium by specific regulators that switch cell fate. (C) During secondary growth, a ring of vascular tissue is generated through formation of cambium that closes the spaces between bundles. Common regulators during primary and secondary growth are indicated. (D) Vascular tissues in the root are organized in a central cylinder. Xylem constitutes a symmetry arch with metaxylem being specified toward the center of the arch and two protoxylem poles toward the outside. Regulators of xylem fate act as multistep switch specifying metaxylem and protoxylem.
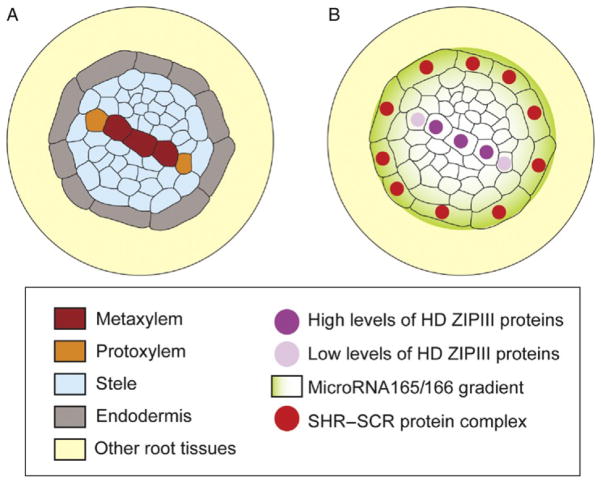
Figure 9.3.
Non-cell autonomous specification of xylem cell types by SHORT ROOT and SCARECROW. (A) Schematic of metaxylem and protoxylem tissues in a transverse section of the Arabidopsis root. Note that the endodermis forms a concentric layer of cells, whereas protoxylem and metaxylem constitute a single row of internal cells surrounded by other cells of the stele. (B) Specification of protoxylem versus metaxylem involves movement of SHORT ROOT to the endodermis where, together with SCARECROW, activates the microRNA165 and 166. The microRNA165/166 then travels back to the stele forming a gradient that targets the transcripts of CLASS III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIPIII) transcription factors. This generates different levels of HD-ZIPIII proteins that determine cell fate in a dose-dependent manner. Low levels of HD-ZIPIII specify protoxylem and high levels specify metaxylem.
Xylem can also be formed during secondary development by the activity of the fascicular and interfascicular cambium (Agusti et al., 2011). Secondary xylem is normally more complex than primary xylem, however, in both xylem types, water conduction is carried out by the tracheary elements: vessels and tracheids (Esau, 1977). In Arabidopsis, secondary vascular growth is observed in the root, hypocotyl, and inflorescence stems (Zhang et al., 2011), although it is more typically associated with perennial plants, like Poplar. Remarkably, recent studies indicate that there are conserved regulatory mechanisms for vascular development in Arabidopsis and Poplar, despite their evolutionary distance (Zhang et al., 2011).
3.1. Specification of procambium and xylem identity
During Arabidopsis postembryonic root development, xylem specification requires a set of five homeodomain leucine zipper III (HD-ZIPIII) TFs: PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), CORONA (CNA), and ATHB8. Seedlings with loss-of-function (LOF) mutations for all the HD-ZIPIIIs TFs fail to form xylem, indicating that these regulators determine de novo xylem formation (Carlsbecker et al., 2010). In addition, the quintuple HD-ZIPIII mutant appears to have broader morphological defects including reduced vascular cell number, which suggests that HD-ZIPIIIs might also regulate maintenance of procambium identity. In agreement with these functions, HD-ZIP III transcripts are accumulated in the root procambium, and their expression patterns overlap in those cells that will give rise to xylem (Carlsbecker et al., 2010; Miyashima et al., 2011). PHB and REV have broad expression in the stele and PHV, CNA, and ATHB8 are more specifically expressed in the xylem precursor cells; however, phv cna athb8 triple mutants still make xylem (Carlsbecker et al., 2010). This suggests that additional regulators participate in xylem cell fate specification or are sufficient for determination of procambium identity.
In the shoot, the HD-ZIPIIIs have been also shown to be involved in the specification of xylem (Ilegems et al., 2010), and ATHB8 has been proposed to have a role in procambium formation during leaf venation and in interfascicular cambium formation (Agusti et al., 2011; Donner et al., 2009; Ohashi-Ito and Fukuda, 2010). ATHB8 and the TF AUXIN RESPONSIVE FACTOR 5/MONOPTEROS (ARF5/MP) are necessary for procambial cell identity during leaf venation and ATHB8 is a direct target of ARF5/MP. These genes are part of a feedback loop that requires localized transport and signaling of the plant hormone auxin (Donner et al., 2009; Ohashi-Ito and Fukuda, 2010). As expected for regulators of pro-cambium specification, mutations in ARF5/MP cause a strong reduction in the number of leaf vascular bundles and athb8 LOF mutants appear to have defects in selection of cells that will acquire preprocambial state. However, the double mutant athb8 arf5/mp exhibits only a slight reduction in vein pattern complexity compared to mp/arf5 single mutants, while single athb8 mutants do not show detectable changes in leaf vein patterns (Donner et al., 2009). Thus, additional regulators redundant with ATHB8 and downstream of MP are likely involved in procambial cell determination. As ATHB8 participates in cambium and/or xylem identity in both shoot and roots, it is possible that other regulators, such as ARF5/MP might be also shared between shoots and roots to specify vascular tissues. Future studies might reveal a role for MP and its downstream targets in establishment of procambial cell identity in root tissues. In support of this, several MP direct targets, such as TARGET OF MP 7 (TMO7) and TMO6, which encode a bHLH and a Dof TF, respectively, are preferentially expressed in procambial cells and their precursors in the embryonic root (Schlereth et al., 2010). It is thus, tempting to speculate that these TFs have a role in regulating root vascular fates.
