Abstract
Plants, being sessile organisms, need to respond to changing environments, and as a result they have evolved unique signalling mechanisms that allow rapid communication between different parts of the plant. The signalling mechanisms that direct plant development include long-range effectors, such as phytohormones, and molecules with a local intra-organ range, such as peptides, transcription factors and some small RNAs. In this Review, we highlight recent advances in understanding plant signalling mechanisms and discuss how different classes of signalling networks can integrate with gene regulatory networks and contribute to plant development. In some cases, we also address the evolutionary context of mechanisms and discuss possible links between the lifestyle of plants and selection for different signalling mechanisms.
In the past few years, several landmark studies, as well as new technological advances, have increased our understanding of the signals required for plant development. The identification of plant hormone receptors and the components that mediate downstream pathways were major steps towards understanding the mechanisms by which phytohormones work. In addition, recent studies have revealed that mobile small RNAs, transcription factors and peptides have important roles in tissue patterning and have promoted our understanding of cellular communication in planta. These breakthroughs would have been hard to achieve without the latest advances in optics, which now allow subcellular and even molecular resolution using different types of visualization techniques, as well as high-throughput methods that facilitate the simultaneous analyses of thousands of genes.
After germination, the physical location of a plant does not normally change during its lifespan. This means that plants live in a ‘noisy’ setting where annual and diurnal environments fluctuate greatly. Consequently, the development of a plant both at the cellular level and at the organismal level must be tightly coordinated with external conditions. To survive in such circumstances, plants have evolved to be highly responsive to the environment and display a high degree of developmental and morphological plasticity1. These features are mainly attributable to their iterative developmental programme and the independent nature of different organs, which can be formed and shed as needed.
Post-embryonically, above-ground and belowground organs are formed from two distinct stem cell populations, which are referred to as meristems (FIG. 1). Each meristem consists of niche cells (namely, the organizing centre in the shoot and the quiescent centre in the root) surrounded by stem cells or initial cells, which produce distinct cell types in each region. Despite their apparent autonomy, roots, stems, leaves and other structures routinely channel information between each other to perceive external conditions and to respond accordingly. The perception, integration and transmission of signals are not dominated by a central nervous system, as is the case in animals. Almost every organ and tissue can generate information and respond to internal and external cues, emphasizing the need for an efficient communication system among cells, tissues and organs.
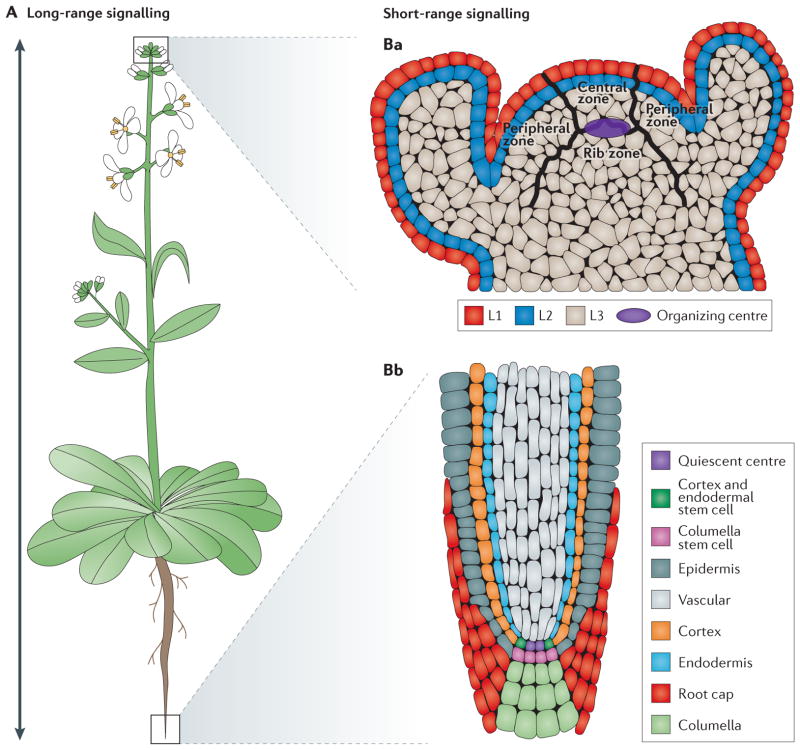
Figure 1. Schematic representation of Arabidopsis thaliana meristems and types of signalling.
Arabidopsis thaliana is a dicotyledonous plant that is used as a model organism for developmental studies. Two classes of signalling are used in A. thaliana: long-range signalling encompassing travel over a considerable distance (for example, shoot to root) (part A); and short-range signalling involving local intercellular movement (parts Ba,Bb). A. thaliana organs originate from two meristem populations, one in the shoot and the other in the root. Both meristems contain niche cells and stem cells or initial cells. However, each meristem has a distinct organization and cell types that arise from it. The shoot apical meristem (SAM) contains three layers (L1, L2 and L3) and three developmental zones (peripheral zone, central zone and rib zone) (part Ba). The niche cells, or organizing centre, of the SAM are specified at the junction of the three developmental zones and function to maintain stem cells in the shoot. The root apical meristem (RAM) (part Bb) is radially symmetric and consists of central niche cells (the quiescent centre) surrounded by stem cells. In the root, each stem cell population gives rise to one or two cell types.
Another unique feature of plants is the presence of a cell wall that surrounds each cell and contributes to its structural integrity but that also prevents cell movement (FIG. 2a). This may well have been a driving force in the evolution of diverse cell–cell communication mechanisms that coordinate growth and development. Owing to the cell wall, two routes of intercellular transport are available to plants: apoplastic and symplastic (FIG. 2b). Apoplastic transport occurs when signals are transmitted through the space between the cell wall and the plasma membrane, whereas symplastic transport occurs when signals are transmitted directly between cells via channels called plasmodesmata.
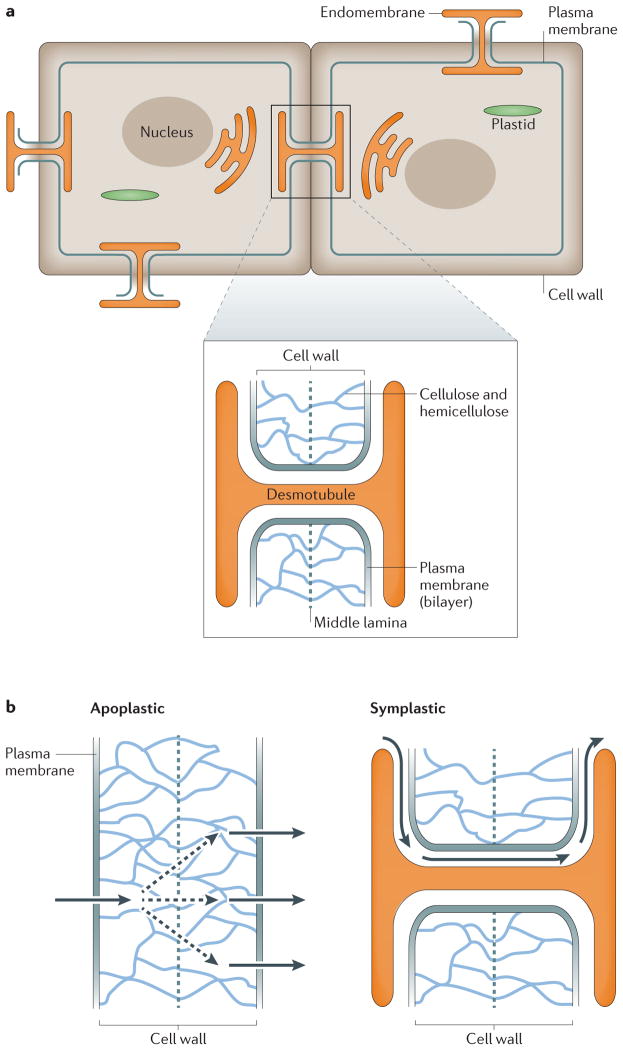
Figure 2. Routes of intercellular movement.
a | Plasmodesmata are cell membrane features that are formed by plasma membrane connections between adjacent cells, which penetrate the cell wall. Plasmodesmata contain a shared portion of the endomembrane system, which is thought to be important for the transmission of intercellular signals. b | Owing to the cell wall, intercellular movement is restricted to two routes: apoplastic and symplastic. An apoplastic route of travel occurs when factors move into the space between the plasma membrane and the cell wall. The cell wall is made up of cellulose and hemicellulose (indicated by blue lines), with a central pectin-based cell plate, called a middle lamella, that connects the cell walls of adjacent cells (grey dashed line). In apoplastic travel, factors move through this cell wall and into neighbouring cells for uptake. This type of movement can be passive (diffusion) or active (export and import). By contrast, a symplastic route of travel occurs when factors move into neighbouring cells through plasmodesmata. Dashed arrows indicate diffusion or random movement, and solid arrows indicate directed movement.
