Abstract
This highlight introduces the development of hydrogelators consisting of nucleobases, amino acids, and glycosides (i.e., molecular trinity), or nucleobases and amino acids (i.e., nucleopeptides). These novel small molecule hydrogelators self-assemble in water to form stable supramolecular nanofibers/hydrogels and exhibit useful biological properties (e.g., biocompatibility, biostability, and the ability to bind and transport DNA into live cells). The approach discussed here not only provides a new strategy to develop soft biomaterials as a form of nanomedicines, but also contributes to the understanding of molecular self-assembly in water by modulating the non-covalent interactions derived from the three basic building blocks used in living organisms.
1. Introduction
This highlight discusses the recent development on the design, synthesis, and applications of the chimeric molecules, made of nucleobases, amino acids, and glycosides, that self-assemble in water to form multifunctional supramolecular nanostructures and hydrogels.1, 2 Since the existence of life on earth 3.5 billion years ago, nature has relied on three classes of molecules, nucleobases, amino acids, and glycosides, to form nucleic acids, proteins, and polysaccharides, as the essential molecular foundation of life. Although polysaccharides and proteins, the most abundant biomacromolecules, consist of a single class of the building blocks (i.e., glycosides, and amino acids, respectively), nature frequently uses more than one class of the building blocks to form functional biomacromolecules. For example, deoxyribonucleic acids (DNA) and ribonucleic acids (RNA) consist of nucleobases and deoxyribose (for DNA) or ribose (for RNA); peptidoglycans are polymers containing glycosides and amino acids; and glycoproteins are proteins that contain oligosaccharide covalently attached to side-chains of the polypeptides. In fact, tRNA, the uniquely important component for translating genes information into proteins, contains amino acids, ribose, and nucleobases.3 Moreover, biomacromolecules also interact with each other to form various supramolecular architectures. Notably, the self-assembly of DNA and proteins condenses millions of base pairs to a chromosome that has a length of less than a few microns.4 These fascinating facts raise intriguing, fundamental questions: What are the minimal sets or combination of the basic biological building blocks needed for sufficient non-covalent interactions that warrant molecular self-assembly in water? What would be the morphologies and/or functions of the resulting supramolecular architectures? How does one optimize the structures of these minimal motifs for desired functions and materials?
Two necessary tasks may help answer the questions raised above: synthesizing small molecules made of nucleobases, amino acids, and glycosides, and testing their self-assembly in water. While the simplest way to integrate nucleobases, amino acids, and glycosides is to connect them covalently (Scheme 1a),1, 2 the easiest assay of molecular self-assembly in water is, probably, hydrogelation by these molecules (though the self-assembly could occur without the formation of a bulk hydrogel). In fact, there are ample examples from literature where each system based on nucleobase, sugar, or amino acids results in self-assembly leading to nanofibers/hydrogel formation. For example, the early works by Shinkai et al. that introduced a uracil-appended cholesterol gelator,5 the studies by Shimizu et al. that reported the hydrogels of bolaamphiphiles consisting of nucleotide (7),6 and the recent works by Barthelemy et al. that described uridine phosphocholine amphiphiles (8) to form hydrogels and organogels,7 clearly, reveal that small molecules containing nucleotides are able to self-assemble in water. Although those molecules require a long alkyl chain to be amphiphilic, their self-assembly in water implies that the replacement of the long chain(s) in those molecules by proper amino acids should be a feasible approach to create a set of new molecules to self-assemble in water and form supramolecular nanostructures. In addition, the recent demonstration of the hydrogelation of small molecules from amino acids8, 9 or different glycoside derivatives10 through intermolecular interactions further supports the notion that integration of nucleobases, amino acids, and glycosides by synthetic manipulations will lead to a novel system for supramolecular self-assembly.
Scheme 1.