In another pathway, the KANADI genes, which are expressed in phloem, repress procambium identity. This has been proposed to occur by regulation of auxin transport, likely mediated by repression of the transporter PIN-FORMED 1 (PIN1). Ectopic expression of KANADI 1 in provascular cells represses the activity and polar localization of PIN1 (Ilegems et al., 2010). Supporting a role for plant hormones in regulation of vascular cell fates, xylem transported auxin and its interaction through a negative feedback loop with cytokinin, which is in turn transported through the phloem, is required to correctly specify vascular tissues (Bishopp et al., 2011a, b). Therefore, specification of procambial cells appears to require the interplay of different types of TFs that not only regulate cell identity but also procambium cell number by promoting differentiation into xylem or phloem.
3.2. Signaling pathways involved in maintenance of cambium identity during primary and secondary growth
During primary growth, the receptor-like kinase PHLOEM INTERCALATED WITH XYLEM (PXY) and the TF WUSCHEL-RELATED HOMEOBOX 4 (WOX4) are expressed in procambium and cambium and respond to CLE41/44 peptides secreted from adjacent phloem tissues. In pxy mutants, procambial cells differentiate into xylem, while in wox4 mutants, procambial cells are reduced to a single layer. These results indicate that procambial cells fail to proliferate and/or self-renew (Hirakawa et al., 2010, 2011). In addition, cle41 mutants have fewer procambial cells and exogenous application of CLE41 inhibits xylem specification from procambial cells. Therefore, CLE41/44-PXY-WOX4 signaling system maintains procambial/cambial identity by using positional information from adjacent (phloem) tissues to activate cell proliferation and repress xylem cell fate.
During secondary growth, differentiated cells are respecified and change their identity to become interfascicular cambium. In the shoot, the interfascicular cambium is formed between the primary vascular bundles generating a ring of cambium that will specify xylem and phloem toward the inside and outside, respectively (Fig. 9.2). This secondary growth coordinately enlarges shoot girth. Genes involved in primary growth also have roles in secondary growth (Agusti et al., 2011; Hirakawa et al., 2011). Likewise, PXY, WOX4, and ATHB8 expression is detected in the fascicular or interfascicular cambium, and pxy mutants fail to establish a closed cambium ring in the stem. In addition, two novel receptor-like kinases have been identified to be involved in secondary growth (Agusti et al., 2011). MORE LATERAL GROWTH 1 (MOL1) has been proposed to repress cambium formation, while REDUCED IN LATERAL GROWTH 1 (RUL1) has been predicted to function as an activator in the process. Genetic and expression analyses showed that MOL1 functions in the same pathway as PXY and WOX4 while RUL1 likely works independently of PXY. Therefore, it appears that a common set of players are required for specification of cambial cells and/or maintenance of their identity during primary and secondary growth.
3.3. Xylem specification requires autonomous and nonautonomous transcriptional regulators
A subsequent step in xylem development involves differentiation into either protoxylem or metaxylem (Fig. 9.3A). Surprisingly, the endodermal regulators SHR and SCR function non-cell autonomously in controlling protoxylem specification in the stele (Carlsbecker et al., 2010; Miyashima et al., 2011). In shr and scr mutants, all xylem cells incorrectly differentiate into metaxylem. SCR is specifically expressed in the endodermis, while SHR is present both in vascular tissues and in the endodermis. However, only endodermis-specific activity of SHR is required for xylem patterning. This was demonstrated by ectopic expression of SHR in the shr mutant followed by examination of protoxylem formation. When SHR was expressed in the ground tissue of shr, protoxylem formation was restored in the stele, whereas SHR expression in the stele did not rescue xylem patterning. In addition, it was shown that this developmental mechanism requires movement of the microRNA165/166 from the endodermis to the stele where it targets the aforementioned HD-ZIP III TFs to specify protoxylem (Fig. 9.3B).
This regulatory loop was unraveled through the identification of a new allele of PHB that has a point mutation in the microRNA165/166 binding site. This gain-of-function mutant fails to form protoxylem. A connection with SHR was made through the observation that the PHB protein encoded by the allele resistant to microRNA165/166 degradation was broadly expressed both in shr and wild-type roots; in contrast to the normal PHB protein, whose expression was reduced in wild-type plants but not in shr mutants. It was also shown that microRNA165a and 166b are direct targets of SHR and SCR and that their endodermal expression largely depended on these two TFs. In agreement with the hypothesized movement and non-cell autonomous function of microRNA165/166, their activity was found to be high in the stele although they are generated in the endodermis. Further, ectopic expression of microRNA165 in the mutant ground tissue layer of shr rescued protoxylem formation in the stele. Thus, SHR activates microRNA165/166 in the endodermis, the microRNA then moves into the stele to restrict PHB protein accumulation to the central part of the vascular cylinder, where metaxylem is specified. In contrast, lower levels of PHB protein in the outer xylem cells specify protoxylem.
However, protoxylem cell fate cannot be explained solely by PHB protein accumulation, as phb LOF mutants do not show defects in protoxylem fate specification. This is because the other four HD-ZIPIIIs (PHV, REV, CNA, and ATHB8) are also targets of the microRNA165/166 and act redundantly with PHB in the specification of metaxylem and protoxylem. All HD-ZIPIIIs have been shown to be directly involved in protoxylem/metaxylem specification through different combinations of LOF mutants that show ectopic formation of protoxylem instead of metaxylem. For instance, athb8 phb as well as any combination of quadruple mutations show ectopic protoxylem formation. Further support for their redundant role in protoxylem/metaxylem specification is provided by the quadruple mutant phb phv rev shr, which rescues the xylem patterning defects of shr (Carlsbecker et al., 2010; Miyashima et al., 2011).