Signalling occurs over long and short distances within the plant. In this Review, short-range signalling refers to mechanisms that are specifically implemented during pattern formation in organs, tissues or between neighbouring cells (FIG. 1). This is usually the ‘final touch’ for shaping and specifying cell function and identity. These short-range signals include peptide signalling and the movement of transcription factors and non-coding small RNAs (sRNAs). By contrast, we define long-range effectors as stimulants that influence many developmental events and that can travel for long distances between cells and organs (FIG. 1). The long- and short-range signalling processes are mechanistically related, but differences in modes of communication are not merely semantic, thus we outline conceptual distinctions, as well as commonalities.
In this Review, we provide an overview of the current state of knowledge regarding how plants channel information within and between different organs, and how this information is integrated to produce desired outputs. Finally, we discuss the fundamental principles that constitute information networks in plants and consider future directions for detecting and analysing these networks.
Short-range: peptides
In animal development, the importance of peptide signalling has long been recognized. However, historically, in plants the focus has been on phytohormones. Only in the past decade or so has the relevance of peptides to plant development begun to be appreciated. In silico analysis of the Arabidopsis thaliana genome for 25–250-amino-acid proteins containing an aminoterminal cleavable signal peptide, but lacking a transmembrane domain and an endoplasmic reticulum retention sequence, predicts thousands of biologically active peptides2. However, the function of only a handful has been identified, and even fewer receptors and downstream components have been characterized. For a comprehensive review on this topic, see REF. 3.
Recent studies have begun to define a general mechanism of peptide signalling in plants. Specifically, two classes of signalling peptides have been identified: cysteine-rich peptides and small post-translationally modified peptides4. For cysteine-rich peptides, a propeptide is produced and the formation of disulphide bonds results in a mature signalling peptide. By contrast, the small post-translationally modified peptides undergo further proteolytic processing of the propeptide, and the addition of post-translational modifications generates a mature signalling peptide. For both classes it is hypothesized that the peptides diffuse through the apoplast to bind target receptors on neighbouring cells5. However, direct evidence of peptide movement and the route of travel remain to be determined.
In plants, the receptors for some signalling peptides are receptor-like kinases (RLKs). Identification of these receptors and their corresponding ligands has been challenging. Genome-wide analysis of A. thaliana has revealed more than 600 putative RLKs6, the largest proportion of which are leucine-rich repeat RLKs (LRR-RLKs)7. This estimate is based on three features: a ligand-binding extracellular domain, a transmembrane domain and a cytoplasmic kinase domain. This is likely to be a conservative approximation, as another class of proteins, called leucine-rich repeat receptor-like proteins (LRR-RPs), lack a kinase domain but form complexes with LRR-RLKs for signal transduction. Despite the identification of putative ligands and receptors, we are only just beginning to appreciate the complexity of these interactions in plants. Currently, fewer than ten ligand–receptor pairs have been proposed and even fewer interactions have been thoroughly validated.
Regulation by CLE peptides in the shoot apical meristem
One class of small post-translationally modified peptides that has provided insights into signalling mechanisms active during plant development is the CLAVATA 3/EMBRYO SURROUNDING REGIONRELATED (CLE) family. A total of 32 CLE peptides have been defined on the basis of a 14-amino-acid conserved CLE motif and a variable N-terminal hydrophobic domain8,9, but many of their functions remain to be uncovered. The first CLE peptide, CLAVATA 3 (CLV3), was identified from a mutagenesis screen in which the loss of CLV3 function resulted in the expansion of the shoot apical meristem (SAM)10,11. CLV3 acts non-cell autonomously and is expressed in a cell layer that is adjacent to the domain in which its receptors are expressed: CLV3 is expressed in stem cells, whereas its receptors are expressed in the underlying niche and surrounding cells11. Three receptor complexes have been identified: CLAVATA 1 (CLV1) homodimers; CLAVATA 2 (CLV2) and CORYNE (CRN) heterodimers; and RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2) homodimers12–14. CLV1 and RPK2 represent typical serine/threonine LRR-RLKs, whereas CLV2 lacks a kinase domain and is a prototypical LRR-RP, requiring CRN for signal transduction13,15,16. CRN is kinase-dead, suggesting that it does not have a role in signal transduction, and it is instead proposed to facilitate CLV2 transport or protein scaffolding17. Interestingly, ~20% of identified RLKs are proposed to be kinase-dead, but the functional importance of these receptors remains to be defined18,19. CLV1 and CLV2–CRN are considered to be the primary CLV3 receptors, as shown by the relatively mild phenotype of RPK2 mutants14.
Developmentally, CLV3 regulates the balance between stem cell proliferation and differentiation in the SAM. Tight regulation of this process is achieved by a feedback loop between CLV3 and a downstream transcription factor, WUSCHEL (WUS), which is expressed in the stem cell niche and which is a key regulator of stem cell maintenance20,21 (FIG. 3a,b). In this pathway, WUS activates CLV3, which in turn represses WUS, generating a controlled balance between differentiation and stem cell maintenance11,22,23. This negative feedback loop has distinct outcomes, including limiting the maximum signal output, which is important for the maintenance of stem cell populations24. It is still unclear how the restriction of WUS expression is coordinated through the three different receptor complexes. Loss of either CLV1 or CLV2–CRN results in expansion of the WUS expression domain; however, loss of RPK2 has only mild effects on WUS expression14,25. Interestingly, the expression domain of the receptors is broader than that of WUS, suggesting additional regulatory mechanisms14,15,21,25– 27. Some signalling intermediates in the CLV3–WUS pathway have been identified (for example, REFS 28–30), but the mechanisms of signal transduction from cell surface receptors to WUS repression remain to be fully elucidated.
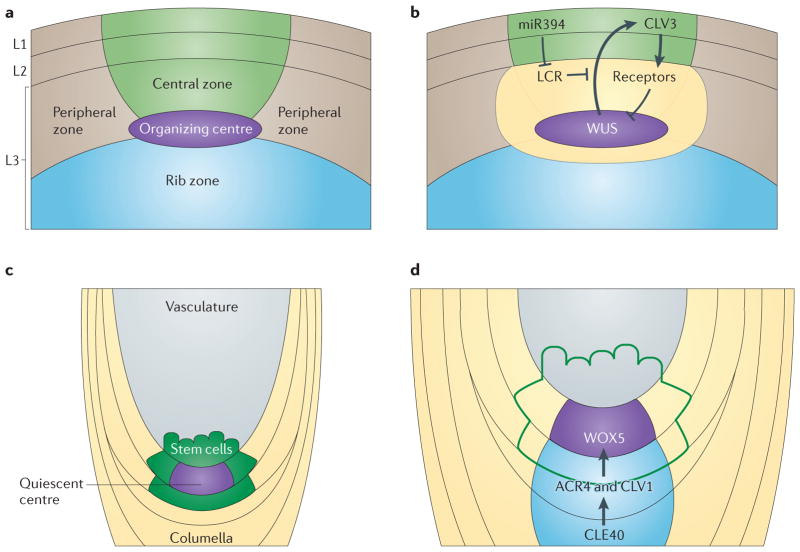
Figure 3. Peptide signalling in meristem maintenance.
Both shoot and root meristems require peptide signalling for their maintenance. Within the central zone of the shoot, expression of the mobile transcription factor WUSCHEL (WUS) originates in the organizing centre and travels to the L1 and L2 layers to directly promote the expression of the peptide CLAVATA 3 (CLV3) (parts a,b). The CLV3 peptide is produced and moves into the underlying L3 layer to bind its receptors (CLV1, CLV2–CORYNE (CRN) and RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2)) and to repress the expression of the WUS transcription factor. Production of a mobile non-coding small RNA (sRNA), microRNA394 (miR394) in the L1 and L2 layers generates an opposing gradient to prevent the feedback of WUS to CLV3 by LCR-mediated degradation of an as-yet-unknown cofactor. In the root, CLE40, which is a homologue of CLV3, is expressed in the columella cells underlying the quiescent centre (parts c,d). CLE40 binds its receptor, ARABIDOPSIS CRINKLY 4 (ACR4) to promote the expression of WUSCHEL-RELATED HOMEOBOX 5 (WOX5) in the quiescent centre. Expression of WOX5 is then necessary to maintain the columella initial cells.
Regulation by CLE peptides in the root apical meristem
In the root apical meristem (RAM), WUSCHEL-RELATED HOMEOBOX 5 (WOX5), which was identified as homologous and interchangeable with WUS, was found to be expressed in the quiescent centre31–33. Thus, it logically followed that a CLE peptide–WOX5 pathway analogous to the CLV3–WUS pathway would also be required for the maintenance of the RAM. Such a pathway has been identified, but it does not seem to function in the same way as CLV3–WUS in the shoot meristem32,33. A close homologue of CLV3, CLE40, is expressed in cells underlying the root stem cell niche and regulates WOX5 expression33,34 (FIG. 3c,d). The receptor for CLE40 has been identified as the non-LRR ARABIDOPSIS CRINKLY 4 (ACR4). ACR4 is expressed in cells adjacent to those expressing CLE40 and it negatively regulates WOX5 expression in the niche33 (FIG. 3c,d). Interestingly, a recent study has shown that CLV1 may act as a co-receptor with ACR4 in the root meristem and could represent an evolutionarily conserved mechanism of meristem maintenance35. However, cle40, acr4, clv1 or wox5 mutants do not globally affect root stem cell populations; alterations are only observed in the distal stem cell population (that is, the columella)32,33,35. There is also no evidence of feedback from WOX5 to CLE40. Together, these results suggest that, although a CLV3– WUS-like pathway does exist in the root, it acts in a more restricted manner than in the shoot.
Regulation by CLE-like peptides in the RAM
A new family of peptides has recently been identified that is required for the maintenance of the root meristem36. The CLE-like (CLEL) peptides (also known as root growth factor (RGF) peptides) were identified through their post-translational modifications36. Despite their name, the CLEL peptides are distinct from CLEs and share only a domain structure37. It was hypothesized that novel peptides could be identified from the analysis of mutants in post-translational modification enzymes36,38, and it was found that tyrosylprotein sulphotransferase (tpst) mutants have severe phenotypes in both roots and shoots38,39. By focusing on the root phenotype of reduced meristem size and expanded niche cells, a candidate peptide (RGF1) was identified bioinformatically, synthesized and applied to tpst-mutant plants36. In the presence of exogenous RGF1, tpst root phenotypes were mostly rescued. The addition of two other previously identified, root-expressed, tyrosine-sulphated peptides — phytosulphokine (PSK) and plant peptide-containing sulphated tyrosine 1 (PSY1) — was sufficient to fully rescue the root phenotype36,40,41. Neither PSK nor PSY1 alone was sufficient, and the application of other peptides from the CLEL family showed varying degrees of rescue, suggesting that RGF1 is the dominant CLEL peptide in the root. No receptors have yet been identified for RGF1; however, it has been shown that RGF1 post-translationally regulates the expression levels of PLETHORA (PLT) transcription factors, which are required for root meristem maintenance36. TPST has been independently shown to regulate the expression of PLT and the localization of biosynthetic and transporter proteins for the phytohormone auxin, possibly through RGF1 (REF. 42). Thus, RAM maintenance seems to rely on the convergence of phytohormone and mobile peptide signalling pathways.
We highlight above a subset of studies on peptide signalling in plants, with a specific focus on meristem maintenance. The studies of ligand–receptor interactions provide some intriguing insights into short-range signalling in plants. In particular, the use of multiple receptor complexes in the SAM is a mechanism by which plants can fine-tune short-range peptide signalling. By contrast, the RAM seems to use multiple ligands with similar post-translational modifications and as-yet-unidentified receptors to regulate meristem size and function. Although some common components exist (for example, the interchangeable WOX5 and WUS transcription factors32, the common requirement of CLV1 in both meristems and shared signalling intermediates43), each stem cell population seems to use unique regulatory mechanisms.
Short range: transcription factors
A second mechanism by which plants transmit local signals is through the intercellular movement of transcription factors. The movement and non-cell-autonomous activity of transcription factors in plants was initially described almost 20 years ago, but our understanding of this process is still evolving44,45. Movement is predicted to occur for 17–29% of transcription factors46,47. This is likely to be a conservative estimate given that this range has arisen from static observation in developmental time. No universal sequence exists to identify the transcription factors that can move, nor has a sequence been identified that is sufficient to mobilize an immobile protein; thus bioinformatic predictions are currently not possible. Instead, mobile transcription factors are often identified on the basis of differences in the expression domains of RNAs and proteins46,47. Transcription factor movement can be restricted either by a point mutation within the protein or by the addition of a cassette containing three copies of the GFP coding region to determine the functional relevance of the movement48,49. Currently, live imaging and quantification of transcription factor movement remain to be fully explored.
Two types of transcription factor movement have been described: non-targeted passive diffusion and active targeted movement. Both types have been proposed to occur by transit through plasmodesmata. There is a size exclusion limit for macromolecules travelling through plasmodesmata. However, this limit is highly dependent on tissue type and age49–51. Plasmodesmata are dynamic features that can expand and contract, and the plasticity of plasmodesmata suggests that size is not the limiting factor for transcription factor movement. In support of this, there is no correlation between size and movement of transcription factors, and mobile transcription factors are a wide range of sizes (from 11 kDa to 70.5 kDa)46,47,52.
Non-targeted movement
Non-targeted movement seems to be the minor mechanism of movement in plants as only one transcription factor has been shown to move via this mechanism in planta. What little we know about non-targeted passive diffusion of transcription factors is from studies of LEAFY (LFY). Developmentally, LFY has two functions: to initiate the formation of the first flower and to activate floral homeotic genes. Three lines of evidence point to a mechanism similar to diffusion: first, there is unregulated movement of LFY53; second, it has not been possible to inhibit movement by the deletion of various sequences; and third, cytoplasmic localization is sufficient to promote movement54. It remains to be seen whether other transcription factors also use this mechanism or whether LFY is unique.
Targeted movement
By contrast, most mobile transcription factors undergo active targeted movement, which has been proposed to occur through association with a cofactor. Cytoplasmic availability was also proposed to regulate protein movement; however, the situation has proved to be more complex, as cytoplasmic localization is not sufficient for transcription factor movement46,54,55. One example of a transcription factor that uses targeted movement is SHORT-ROOT (SHR), which moves from the root vascular tissue into the surrounding cell layer to regulate cell fate choices56 (FIG. 4a–c). SHR was classified as engaging in active targeted movement because a single point mutation was sufficient to inhibit movement, which is thought to occur via plasmodesmata48,55,57. Recently, a cofactor, SHORT-ROOT INTERACTING EMBRYONIC LETHAL (SIEL), was identified that facilitates SHR movement58 (FIG. 4c). Interestingly, siel mutants also disrupt the movement of other transcription factors, suggesting that there may be common cofactors involved in this process58. SIEL colocalizes with endosome markers58,59, and because the endomembrane is a key component of viral intercellular movement, this suggests that a conserved mechanism for macromolecule movement may exist59.