HD-ZIPIIIs are not the only regulators of xylem fate that have been identified. Other regulators of protoxylem/metaxylem specification include WOODENLEG (Mahonen et al., 2000) and ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (Mahonen et al., 2006) in the cytokinin signaling pathway; AUXIN RESISTANT 3 (Bishopp et al., 2011a), in the auxin signaling pathway, as well as a group of seven TFs designated as VASCULAR-RELATED NAC-DOMAIN (VND; Kubo et al., 2005; Zhang et al., 2011). The VND TFs were identified through microarray analyses of cells induced to differentiate into xylem tracheary elements under specific in vitro conditions. Expression analyses of these TFs showed that they were specifically expressed in vascular tissues. Intriguingly, VND6 expression was restricted to root metaxylem precursor cells, while VND7 was expressed in immature protoxylem cells. The ectopic expression of VND6 and VND7 switched the fate of various cell types into metaxylem and protoxylem, respectively. Based on morphology, VND6 specified xylem elements with reticulate and/or pitted wall thickening similar to metaxylem, whereas VND7 produced xylem cells with annular and/or spiral wall thickening similar to protoxylem. However, vnd6 and vnd7 mutants did not show obvious morphological defects, which may be attributed to genetic redundancy with other VNDs or other regulators (Kubo et al., 2005).
Transcriptional analyses revealed that VND6 and VND7 regulate genes involved in tracheary element specification, such as secondary cell wall formation and programmed cell death genes. Additionally, VND7 also regulates proteolytic enzyme encoding genes (Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Further examination of these data might reveal the molecular basis of protoxylem and metaxylem identity and their subsequent differentiation. In addition, a number of secondary cell wall formation genes identified as regulated by VND6 and VND7 were different from those downstream of the xylem fiber regulators, SECONDARY WALL-ASSOCIATED NAC-DOMAIN PROTEIN 1 (SND1) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR 3 (NST3; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Fibers and tracheary elements constitute distinct xylem types; however, both require secondary cell wall formation during their differentiation process. The VND6/7 transcriptomic analyses suggest that unique genes or pathways may be functioning in secondary cell wall formation in these two xylem types. Finally, ASYMMETRIC LEAVES 2 LIKE (ASL) 19/LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 30 and ASL20/LBD18 are TFs downstream of VND6 and VND7 (Soyano et al., 2008). ASL19 and ASL20 are expressed in immature tracheary elements and their expression depends on VND6 and VND7. Ectopic expression of ASL19 and ASL20 specifies tracheary elements similar to VND6 and 7. For ASL20, this appears to occur not only by activation of a number of VND downstream targets but also, unexpectedly, by activation of VND7 expression itself. Thus, ASL20 and VND7 appear to function in a regulatory feedback loop that is able to specify xylem fate.
In conclusion, specification of different xylem cell fates is regulated by many transcriptional regulators functioning in different pathways and linked, in some cases, to non-cell autonomous regulators signaling from adjacent tissues. This tight regulation likely ensures proper patterning of the water-conducting tissues, which are vital for plant survival.
4. Transcriptional Regulation of Apical–Basal Cell Fate Determination after Zygotic Division in Arabidopsis
Embryogenesis is the process by which a unicellular zygote undergoes elaborate changes in cell number, fate, and morphology to ultimately form the mature embryo of a multicellular organism. Initial establishment of polarity is a critical step in embryogenesis in many organisms and typically involves activation of distinct transcriptional programs to drive differential development of the two embryonic axes. In many organisms, the single-celled zygote undergoes an asymmetric division that is critical in establishing embryo polarity. Initial embryo polarity is typically anterior–posterior (head–tail) in animals and apical–basal (shoot–root) in plants.
In Arabidopsis, the egg cell is polarized with the nucleus and the majority of cytoplasm localized apically and vacuoles localized basally (Fig. 9.4A). After fertilization, the zygote dramatically elongates and, like the egg cell, the nucleus is localized apically and a large vacuole forms basally (Faure et al., 2002). The zygote then undergoes an asymmetric division to produce two daughter cells with distinctly different sizes and developmental fates. The smaller apical cell will give rise to a majority of the embryo (Fig. 9.4A). The basal cell will generate an extraembryonic support structure, the suspensor, which connects the embryo to the maternal tissue (Jurgens, 2001; Scheres et al., 1994). Given the similar polarity of the egg cell and zygote, it was unclear whether egg cell polarity was required for zygotic polarity. Additionally, the transcriptional mechanisms regulating specification of cell fate after the first zygotic cell division were unknown. Recent findings have begun to clarify some of these questions.
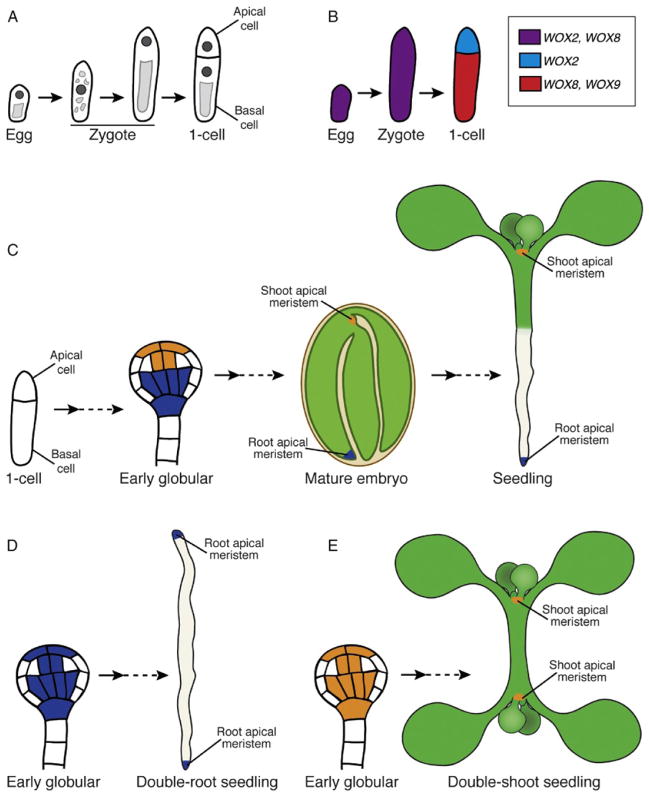
Figure 9.4.