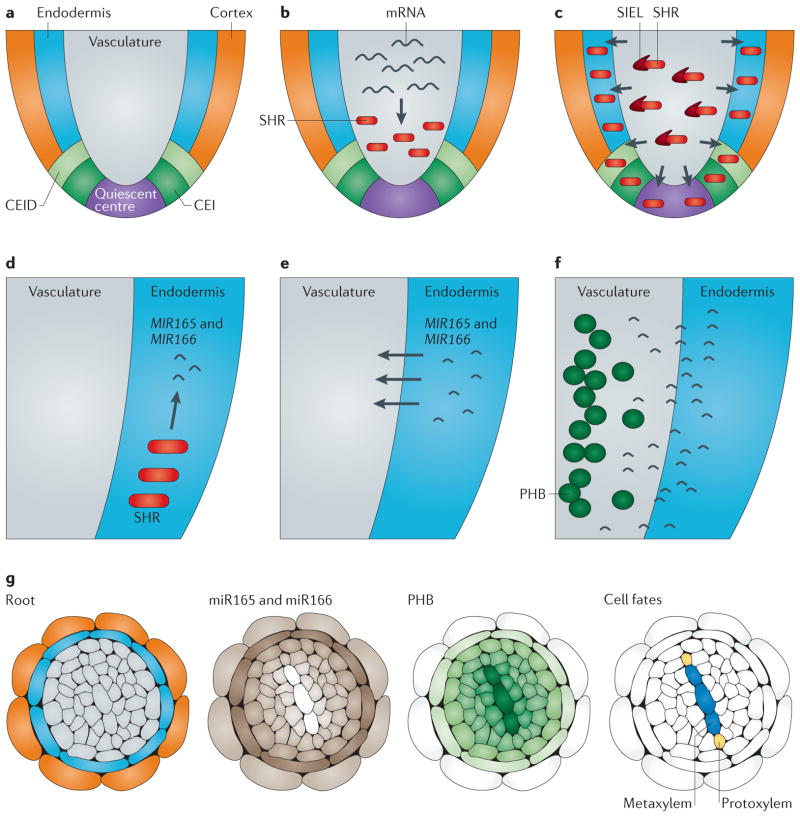
Figure 4. Transcription factor and small non-coding RNA movement in the root meristem.
Cell fate specification in the root is a tightly coordinated process that uses the reciprocal movement of transcription factors and non-coding small RNAs (sRNAs). a | In the root, a central vascular cylinder is surrounded by the endodermis and cortex. The endodermis and cortex originate from a common progenitor called the cortex/endodermal initial (CEI) and its resultant daughter cell, the CEI daughter (CEID). Cortex and endodermal cell fate is regulated by the mobile transcription factor SHORT-ROOT (SHR). b | SHR is transcribed and translated in the vasculature. c | The SHR transcription factor then moves outwards into the CEI, CEID, quiescent centre and endodermis by association with a movement cofactor, SHORT-ROOT INTERACTING EMBRYONIC LETHAL (SIEL). d | In addition to the role of SHR in endodermal and cortex cell fates, SHR activates mobile sRNAs, microRNA165 (MIR165) and MIR166, which are important for vascular cell fate identity. SHR induces the expression of MIR165 and MIR166 in the endodermis. e | MIR165 and MIR166 then move into the vasculature, generating a gradient of sRNA. f | The target of miR165 and miR166, PHABULOSA (PHB) mRNA, is degraded in a concentration-dependent manner. g | A cross-section schematic of the root depicts the cylindrical gradient of miR165 and miR166 and PHB expression. The graded expression of PHB is responsible for cell fate decisions in the xylem. Specifically, high PHB and low miR165 and miR166 expression results in metaxylem formation and low PHB and high miR165 and miR166 expression results in protoxylem formation.
Transcription factor movement is involved in many developmental processes and is integrated with other signalling mechanisms. For example, in the SAM, WUS moves into adjacent cells to activate CLV3 transcription, although the mechanism by which it moves remains to be determined60. Although the mobility of transcription factors is predicted to be widespread in A. thaliana, we currently lack the ability to rapidly identify moving proteins. With the continued discovery of cofactors that facilitate movement and routes of travel, we may be able to identify the motifs required for protein shuttling or even three-dimensional structures that will facilitate the high-throughput identification of mobile transcription factors.
Short-range: small RNAs
Around the time that transcription factor movement was first described, a mobile silencing signal was discovered that was later found to consist of sRNAs61,62. sRNAs in A. thaliana can be broadly divided on the basis of biogenesis and size into two classes: small interfering RNAs (siRNAs) and microRNAs (miRNAs)63. Mobile silencing is not associated with any particular class of sRNA and instead seems to be a general feature of specific plant sRNAs. As with transcription factors, the movement of sRNAs mostly occurs through plasmodesmata, as the disruption of plasmodesmata inhibits movement57. Also in common with transcription factors, the movement of sRNAs is thought to require cofactor association, but no such cofactors have yet been identified. Further, no sequence has been identified that predicts which sRNAs are mobile.
In general, sRNAs are generated from a double-stranded RNA (dsRNA) precursor and processed by the Dicer-like ribonucleases (DCL1–4) into single-stranded RNA (ssRNA), which associates with the ARGONAUTE proteins (AGO1–10). There is conflicting evidence as to which of these forms moves between cells. Originally, microinjection studies demonstrated intercellular mobility of ssRNA but not of dsRNA64. However, a series of microparticle bombardment experiments, in which RNA was precipitated onto micron-sized metal particles and accelerated to locally penetrate plant tissue, established that dsRNA but not ssRNA can move between cells65,66. These contradictory results could indicate multiple forms of the mobile sRNA signal or could be due to an artefact of the experimental approach.
Insights into sRNA movement are being revealed by manipulations of the sRNA biogenesis pathways67. Specifically, non-vasculature sRNA movement is limited to 10–15 cells68. Over this distance the sRNA is continually diminished to generate a signal gradient that has been likened to that of a morphogen in animals69. One example of this is miR165a and miR166b, which generate a gradient to define vascular cell types in the root (FIG. 4d–g). Specifically, the mobile transcription factor SHR moves from the vasculature into the surrounding endodermis to activate MIR165 and MIR166 transcription70 (FIG. 4d). miR165 and miR166 expression then radiates from the endodermis through the plasmodesmata and into the vasculature to regulate the expression of PHABULOSA (PHB), a homeodomain leucine zipper (HD-ZIP)57,70 (FIG. 4e–g). The level of expression of PHB in different vascular cells then influences the decision to become protoxylem versus metaxylem70,71 (FIG. 4g).
In the SAM, the sequestration and absence of miR165 and miR166 are important for meristem maintenance. The first indication that sRNAs were important for meristem maintenance came from the identification of an AGO protein, AGO10 (also known as ZLL)72,73. In the absence of AGO10, CLV3 expression is lost despite increased expression of WUS73 (discussed above). AGO proteins typically mediate repression, but AGO10 has been shown to act uniquely as a decoy to block the accumulation of miR165 and miR166 in the meristem. Specifically, miR165 and miR166 target a set of HD-ZIP transcription factors that are important for maintaining pluripotency in the SAM. AGO10 is expressed early in the presumptive SAM and binds miR165 and miR166 to prevent its association with the broadly expressed AGO1 and thus prevent the degradation of HD-ZIP transcription factors in the SAM74,75. Interestingly, the role of AGO10 may be dependent on developmental context, as demonstrated by a study in the floral meristem, where AGO10 seems to degrade miR165 and miR166 through an unknown mechanism76.
Another miRNA, MIR394, was identified through a forward genetics screen for enhancers of a weak ago10 mutant and it is required for the specification of the shoot niche cells (organizing centre)77. miR394 is generated in layer 1 (L1) of the SAM and moves proximally, through approximately three cell layers, to act in an opposing gradient to WUS (FIG. 3b). In MIR394;ago10-1 mutants, CLV3 expression is lost, despite the WUS expression domain being maintained and even expanded. As WUS movement activates CLV3 transcription (discussed above), this suggests that WUS function is impaired, presumably owing to the loss of a cofactor. Functionally, miR394 targets the expression of a protein called LEAF CURLING RESPONSIVENESS (LCR), which putatively functions to direct proteins for degradation through the 26S proteasome. Thus, in the absence of miR394, LCR accumulates and targets an as-yet-unknown protein for degradation, preventing the feedback of WUS to CLV3 (REF. 77).
We are now beginning to understand the general mechanism of sRNA movement and are gaining a cursory understanding of how mobile sRNAs contribute to plant development. Future studies will help to define whether sRNA movement might be dependent on secondary RNA structure and/or interactions with transporter proteins. Despite recent progress, our understanding of how the mechanism of movement for specific sRNAs influences their developmental function is still limited. At present too few mobile sRNAs have been identified to draw broad conclusions about this type of signalling and the part it plays in plant development. The biggest challenge remains the identification of mobile sRNAs and their targets. Forward genetics screens for enhancers of a general sRNA mutant phenotype (as described above) are likely to yield additional factors; however, these approaches are limited by the sRNA mutant that is selected.
Long-range signalling: overview
There are several forms of long-range signalling mechanisms, which usually cause pleotropic effects that are produced by molecules travelling between organs. Accordingly, we consider phytohormones (discussed below), florigen (BOX 1) and a subset of sRNAs (BOX 2) in this category. Because a full description of plant hormones is beyond the scope of this Review and as auxin is arguably the best-studied plant hormone, we use it as an example to demonstrate the key principles of hormone signalling. We highlight below prominent themes, rather than comprehensive mechanistic details, that are crucial for balanced and efficient spatiotemporal control during plant response and development.
Box 1. Long-range movement of proteins: the odyssey of florigen.
During the nineteenth126 and twentieth centuries, experimental data showed that the transition from vegetative phase to reproductive phase in flowering plants is mediated by a light signal127, which is perceived in the leaves128–132. In the mid-1930s, the Russian botanist Mikhail Chailakhyan, probably influenced by Frits Went’s studies on auxin, dubbed this mobile signal ‘florigen’ and declared it to be a hormone-like molecule that is produced in the leaves and transported to the shoot meristem to induce flowering. The most convincing evidence came from intraspecies and interspecies graft experiments128,132,133 in which a photo-induced donor could initiate flowering in a recipient plant. Initially, it was thought that in Arabidopsis thaliana FLOWERING LOCUS T (FT) mRNA was the mobile signal, but this result could not be reproduced in the tomato134. The results from A. thaliana proved to be incorrect, and this publication was retracted. On the basis of strong circumstantial evidence, Lifschitz et al.134 proposed that the protein product of SINGLE FLOWER TRUSS (SFT), the tomato orthologue of A. thaliana FT, was the mobile florigen or at least part of the systemic signal. Indeed, most data are consistent with this hypothesis: overexpressing SFT donor shoots can induce flowering not only in sft and unfolded (uf; a late-flowering mutant) receptor shoots, but also in late-flowering tobacco. Similar observations have also been made in other species135–138. In a follow-up study139 it was shown that SFT functions as a general growth regulator as part of a system that keeps the balance between the vegetative phase and the reproductive phase. In the current model, FT is produced in the leaves and the protein moves to the shoot apex through the phloem, where it interacts with FLOWERING LOCUS D (FD) and other proteins to induce flowering. Nevertheless, one crucial observation is still lacking — visual detection of FT that overlaps with FD expression in incipient inflorescence primordia, either by a fluorescent protein fusion or by immuno-hybridization. Currently, FT is the only example of long-range signalling by a protein.
Box 2. Long-range signalling of small non-coding RNAs.
Although many non-coding small RNAs (sRNAs) have been found to signal over short distances, it has recently been appreciated that sRNAs can also act as long-range signals140,141. For more than 40 years, RNA has been identified in the vasculature conduits, but its presence was always attributed to experimental contamination. There is now evidence that the long-range activity of sRNA is important for many aspects of plant homeostasis. As sRNA species are involved in defence mechanisms and in the elimination of foreign nucleic acids, it was important to demonstrate that endogenous rather than transgenic (for example, GFP) sRNAs are capable of moving long distances and regulating target genes. Two recent studies66,142 on sRNA have demonstrated that 23–24-nucleotide-long RNAs can cross a graft union and function in recipient tissues to induce genomic hypermethylation. Two models of sRNA long-range signal transduction have been proposed. The first is through the specialized, enucleated phloem cells of the vasculature66,143. A second mechanism is through relay amplification of short-range signals, which requires the combined activation of a putative RNA-dependent RNA polymerase (SILENCING DEFECTIVE 1 (SDE1; also known as RDR6)) and a putative RNA helicase (SDE3)68. A combination of these two mechanisms is likely to be responsible for the long-range activity of sRNAs. Interestingly, grafting experiments have shown the long-range movement of sRNA to be bidirectional, although sRNA movement seems to be more efficient from shoot to root, following the directional flow of the phloem sap66,142. Long-range sRNA movement presents an intriguing process by which plants can control signals at a distance and can integrate developmental and environmental cues on a global scale. However, the specific signals that are propagated through this mechanism remain to be fully defined.
Plant hormones
Phytohormones are small molecules that act at low concentrations and that are involved in almost every developmental process, as well as in responses to external signals. They are distinct from mammalian hormones, which are transported from sites of production in specific organs (glands) to target tissues. Instead, phytohormones are produced in multiple tissues and flow between organs via the vasculature or through the use of special transporters. Each organ and, in many cases, tissues or groups of cells can produce and perceive cues that are mediated by hormones.
In plants, there are several phytohormones, each with distinct signalling mechanisms and downstream outcomes; however, some common themes have emerged. For example, four plant hormones — auxin78,79, gibberellin80, jasmonate81 and salicylic acid82 — transmit their signal via the SKP, cullin, F-box (SCF) E3 ubiquitin ligase complex83,84, and signal transduction of abscisic acid and ethylene also requires E3 ligase activity85.
Long-range signalling: auxin
As noted above, auxin is the best-studied phytohormone and is involved in many developmental processes, so here we focus on auxin as a case study. It is noteworthy that the auxin compound family has multiple derivatives that might function differently in signal transduction86,87. We discuss below the most abundant form, indole-3-acetic acid (IAA) (FIG. 5).
Figure 5. Dynamics of auxin responses.
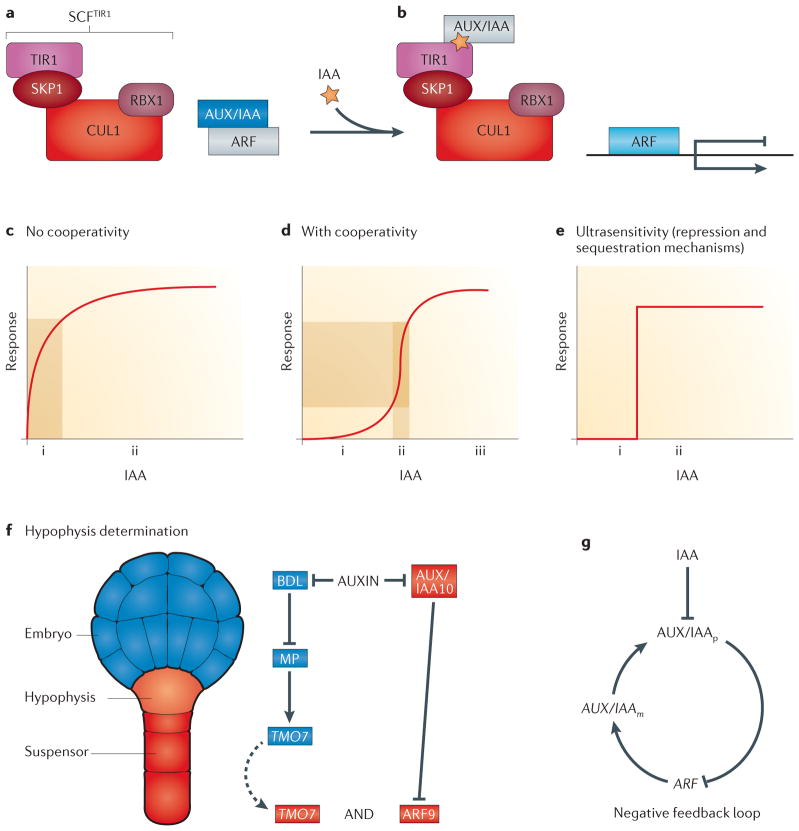
a | Shown on the left-hand side is a schematic of the auxin receptor complex SCFTIR1, which consists of the E3 ubiquitin-protein ligase RING-BOX PROTEIN 1 (RBX1), S PHASE KINASE-ASSOCIATED PROTEIN 1 (SKP1), CULLIN 1 (CUL1) and TRANSPORT INHIBITOR RESPONSE 1 (TIR1). On the right-hand side is a heterodimer formed of an auxin response factor (ARF) and an AUX/IAA protein. ARF and AUX/IAA are shown in an inactive form (shown in grey) or an active form (coloured). b | Auxin enhances the association of AUX/IAA with the F-box protein TIR1 within the SCFTIR1 complex, which leads to degradation of AUX/IAA. This releases ARF proteins so that they can function as transcription activators or repressors. c | The response to simple activation of a single protein by a stimulus (auxin) is shown. Minor fluctuations in auxin concentrations will immediately cause a response (i) (downstream gene expression) that rapidly reaches a steady state (ii). d | When several proteins work as a complex, the input–output graph has a Hill function shape. If hormone levels are low as a result of natural features of the system but not necessarily as a consequence of a real stimulus, the response is minimal (i). Subsequently, when sufficient amounts of auxin have accumulated owing to a true signal, gene expression can rapidly reach a steady state (iii). A desired and well-known characteristic of hormonal signalling is that the fold change in input signal is amplified (ii). e | An ultrasensitive bistable (on and off (i and ii, respectively)) response occurs when repressive mechanisms sequester a protein and release it in response to a stimulus or when the Hill coefficient is very high. These models have been generated from non-plant systems, but the concepts are applicable in this context. Two postulated network motifs in auxin signalling are shown. f | Hypophysis determination during embryogenesis is shown. A schematic structure of an Arabidopsis thaliana embryo in the globular stage is shown (left-hand side). A postulated feedforward loop during hypophysis specification is shown (right-hand side). The dashed arrow represents the movement of TARGET OF MONOPTEROS 7 (TMO7) from its site of transcription and translation in the provascular cells to the adjacent pro-hypophysis. g | A negative feedback loop motif between the AUX/IAAs and the ARFs can stabilize a system; for example, when a constant level of gene expression is required in the root tip.