Apical–basal polarity and specification of meristem fate in the Arabidopsis embryo through establishment of distinct transcriptional domains. (A) Schematic of early embryo development from the unfertilized egg cell through the first zygotic division. (A, left to right) The egg cell is polarized with the nucleus (dark gray) at the apical end and a vacuole at the basal end (light gray). After fertilization, the zygote is transiently symmetrical, then it elongates and is repolarized. The first zygotic division is asymmetric and producing an apical and basal cell with distinct fates. Note that embryo stages are based on the cell number in the apical domain, thus the first zygotic division results in one-cell stage embryo. (B) Schematic of the WOX2/8/9 gene expression patterns in the egg cell through the first zygotic division. (C) Schematic of developmental snapshots from the one-celled embryo to the seedling focused on specification of the root and shoot apical meristems. (C, left to right) Apical–basal polarity is established after the first zygotic division. By the early globular embryo stage, PLETHORAs (PLTs; blue) and the HD-ZIPIIIs (orange) expression is restricted to the domains that will give rise to the root (blue) and shoot (orange) apical meristems, respectively. In the mature embryo, these meristems are quiescent; however, become active after germination. The root meristem (blue) will give rise to all the cells of the primary root and the shoot meristem (orange) will give rise to all the cells of the aerial organs. (D) In the topless-1 (tpl-1) mutant, the PLT expression domain has expanded into more apical regions and HD-ZIPIII expression is absent leading to a double-root phenotype. (E) Expansion of HD-ZIPIII expression into more basal embryo regions represses PLT expression leading to a double-shoot phenotype.
4.1. Homeodomain TFs establish apical–basal polarity after the asymmetric zygotic division
WUSCHEL-RELATED HOMEOBOX (WOX) genes are a plant-specific family of TFs that demarcate distinct transcriptional domains along the apical–basal embryo axis starting with the first zygotic division. Expression of WOX2 and WOX8 is found in both the egg cell and single-celled zygote. Following the first asymmetric division of the zygote, expression of WOX2 and WOX8 becomes restricted to the apical and basal cells, respectively (Fig. 9.4B; Haecker et al., 2004). Expression of another WOX gene, WOX9, is then activated in the basal cell (Haecker et al., 2004; Wu et al., 2007). Consistent with their expression patterns, these WOX genes have critical roles in development of the apical and basal lineages. Embryos of wox2 mutants show cell division defects in the apical domain, while wox8 wox9 double mutants, have abnormal cell divisions in both the apical and basal cell lineages. Additionally, several markers of apical cell fate, including WOX2, are undetectable in wox8 wox9 embryos (Breuninger et al., 2008; Haecker et al., 2004; Wu et al., 2007). These results indicate WOX8/ WOX9 function in the basal lineage is required for apical lineage development via WOX2 expression and also suggests signaling between the lineages is important for early embryo patterning (Breuninger et al., 2008). Ectopic WOX2 expression in wox8 wox9 partially rescued later developmental defects in both the apical and basal lineages, indicating the apical defects in wox8 wox9 embryos can be attributed to loss of WOX2 expression. Unexpectedly, these zygotes showed defects earlier in development, they failed to elongate and the first zygotic division was more symmetrical. Together, these results reveal a critical role for these WOX genes in the establishment of distinct transcriptional domains that determine differential fate decisions in the apical and basal cells.
4.2. Transcriptional activation of WOX8/9 is required to break zygotic symmetry and specify the basal cell lineage
In animals, transcription is often inhibited during early embryogenesis with important developmental transcripts inherited from the gametes or maternally supplied after fertilization. In addition, transcript localization within the egg or zygote can be important for embryo polarity. To address whether plant embryos utilize similar mechanisms to regulate zygotic polarity, the mechanisms directly regulating WOX8 expression were investigated. This revealed a candidate transcriptional regulator of WOX8 expression called WRKY2. WRKY2 is a member of the plant-specific family of WRKY proteins, which are zinc finger-containing proteins. WOX8 and WRKY2 have overlapping expression patterns in the early embryo and wrky2 mutants show reduced expression of a WOX8 transcriptional reporter. The WOX8 promoter contains a known WRKY binding site, the W-box, and mutations in the W-box reduced expression of reporter constructs containing this cis-element. The WOX9 promoter also contains a W-box and, like WOX8, its expression is significantly reduced in wrky2 mutant embryos (Ueda et al., 2011). Together these results indicate that WRKY2 is required to activate or maintain expression of WOX8 and WOX9 in the basal cell lineage, as well as in the single-celled zygote.