The auxin receptor is the F-box protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1), which is a part of the SCFTIR1 complex. Auxin perception occurs by binding of AUX/IAA proteins to TIR1, targeting them for degradation by the proteasome and releasing AUXIN RESPONSE FACTOR (ARF) transcription regulators from inhibition by AUX/IAA (FIG. 5a,b). Accumulation of auxin can be stable or dynamic. For example, in the root meristem, high and stable levels of auxin are consistently present around the quiescent centre. Interestingly, although the accumulation of auxin is considered to be stable, auxin molecules are constantly flowing in and out of this region88–90. A second pattern of auxin accumulation is dynamic and often arises in newly emerging structures such as lateral root, leaf and flower primordia. Auxin dynamics, accumulation and outcomes are accomplished through interplay among its metabolism, transport and perception.
Auxin metabolism
Auxin metabolism can be divided into three main parts: de novo biosynthesis, conjugation and degradation86,87. Biosynthesis takes place in various tissues91,92 through multiple biosynthesis pathways (six have been postulated) that produce dozens of conjugates with amino acids, sugars and proteins93. These conjugates are made and unmade by actively coupling and uncoupling auxin, suggesting that they serve as readily available reserves of auxin. This mechanism provides the opportunity for the rapid regulation of developmental processes, which can be turned on by the conversion of conjugates into IAA when other components of the system (such as carriers and receptors) are already present. Two recent studies have shown that auxin biosynthesis is dependent on the availability of some sugar compounds94 and that the IAA–Trp conjugate antagonizes IAA accumulation and affects both root growth and the number of lateral roots through an undefined mechanism95. These results suggest that the balance between IAA and its precursors and conjugates is important for proper development. Interestingly, during the evolution of plants and the increase in complexity of their body plan, the overall balance has shifted from high free IAA levels and slow conjugation rates to lower levels of free IAA and fast conjugation96. A mechanism involving conjugation might provide higher precision in controlling developmental events through spatial and temporal changes to IAA96.
Auxin transport
Auxin has two primary modes of transport: rapid movement through the vasculature and short-range, accurate intercellular movement by means of the polar distribution of special transporters90,97. Four major gene families divided into two groups control polar auxin distribution: influx carriers (namely, the AUX-LAX1 family) that pump auxin into the cell98 and export carriers90,99 (namely, the PIN-FORMED (PIN), ABC TRANSPORTER B (ABCB) and PIN-LIKES (PILS) families) that pump auxin from cells into the apoplast. One of the most interesting features of some of the auxin carriers — for example, PIN1 and AUX1 — is their polar subcellular localization in the plasma membrane. This localization pattern in root cells is necessary for accurate canalization of auxin to sink regions to initiate and/or maintain patterning90.
One remarkable example of auxin localization as an essential factor in morphogenesis is the specification of the hypophysis, the founder cell of the quiescent centre (that is, the root organizing centre) and the columella tissue during embryogenesis. This specification event starts with the accumulation of auxin in the proximal part of the suspensor (near the embryo proper). It requires precise auxin build-up in time and space, as shown by polar and opposing localization of PIN1 and PIN7 facing the embryo–suspensor junction, expression of an auxin-responsive marker in the hypophysis (and its adjacent basal cell), and basal-pole defects in pin1 and pin7 mutants100. To our knowledge there is no other similar example of spatiotemporal specification of a single cell by precise canalization of a plant (or animal) hormone. Local accumulation of auxin does occur before patterning of other organs such as lateral roots, floral and leaf primordia101, albeit not in a single cell.
Auxin perception
The auxin signal is perceived by a four-protein receptor complex (known as SCFTIR1) that consists of the E3 ubiquitin-protein ligase RING-BOX PROTEIN 1 (RBX1), S PHASE KINASE-ASSOCIATED PROTEIN 1 (SKP1; also known as ASK1), CULLIN 1 (CUL1) and TIR1 (REFS 102,103). Binding of auxin to the F-box protein TIR1 attaches AUX/IAA ARF inhibitors to the SCFTIR1 complex, directing AUX/IAA proteins for degradation in the proteosome and releasing the ARFs so that they can act as transcription factors (FIG. 5a,b). The A. thaliana genome encodes multiple homologues of each SCF subunit — 23 ARFs, 29 AUX/IAA family members and ~700 predicted F-box genes (in comparison to ~100 in the human genome) — each with potential multiprotein-binding domains104. Thus, combinatorial interactions between these components could provide an explanation for the multiple outputs generated solely by auxin.
An intriguing question is why the auxin pathway operates through a release of repression mechanism (that is, the release of ARFs from AUX/IAA proteins). Mathematical models, as well as experimental data in budding yeast, suggest that molecular titration by protein sequestration can generate an ultrasensitive, all-or-none output (FIG. 5e) that is similar to the cooperativity described below105,106. Using a synthetic regulatory circuit, it has been shown that linear rather than sigmoidal responses are obtained in the absence of an inhibiting protein. Another advantage of repressive mechanisms is that molecular components of a given developmental path are available and readily functional by removal of repression, thereby generating rapid responses.
As described above, precise auxin accumulation is necessary for the specification of the hypophysis. This, in turn, leads to BODENLOS (BDL; also known as IAA12) degradation and derepression of MONOPTEROS (MP; also known as ARF5)107 proximal to the future hypophysis, which promotes transcription of several TARGET OF MONOPTEROS (TMO) proteins108. Movement of TMO7 (also known as BHLH135) from its site of transcription to the hypophysis precursor is required for patterning the hypophysis non-cell autonomously (FIG. 5f), whereas auxin accumulation in the suspensor (but not necessarily in the prohypophysis cell itself) is necessary for the regulation of the ARF9–IAA10 module and the specification of the hypophysis109.
Auxin action during the specification of the hypophysis has similar characteristics to those described by feedforward loops (FFLs)110 (FIG. 5f). Although these network motifs are primarily found in transcriptional networks, the principles are similar. Because the input of both MP–BDL and ARF9–IAA10 are needed, the FFL must have an AND logic gate (FIG. 5f). ARF5 and ARF9 are likely to act as the transcriptional activator and repressor, respectively111,112. Additionally, these two ARFs have antagonistic roles during hypophysis specification109, suggesting that in this case the FFL is an incoherent type 1 FFL. Thus, we hypothesize that the output of this mechanism is a pulse response; that is, specification of the hypophysis is followed by exponential decay of the signal, which eventually reaches a steady state involving stable expression of meristem-specific genes, such as those that encode the PLETHORA (PLT) transcription factors, WOX5 and other factors (including auxin itself).
A second mechanism that might contribute to accurate and stable auxin response is the presence of a negative feedback loop113 (FIG. 5g). This type of regulatory motif is common in animals and bacteria and can be used to increase the stability of a system. Concurrent with AUX/IAA degradation in response to auxin, the mRNA levels of several AUX/IAAs increase within minutes after ARF-mediated transcription114–117, creating a negative feedback loop. These examples of multilevel regulation on auxin perception can generate a bistable switch in which auxin initiates a developmental response and immediately shuts it down to form a new stable pattern. It seems that auxin is not sufficient to activate cellular events but that it has a role in releasing from inhibition processes for which molecular paths are already set.
The SCF complex functions as a molecular hub
It is now clear that the SCF complex has a central role in hormonal signalling. Thus, it might have been evolutionarily selected as a network hub to integrate signals from multiple hormones. Network topology using a central hub has been shown to contribute to robustness and stability in changing environmental conditions in Escherichia coli118. Thus, environmental variations, as well as intrinsic differences between cells, can be moderated by such a mechanism119.
Cooperativity among the SCF subunits can also contribute to the robustness of hormonal signalling. When several proteins act cooperatively, the input–output relationship can be modelled using a Hill function119–121 (FIG. 5c,d), or it can even generate an all-or-none response (FIG. 5e). It is clear that a single discrete output is beneficial, especially in cases in which developmental changes are a primary response to changing environmental conditions. It is possible that the SCF complex acts as a molecular capacitor to buffer inputs from developmental and environmental cues when these are at low levels and to accelerate and amplify them only when they cross a certain threshold. The types of variations in the conditions to which plants are exposed require buffering mechanisms that stabilize physiology and maintain homeostasis.
Each of the SCF subunits possesses several homologues. Thus, various types of interactions can be attained by the modular composition of this complex. For example, both auxin and jasmonate signalling are mediated by the SCF complex with a single difference in the F-box protein: auxin uses TIR1 as a receptor, whereas jasmonate conjugated to isoleucine (the active form) is perceived by the F-box protein CORONATINE-INSENSITIVE PROTEIN 1 (COI1)81.
One controversial issue is whether auxin functions as a morphogen. Currently, there is no evidence to support a model by which discrete auxin concentrations are spatially translated into different developmental outputs. Although the term ‘gradient’ is commonly used to describe auxin distribution92, it has not been shown that it functions in a manner similar to animal morphogens. A recent study on the Bicoid (Bcd) morphogen in Drosophila melanogaster122 shows that it is combinatorial interplay between the anterior-expressed Bcd and the antagonistic posterior repressors that defines the spatial boundaries of downstream genes. According to the authors of this study, these results, as well as others, put considerable limits on the morphogen theory123. Our current understanding is that the data on auxin regulation support various mechanisms other than morphogen gradients that can better explain its mode of action.
Future perspective
The division of this Review into short- and long-range signalling is overly simplistic, as short-range signals can act as long-range signals and vice versa. Although peptides, transcription factors and sRNAs typically function at short ranges, there are examples of each acting more globally. For example, peptides have been suggested to act globally in response to injury124. Similarly, phytohormones can mediate short-range signals and, indeed, often exhibit a global effect through a series of short-range signals.
We are beginning to understand how each of these signalling pathways functions independently, but the current challenge is to develop high-throughput and comprehensive assays to uncover the mechanisms of complex networks. Specifically, we lack straightforward methods for the identification of small peptides and their receptors. Moreover, despite the large-scale bioinformatic identification of putative peptides and receptors, the validation of these potential factors is still very slow. Studies on protein and sRNA movement lag even further behind; without a conserved sequence or mechanism, it is difficult to predict which factors are mobile and to study the mechanism of movement. Another challenge is to develop visualization techniques that allow simultaneous time-lapse imaging of multiple cellular components. Although the current era provides sequencing and bioinformatics tools that can identify thousands of affected genes in different conditions and genotypes, a full understanding of biological processes cannot be achieved without experimental data. An elegant approach that is currently being taken is to label multiple mRNAs, which can be measured concurrently in single cells125. This method, together with others, will allow rapid and accurate quantification of cellular components that together can give a better picture of signalling during developmental processes.
Acknowledgments
Work in the Benfey laboratory is funded by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through grant GBMF3405) to P.N.B., as well as by grants from the US National Institutes of Health (R01-GM043778), the US National Science Foundation and the Defense Advanced Research Projects Agency.
Glossary
- Phytohormones
Signal molecules produced in plants that elicit diverse effects depending on context
- Meristems
Groups of dividing cells (in the root and shoot), which include the stem cells that give rise to new tissues and organs
- Organizing centre
A group of meristem cells in the shoot that have low mitotic activity and that are required for the maintenance of the stem cells
- Quiescent centre
A group of meristem cells in the root that have low mitotic activity and that are required for maintenance of the stem cells
- Initial cells
Progenitor cells of a tissue or an organ
- Cell wall
A structure composed of cellulose, hemicellulose and pectin that surrounds plant cells (and some other organisms such as fungi), contributes towards the overall firmness of the organism and prevents cellular movement
- Kinase-dead
Contains a kinase domain identified by its sequence, but without kinase activity; that is the ability to add phosphate to a substrate
- Morphogen
A signal gradient that has a single source that results in differential output (that is, cell fate) as a readout of the local concentration
- Florigen
A universal mobile signal, originating from leaves, that is necessary for the initiation of flowering in plants
- Hypophysis
The most proximal cell of the suspensor that will initiate the columella and quiescent centre
- Suspensor
A cell population that connects the embryo proper to the endosperm feeding tissue. It forms as a result of asymmetric division of the zygote and functions similarly to the placenta in animals
- Feedforward loops
(FFLs). Gene network motifs in which a protein, X (usually a transcription factor), regulates a target gene, Z, directly as well as indirectly through another regulator, Y. The input of X and Y can be either positive or negative. X and Y might both be required (AND gate) or either one might be sufficient on its own (OR gate)
- Hill function
A function used in biochemistry to describe cooperative binding. It usually reflects the enhanced efficiency of ligand binding as a result of other ligand molecules that are already bound to a receptor
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Bradshaww AD. Evolutionary significance of phenotypic plasticity in plants. Adv Genet. 1965;13:115–155. [Google Scholar]
- 2.Lease KA, Walker JC. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006;142:831–838. doi: 10.1104/pp.106.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy E, Smith S, De Smet I. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell. 2012;24:3198–3217. doi: 10.1105/tpc.112.099010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsubayashi Y. MBSJ MCC Young Scientist Award 2010. Recent progress in research on small posttranslationally modified peptide signals in plants. Genes Cells. 2012;17:1–10. doi: 10.1111/j.1365-2443.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 5.Hirakawa Y, et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiu S-H, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci Signal. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 7.Torii KU. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol. 2004;234:1–46. doi: 10.1016/S0074-7696(04)34001-5. [DOI] [PubMed] [Google Scholar]
- 8.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oelkers K, et al. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- 11.Fletcher JC. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294–294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinoshita A, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 15.Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- 16.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell. 2011;23:851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier D, et al. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:9074–9079. doi: 10.1073/pnas.0503526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castells E, Casacuberta JM. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot. 2007;58:3503–3511. doi: 10.1093/jxb/erm226. [DOI] [PubMed] [Google Scholar]
- 20.Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 21.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 22.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 23.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 24.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Sci Signal. 2008;20:934. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy GV. Live-imaging stem-cell homeostasis in the Arabidopsis shoot apex. Curr Opin Plant Biol. 2008;11:88–93. doi: 10.1016/j.pbi.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 28.Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development. 2006;133:4691–4698. doi: 10.1242/dev.02652. [DOI] [PubMed] [Google Scholar]
- 31.Haecker A. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 33.Stahl Y, Wink RH, Ingram GC, Simon RA. Signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 34.Hobe M, Müller R, Grünewald M, Brand U, Simon RD. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol. 2003;213:371–381. doi: 10.1007/s00427-003-0329-5. [DOI] [PubMed] [Google Scholar]
- 35.Stahl Y, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol. 2013;23:362–371. doi: 10.1016/j.cub.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in. Arabidopsis Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. This article takes a unique approach to identifying peptide signals involved in plant development by examining a mutant of a protein that modifies peptide signals. [DOI] [PubMed] [Google Scholar]