Mutation of WRKY2 resulted in a more symmetrical first zygotic division although the wrky2 mutant egg cell maintained its polarity (Ueda et al., 2011). This phenotype suggested that characterizing WRKY2 function would provide insight into the relationship between egg cell and zygotic polarity. Careful examination revealed that after fertilization, the wild-type zygote is transiently symmetrical with the nucleus at the center and small vacuoles distributed throughout the cell. The single-celled zygote then expands and repolarizes with an apically localized nucleus and one large vacuole localized basally (Ueda et al., 2011). Similar to wild type, wrky2 mutants maintain egg cell polarity and the zygote shows transient symmetry. However, wrky2 zygotes fail to repolarize and the majority does not undergo an asymmetric division. These defects were attributed to WOX8 misregulation after fertilization as WOX8 expression independent of WRKY2 partially restores asymmetric division of wrky2 zygotes (Ueda et al., 2011). This indicates that polarity of the egg cell and zygote can be uncoupled and that zygotic polarity depends on the transcriptional activity of WRKY2 and WOX8.
WRKY2 is expressed in both gametes but appears to be required only zygotically. This raised questions about whether zygotic activity of WRKY2 is sufficient for normal embryogenesis. Reciprocal crosses between wrky2 and wild type revealed that normal embryogenesis occurred when functional WRKY2 was inherited from either gamete, indicating that its activity post-fertilization is sufficient for embryo patterning (Ueda et al., 2011). However, WRKY2 expression in both gametes suggests that WRKY2 protein or transcript may be inherited by the zygote. Because only the female gamete expresses WOX8, this expression can be used to determine if WRKY2 activates WOX8 transcription immediately after fertilization or whether WOX8 transcripts are maternally inherited. In zygotes from wild type or wrky2 egg cells fertilized with wrky2 pollen, carrying a WOX8 reporter, WOX8 expression was higher when a functional copy of WRKY2 was inherited from the egg cell (Ueda et al., 2011). This result indicates that WOX8 transcript inheritance is not necessary; instead WRKY2-driven transcriptional activation of WOX8 in the zygote is sufficient for normal embryogenesis. Thus, WRKY2 directly activates a transcription switch required for zygotic polarization and asymmetric division leading to differential cell fate decisions in the resulting daughter cells. How the apical and basal lineages shutdown expression of WOX8/9 and WOX2, respectively, remain open questions in the mechanics of this switch.
4.3. The impact of cell-to-cell signaling networks on early embryo patterning and its relationship to WOX transcriptional regulation
Despite the importance of the WOX transcriptional domains in determination of distinct apical and basal lineage development in the Arabidopsis embryo, plant cell fate decisions often rely more on positional cues. There are two main signaling pathways known to function in early embryogenesis in plants, a mitogen-activated protein (MAP) kinase cascade and a plant hormone signaling pathway. Mutation of the MAPKK kinase, YODA (YDA), results in embryos that do not elongate and the first zygotic division is more symmetrical, closely resembling embryos ectopically expressing WOX2 in a wox8 wox9 mutant background (Lukowitz et al., 2004). This similarity suggests that YDA and WOX8/9 function in a common pathway. However, yda wox8 wox9 triple mutant embryos arrest development after the first nearly symmetrical zygotic division (Breuninger et al., 2008). These data indicate that there are at least two independent pathways regulating asymmetry in the first zygotic division: a kinase cascade including YDA and a transcriptional mechanism via WOX8/9.
The plant hormone auxin has roles in a broad range of developmental processes (see Jenik et al., 2007; Leyser, 2006 for recent reviews) including embryo patterning. Directional movement of auxin from cell-to-cell is a key feature of this signaling pathway; a family of transmembrane efflux carriers, called PINs, mediates auxin movement through specific membrane localization (Galweiler et al., 1998; Petrasek et al., 2006). PIN7 is localized on the apical membrane of cells in the basal lineage indicating auxin movement toward the apical cells. Additionally, expression of a synthetic auxin-dependent transcriptional reporter, DIRECT REPEAT5 (DR5), occurs only in the apical lineage during early embryo stages (Friml et al., 2003). However, at early embryo stages the role for auxin is not entirely clear, as mutants with defects in auxin transport generally have phenotypes that are very weak and not fully penetrant (Friml et al., 2003), while wox8 wox9 mutants exhibit more severe embryo defects. Additionally, neither WOX2 nor WOX8 expression in early embryos is affected by exogenous auxin or by mutants affecting auxin transport suggesting that WOX2/8 expression is independent of auxin distribution (Breuninger et al., 2008; Ueda et al., 2011). These data suggest that regulation of apical/basal cell fate specification via the WOX genes occurs earlier and upstream of auxin in the early embryo.
5. Antagonism Between Transcriptional Regulators Specifies Two Distinct Stem Cell Populations in the Embryo
In plant embryos, apical–basal polarity is particularly important as the two main stem cell populations (meristems) are formed at opposite ends of this axis (Fig. 9.4C). Because plant growth and development is indeterminate, formation of the root and shoot apical meristems is essential for growth of all plant organs below and above ground, respectively. Two classes of transcriptional regulators have been identified as key regulators of root and shoot meristem fate specification in the embryo: the PLETHORA (PLT) family of AP2-domain proteins and the previously mentioned HD-ZIPIII TFs. There are four related PLT genes expressed in the root meristem of the embryo and mature root. Mutation of multiple PLTs results in rootless seedlings and embryo lethality; importantly, PLT overexpression induces formation of ectopic root meristems, indicating that PLTs have a master regulatory role in embryonic root meristem formation (Aida et al., 2004; Galinha et al., 2007). HD-ZIPIII proteins participate in various shoot developmental processes such as specification of the central domain of the shoot apical meristem and lateral organ polarity (reviewed in Engstrom et al., 2004). Similar to the PLT genes, only mutation of multiple HD-ZIPIII genes results in defective embryogenesis (Prigge et al., 2005). The crucial role for HD-ZIPIIIs in shoot fate specification and their antagonistic relationship with PLTs in the embryo was recently revealed through functional characterization of the TOPLESS (TPL) protein.