- 37.Meng L, Buchanan BB, Feldman LJ, Luan S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:1760–1765. doi: 10.1073/pnas.1119864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:15067–15072. doi: 10.1073/pnas.0902801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem. 2003;278:24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 40.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, et al. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell. 2010;22:3692–3709. doi: 10.1105/tpc.110.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song SK, Hofhuis H, Lee MM, Clark SE. Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell. 2008;15:98–109. doi: 10.1016/j.devcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucas WJ, et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 45.Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- 46.Lee JY, et al. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rim Y, et al. Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol Cell. 2011;32:519–526. doi: 10.1007/s10059-011-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 2009;57:785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim I, Kobayashi K, Cho E, Zambryski PC. Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2005;102:11945–11950. doi: 10.1073/pnas.0505622102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford KM, Zambryski PC. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001;125:1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J-Y, Zhou J. In: Short and Long Distance Signaling. Kragler F, Hülskamp M, editors. Springer; New York: 2011. pp. 61–86. [Google Scholar]
- 53.Sessions A, Yanofsky MF, Weigel D. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289:779–782. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, et al. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 55.Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 57.Vatén A, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21:1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 58.Koizumi K, Wu S, MacRae-Crerar A, Gallagher KL. An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr Biol. 2011;21:1559–1564. doi: 10.1016/j.cub.2011.08.013. One of the first identifications of a cofactor that facilitates the movement of transcription factors through the plasmodesmata. [DOI] [PubMed] [Google Scholar]
- 59.Harries PA, Schoelz JE, Nelson RS. Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol Plant Microbe Interact. 2010;23:1381–1393. doi: 10.1094/MPMI-05-10-0121. [DOI] [PubMed] [Google Scholar]
- 60.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgenespecific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 63.Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoo BC, et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunoyer P, et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 66.Dunoyer P, et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Brosnan CA, Voinnet O. Cell-to-cell and longdistance siRNA movement in plants: mechanisms and biological implications. Curr Opin Plant Biol. 2011;14:580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skopelitis DS, Husbands AY, Timmermans MCP. Plant small RNAs as morphogens. Curr Opin Cell Biol. 2012;24:217–224. doi: 10.1016/j.ceb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 70.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. This study identified mobile sRNAs, miR165 and miR166, that are required for specific cell fates in the vasculature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dosedependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 72.Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tucker MR, et al. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- 74.Liu Q, et al. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 75.Zhu H, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. This study identified a unique role for an ARGONAUTE protein, AGO10, which acts as a decoy in the shoot meristem to sequester miR165 and miR166 from the meristem and maintain pluripotency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ji L, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Knauer S, et al. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell. 2013;24:125–132. doi: 10.1016/j.devcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 78.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 79.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. These two studies reported the identification of TIR1 as the auxin receptor. [DOI] [PubMed] [Google Scholar]
- 80.Ueguchi Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 81.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fu ZQ, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santner A, Calderon-Villalobos LIA, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nature Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 84.Lumba S, Cutler S, McCourt P. Plant nuclear hormone receptors: a role for small molecules in protein-protein interactions. Annu Rev Cell Dev Biol. 2010;26:445–469. doi: 10.1146/annurev-cellbio-100109-103956. [DOI] [PubMed] [Google Scholar]
- 85.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 86.Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- 87.Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol. 2010;2:a001594. doi: 10.1101/cshperspect.a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 89.Petrášek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 90.Wisniewska J, et al. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- 91.Ljung K. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petersson SV, et al. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 2009;21:1659–1668. doi: 10.1105/tpc.109.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ludwig-Muller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- 94.Sairanen I, et al. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2013;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Staswick PE. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009;150:1310–1321. doi: 10.1104/pp.109.138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cooke TJ, Poli D, Sztein AE, Cohen JD. Evolutionary patterns in auxin action. Plant Mol Biol. 2002;49:319–338. [PubMed] [Google Scholar]
- 97.Dharmasiri S, et al. AXR4 is required for localization of the auxin influx facilitator AUX1. Science. 2006;312:1218–1220. doi: 10.1126/science.1122847. [DOI] [PubMed] [Google Scholar]
- 98.Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 99.Barbez E, et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485:119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- 100.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 101.Benkova E, Michniewicz M, Sauer M, Teichmann T. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 102.Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 103.Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception-structural insights. Cold Spring Harb Perspect Biol. 2010;2:a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA & ARF gene families. Plant Physiol. 2004;135:1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buchler NE, Louis M. Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 106.Cross FR, Buchler NE. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Systems Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weijers D, et al. Auxin Triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 108.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 109.Rademacher EH, et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell. 2012;22:211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 110.Alon U. Network motifs: theory and experimental approaches. Nature Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 111.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alon U. An Introduction to Systems Biology — Design Principles of Biological Circuits. Chapman & Hall/CRC Mathematical & Computational Biology; 2006. [Google Scholar]
- 114.Paponov IA, et al. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant. 2008;1:321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- 115.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- 116.Middleton AM, King JR, Bennett MJ, Owen MR. Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol. 2010;72:1383–1407. doi: 10.1007/s11538-009-9497-4. [DOI] [PubMed] [Google Scholar]
- 117.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 118.Cooper TF, Morby AP, Gunn A, Schneider D. Effect of random and hub gene disruptions on environmental and mutational robustness in Escherichia coli. BMC Genomics. 2006;7:237. doi: 10.1186/1471-2164-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:1–7. [Google Scholar]
- 121.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:1–7. [Google Scholar]
- 122.Chen H, Xu Z, Mei C, Yu D, Small SA. System of repressor gradients spatially organizes the boundaries of bicoid-dependent target genes. Cell. 2012;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turing AM. The chemical basis of morphogenesis. Phil Trans R Soc Lond B. 1952;237:37–72. [Google Scholar]
- 124.Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu Rev Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- 125.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sachs J. Handbuch der experimental-physiologie der Pflanzen: Untersuchungen uber die allgemeinen Lebensbedingungen er Pflanzen und die Functionen ihrer Organe. W. Engelmann; 1865. (in German) [Google Scholar]
- 127.Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Mon Wea Rev. 1920;48:415–415. [Google Scholar]
- 128.Melchers G, Die Wirkungvon Genen S, tiefen Temperaturen und blühenden Propfpartnern auf die Blühreife von Hyoscyamus. Biol Zbl. 1937;57:568–614. (in German) [Google Scholar]
- 129.Chailakhyan MK. About the mechanism of the photoperiodic response. Dokl Akad Nauk SSSR. 1936;1:85–89. (in Russian) [Google Scholar]
- 130.Chailakhyan MK. New facts supporting the hormonal theory of plant development. Dokl Akad Nauk SSSR. 1936;4:77–81. in Russian. [Google Scholar]
- 131.Chailakhyan MK. Bull Acad Sci URSS. 1937. Hormonal Theory of Plant Development; p. 198. (in Russian) [Google Scholar]
- 132.Zeevaart JAD. Flower Formation As Studied By Grafting. H. Veenman; 1958. [Google Scholar]
- 133.Lang A. Physiology of flowering. Annu Rev Plant Physiol. 1952;3:265–306. [Google Scholar]
- 134.Lifschitz E, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. This study made the first correlation between tomato SINGLE FLOWER TRUSS (SFT; the orthologue of FLOWERING LOCUS T in A. thaliana) and the florigen signal originating from the leaf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Notaguchi M, et al. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 2008;49:1645–1658. doi: 10.1093/pcp/pcn154. [DOI] [PubMed] [Google Scholar]
- 136.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 137.Lin MK, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 139.Shalit A, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kehr J. In: Short and Long Distance Signaling. Kragler F, Hülskamp M, editors. Springer; 2011. pp. 131–149. [Google Scholar]
- 141.Ruiz-Medrano R, Kragler F, Wolf S. In: Short and Long Distance Signaling. Kragler F, Hülskamp M, editors. Springer; 2011. pp. 151–177. [Google Scholar]
- 142.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 143.Buhtz A, Pieritz J, Springer F, Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010;10:64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]