The key role for the transcriptional corepressor TPL in apical–basal embryo patterning is evident as tpl-1 mutants conditionally produce embryos in which the shoot meristem is replaced by a second root meristem (Fig. 9.4D; Long et al., 2006, 2002). In these tpl-1 embryos, PLT transcripts ectopically accumulated in the apical embryo domain, suggesting a role for TPL in repressing PLT expression apically. The double-root phenotype was not observed in tpl-1 plt1 plt2 triple mutants consistent with the master regulatory role of the PLTs in root meristem formation (Fig. 9.4D). Additionally, PLT1 and PLT2 were identified as direct targets of TPL indicating that PLT repression in the apical domain is required for normal embryogenesis (Smith and Long, 2010). It seemed plausible from tpl-1 phenotype that TPL or TPL-related proteins may also regulate genes specifically involved in shoot meristem fate in the embryo, which could lead to a double-shoot phenotype. In a screen for suppressors of the tpl-1 double-root phenotype, a mutation in an HD-ZIPII TF gene was identified. This mutation disrupts a microRNA-binding site leading to excess transcript accumulation. Genetic analyses of multiple mutant embryos revealed that PLTs act as negative regulators of HD-ZIPIII expression in the basal domain and that HD-ZIPIIIs block ectopic PLT expression in the apical domain. These data indicate that the PLT and HD-ZIPIII pathways act antagonistically in embryonic meristem formation. Finally, a second shoot meristem was formed in place of a root meristem as a consequence of HD-ZIPIII expression in the basal embryo domain (Fig. 9.4E; Smith and Long, 2010). These results indicate that transcriptional activity of these factors are necessary and sufficient for meristem formation and their mutual antagonism is also central to the formation of two distinct stem cell populations.
6. Specification and Positioning of Organs Forming Postembryonically
Plant postembryonic development initially relies on the activity of primary meristems, which are already present in the embryo. Primary meristems generate the main stem, leaves, flowers, and the primary root. Positioning of leaves and flowers takes place in the shoot apical meristem and ultimately requires that subsets of meristematic cells are specified to become new organs emerging from the flanks of the primary meristem (Hamant et al., 2010). In contrast, subsequent growth of branches and lateral roots (LRs) is coordinated by the activity of lateral meristems. Because lateral meristems generating LRs are specified de novo, proper patterning requires not only cell fate specification but also correct positioning of the newly specified populations of cells along the primary root. Because primary root growth is continuous, positioning of LRs has to integrate spatial and temporal information. Recent findings in Arabidopsis show that positioning of LRs is mediated by a time-keeping mechanism that appears to involve oscillating gene expression (Moreno-Risueno et al., 2010).
6.1. Positioning of leaves and flowers
In Arabidopsis, new aerial organ positioning and determination requires the hormone auxin. Leaves and flowers are positioned around the growing stem with a certain angle relative to the previous one (phyllotaxis), which likely maximizes light harvesting or pollination. Phyllotaxis takes place in the shoot apical meristem and involves formation of local gradients of auxin (Reinhardt et al., 2003). These gradients form through the activity of intercellular transporters, such as PIN1, whose expression is, in turn, activated by auxin. Models of this feedback mechanism indicate that different auxin maxima may be formed in specific subsets of cells at precise angles, normally 137.5°, following a helical curve around the main axis. Live confocal imaging revealed that these subsets of cells reporting an auxin maximum initiate new lateral organs and showed temporal correlations between expression of PIN1 and known regulators of meristem identity and function (Hamant et al., 2010; Heisler et al., 2005). Additionally, differential growth of cells during phyllotaxis appears to generate biomechanical signals. These signals, in turn, feed back into this morphogenetic process (Hamant et al., 2008) and may mediate the link between auxin and its transport (Heisler et al., 2010).
6.2. Oscillating gene expression is involved in positioning LRs
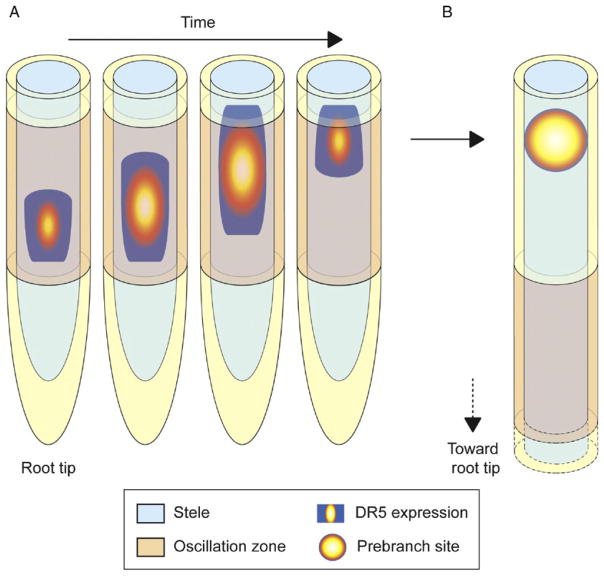
In roots, evidence for oscillating gene expression playing a role in positioning lateral organs came from the observation of the dynamic expression of the DR5 marker gene (De Smet et al., 2007; Moreno-Risueno et al., 2010). Live imaging of DR5 fused to a Luciferase reporter allowed real-time expression analyses and showed a temporal and spatial relationship between periodic pulses of DR5 expression and the subsequent generation of LRs (Moreno-Risueno et al., 2010). These periodic pulses of expression take place over a region of the Arabidopsis root tip termed the oscillation zone (OZ). During an oscillation cycle, DR5 expression is first observed at the more proximal or rootward region of the OZ, and over time, its expression increases and moves shootward within the OZ. Then, DR5 expression shuts down and a new oscillation begins. Growth of the root continuously displaces the OZ further from the shoot but coincident with the physical location where a DR5 oscillation occurred, a prebranch site is observed outside of the OZ (Fig. 9.5). Prebranch sites are marked by static points of DR5 expression, and subsequently, LR primordia are formed at prebranch sites as shown by lineage analyses and microscopy. Selection of subsets of cells in the primary root that become competent to generate a new organ (prebranch sites) is therefore reported by the DR5 oscillation. A remarkable feature of prebranch site formation is its capacity to compensate for variation in temperature and other environmental conditions (Moreno-Risueno et al., 2010). This indicates that formation of prebranch sites acts as a biological clock and/or time-keeping mechanism, and it was consequently named the lateral root clock.
Figure 9.5.
The DR5 oscillation marks position of future lateral roots through establishment of prebranch sites. (A) DR5 expression in the oscillation zone (OZ) increases and moves shootward over time. At the beginning of one oscillation cycle, DR5 is expressed at the proximal part of the OZ (rootward). Over time DR5 expression increases and moves toward the distal or shootward part of the OZ. At the end of the cycle, DR5 expression is turned off. A new oscillation then begins in the OZ. Similar expression patterns have been observed for genes oscillating in phase with DR5. (B) Following a DR5 oscillation in the OZ, a prebranch site is established. Prebranch sites are observed outside the OZ but their locations coincide with the region of the root where an oscillation was observed. Prebranch sites mark the position of future lateral roots.
DR5 is also used as a marker for the transcriptional readout of auxin signaling. Thus, it was initially proposed that the changing expression of DR5 in the OZ was due to formation of a local auxin maximum or to increased auxin sensitivity in this region (De Smet et al., 2007). This would lead to the prediction that priming of cell populations to become LRs is determined by local accumulation of auxin acting as a switch of cell fate, in a fashion somewhat analogous to phyllotaxis (Heisler et al., 2005). However, other auxin-responsive promoters did not oscillate in the OZ and some genes that showed oscillating expression did not respond to auxin. This indicates that oscillations in DR5 and in other genes cannot be entirely explained by changing auxin levels in the OZ. Furthermore, exogenous auxin treatments localized to the OZ did not induce prebranch site formation through the initiation of a DR5 oscillation (Moreno-Risueno et al., 2010). Intriguingly, a gain-of-function mutation of INDOLE ACETIC ACID FACTOR 28 (IAA28) that confers resistance to auxin, likely because the mutant protein is not degraded by auxin, appeared to affect the DR5 oscillation and is required for normal LR formation (De Rybel et al., 2010; Dreher et al., 2006; Rogg et al., 2001). However, IAA28 expression has not been shown to oscillate, and appears to be complementary to the graded distribution of auxin at the root tip (Petersson et al., 2009). Thus, auxin appears to be required for positioning of LRs but appears insufficient to trigger this developmental mechanism independently. Future studies might reveal a connection between auxin and oscillating gene expression in recruiting specific cell populations in time and space.
Further insight into the morphogenetic mechanism positioning lateral organs in the Arabidopsis root was obtained by microarray analyses of the OZ at various discrete points during the DR5 oscillation (Moreno-Risueno et al., 2010). Approximately 2000 genes showed a similar oscillatory pattern as DR5, and about 1400 were shifted one phase and therefore showed an antiphase oscillatory pattern. Gene expression in two different oscillatory phases was confirmed by real-time imaging of predicted oscillating TF genes fused to a Luciferase reporter. In addition, the expression of these oscillating genes also propagated along the OZ and in some cases passed outside this developmental region. LOF mutants for TFs oscillating in both phases showed reduced numbers and irregular positioning of prebranch sites and LRs. This indicates that both phases are required for lateral organ positioning, suggesting that this mechanism operates as a complex and interconnected network.
6.3. A developmental switch might operate in combination with oscillating gene expression to position LRs
Oscillating gene expression is involved in another developmental mechanism in which repeating units are specified along an elongating axis: the segmentation clock of vertebrates (Krol et al., 2011). In both the segmentation and the LR clocks, there are two sets of genes oscillating in opposite phases and their expression propagates along an elongating axis (the presomitic mesoderm and the primary root). In addition, based on their periodic and compensatory nature, both mechanisms can be described as biological clocks that convert time into precise spatial developmental patterns (Moreno-Risueno and Benfey, 2011). During vertebrate segmentation, a model for how cells can be recruited to form somites (vertebrae precursors) in a succession of discrete groups in time and space is given by the clock and wavefront model (Cooke and Zeeman, 1976). In this model, the clock is defined as an oscillator shared by all presomitic cells to which they are entrained and synchronized on a developmental time scale. The wavefront is defined as a point of irreversible, rapid cell change that moves down the longitudinal axis of the embryo. The interaction between the oscillator and the wavefront creates a pattern by selecting (in time) cells oscillating between a permissive and nonpermissive phase to undergo rapid alteration.
Further development of the clock and wavefront model has shown that mutually inhibitory gradients of retinoic acid (RA) and fibroblast growth factor (FGF) along the presomitic mesoderm can generate and position a sharp morphogen threshold (Goldbeter et al., 2007). This threshold separates two stable steady states based on abrupt changes in levels of FGF and RA. The segmentation clock (the oscillating genes), in combination with two different developmental states, has been proposed to synchronously activate segmentation genes in successive discrete populations of cells. This mechanism, thus, combines a developmental switch with oscillating gene expression (segmentation clock) to precisely pattern somites during embryogenesis. In the root clock, oscillating gene expression is required for proper LR positioning along the primary root (Moreno-Risueno et al., 2010). However, it is unknown how oscillating gene expression in the root clock successively selects populations of cells to form prebranch sites. Given the similarity with somitogenesis, it is tempting to speculate that a developmental switch is working in combination with the oscillating genes of the root clock. Future studies may demonstrate how similar or disparate these clock mechanisms are, despite the obvious evolutionary distance.
7. Concluding Remarks
Plants specify and pattern new organs both embryonically and post-embryonically as part of normal development. This is largely achieved by transcriptional regulators functioning as switches for cell fate specification. Over the past few years, a number of transcriptional regulators have been identified in Arabidopsis that integrate positional information and cues relative to cell ancestry or lineage to coordinate patterning and development. In addition, different regulators act in parallel pathways to specify cell fates in response to various endogenous and, in some cases, environmental stimuli. This tight regulation ensures proper patterning under a wide range of conditions, which can help explain the developmental plasticity observed in plants.
Despite recent progress, there are still numerous unresolved developmental and mechanistic questions: How do the transcriptional switches work at the molecular level? What cis-motifs are responsible for specific protein–DNA interactions? And what is the nature of the transcriptional changes produced by these cell fate regulators? Some studies have begun to address these questions by molecular, genetic, and genome-wide approaches. For instance, WRKY2 is known to bind to the W-box in the regulatory sequences of its downstream target WOX8 to establish embryo polarity, and SHR binds to the promoter of the cell cycle regulator CYCD6;1 to activate an asymmetric division required for ground tissue patterning and specification. Future work might address how different pathways involved in cell fate determination function at the molecular level in response to variable inputs both from endogenous and exogenous sources.
Acknowledgments
We apologize to those researchers whose work we could not cover due to space limitations. We thank H. Cederholm, A. Iyer-Pascuzzi, R. Sozzani, C. Topp, and C. Winter for critical reading of this chapter. J. M. V. N. is supported by a NIH NRSA postdoctoral fellowship. Work in the Benfey lab is supported by grants from the NIH, NSF, and DARPA.
References
- Agusti J, Lichtenberger R, Schwarz M, Nehlin L, Greb T. Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet. 2011;7:e1001312. doi: 10.1371/journal.pgen.1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benkova E, Mahonen AP, Helariutta Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011a;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vaten A, Help H, El-Showk S, Scheres B, Helariutta K, Mahonen AP, Sakakibara H, Helariutta Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol. 2011b;21:927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical–basal axis formation in the Arabidopsis embryo. Dev Cell. 2008;14:867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A, Lee JY, Demura T. Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu Rev Cell Dev Biol. 2010;26:605–637. doi: 10.1146/annurev-cellbio-100109-104107. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Bilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. A novel aux/ IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development. 2009;136:3235–3246. doi: 10.1242/dev.037028. [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom EM, Izhaki A, Bowman JL. Promoter bashing, microRNAs, and Knox genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol. 2004;135:685–694. doi: 10.1104/pp.104.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. Wiley; New York: 1977. [Google Scholar]
- Faure JE, Rotman N, Fortune P, Dumas C. Fertilization in Arabidopsis thaliana wild type: Developmental stages and time course. Plant J. 2002;30:481–488. doi: 10.1046/j.1365-313x.2002.01305.x. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT–ROOT movement. Plant J. 2009;57:785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT–ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Gonze D, Pourquie O. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and FGF signaling. Dev Dyn. 2007;236:1495–1508. doi: 10.1002/dvdy.21193. [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- Hamant O, Traas J, Boudaoud A. Regulation of shape and patterning in plant development. Curr Opin Genet Dev. 2010;20:454–459. doi: 10.1016/j.gde.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jonsson H, Traas J, Meyerowitz EM. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010;8:e1000516. doi: 10.1371/journal.pbio.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung JW, Sena G, Hauser MT, Benfey PN. The SHORT–ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. Establishment and maintenance of vascular cell communities through local signaling. Curr Opin Plant Biol. 2011;14:17–23. doi: 10.1016/j.pbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development. 2010;137:975–984. doi: 10.1242/dev.047662. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- Jurgens G. Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO J. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol AJ, Roellig D, Dequeant ML, Tassy O, Glynn E, Hattem G, Mushegian A, Oates AC, Pourquie O. Evolutionary plasticity of segmentation clock networks. Development. 2011;138:2783–2792. doi: 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Bilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. Whole-genome analysis of the SHORT–ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. Dynamic integration of auxin transport and signalling. Curr Biol. 2006;16:R424–R433. doi: 10.1016/j.cub.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development. 2002;129:2797–2806. doi: 10.1242/dev.129.12.2797. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116:109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Benfey PN. Time-based patterning in development: The role of oscillating gene expression. Transcription. 2011;2:124–129. doi: 10.4161/trns.2.3.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H. Transcriptional regulation of vascular cell fates. Curr Opin Plant Biol. 2010;13:670–676. doi: 10.1016/j.pbi.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 2009;21:1659–1668. doi: 10.1105/tpc.109.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Van Norman JM, Benfey PN. Symmetry breaking in plants: Molecular mechanisms regulating asymmetric cell divisions in Arabidopsis. Cold Spring Harb Perspect Biol. 2009;1:a000497. doi: 10.1101/cshperspect.a000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA. Control of Arabidopsis apical–basal embryo polarity by antagonistic transcription factors. Nature. 2010;464:423–426. doi: 10.1038/nature08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/ LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell. 2008;20:3359–3373. doi: 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norma JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell. 2011;20:264–270. doi: 10.1016/j.devcel.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT–ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chory J, Weigel D. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol. 2007;309:306–316. doi: 10.1016/j.ydbio.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Elo A, Helariutta Y. Arabidopsis as a model for wood formation. Curr Opin Biotechnol. 2011;22:293–299. doi: 10.1016/j.copbio.2010.11.008. [DOI] [PubMed] [Google Scholar]