The molecular structures of the hydrogelators based on (a) the conjugates of nucleobase, amino acid, and glycoside or the conjugate of nucleobase and amino acid; (b) bolaamphiphiles of nucleotides and uridine phosphocholine amphiphiles
The objective of this highlight is to illustrate that the small molecules made of nucleobases, amino acids, and glycosides promise a new opportunity to generate the self-assembly of functional molecules in water, including forming new hydrogels as soft nanomaterials.9, 11 That is, the conjugation of nucleobase, amino acid, and glycoside leads to a new supramolecular system that exhibits diverse structural morphologies in nanoscale (e.g., nanoparticles, nanofibers, or other nanostructures) and has useful biological properties (e.g., biostability, biocompatibility, and the ability to bind and transport DNA into live cells). Because it is possible to incorporate other functional peptides or glycoside into the chimeric molecules to interact with multiple proteins, protein and nucleic acid, or only nucleic acids for target(s)-specific functions, this approach provides a wonderful opportunity for developing biomaterials. Moreover, since life itself is a phenomenon at nanoscale,12 this approach will facilitate the design of supramolecular assemblies that mimic the molecular organization observed in nature, a fundamental aspect of the research on soft nanomaterials.
2. Molecules for self-assembly and hydrogelation
Molecular trinity
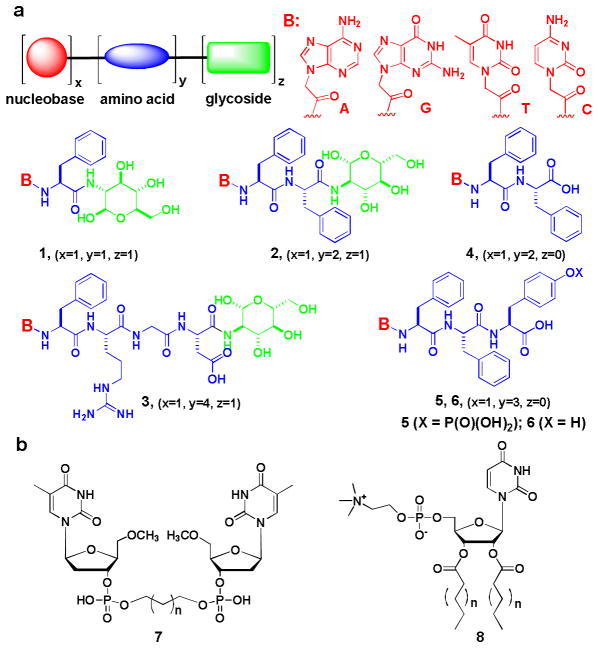
Due to the unique importance of nucleobase, amino acid, and glycoside in living organisms, we term the chimeric molecules consisting of them as “molecular trinity”. We connected a nucleobase (e.g., adenine, guanine, thymine, or cytosine), an amino acid, a dipeptide, or a tetrapeptide (e.g., phenylalanine, diphenylalanine, or Phe-Arg-Gly-Asp), and a glycoside (e.g., D-glucosamine) via covalent bonds and successfully obtained a new class of hydrogelators (1, 2, or 3 in Scheme 1a) that self-assemble in water to afford molecular nanofibers and hydrogels. For example, compound 1T derived from the conjugation of thymine, phenylalanine, and D-glucosamine forms molecular nanofibers (12 nm in width) in water and results in a hydrogel at pH 7.0 and concentration of 3.0 wt% (Figure 1). When it has two phenylalanine, another hydrogelator 2T self-assembles to generate molecular nanofibers, with the width of 15 nm, that appear to bend easily and to crosslink relatively efficiently, thus forming a fine network to support the hydrogel at 3.0 wt% and pH 8.5 (Figure 1). The replacement of thymine by other nucleobases affords a series of novel hydrogelators (1A, 1G, 2C, 2A, and 2G) that self-assemble in water to form nanostructures/hydrogels at 3.0 wt% and proper pHs. The introduction of RGD, a well-established epitope that promotes cell-adhesion,13 into 2T gives 3, which forms nanofibers with the diameter of 25 nm and results in a hydrogel at 3.0 wt% and pH 4.0. The characterization of these hydrogelators (by TEM and CD) suggests that the incorporation of glycoside in these molecular trinity hydrogelators results in quite diverse morphologies of the self-assembled structures in nanoscale, which likely originates from the unique nature of sugar molecules.
Figure 1.
The chemical structure (A) of 1T, (B) of 2T; (C) optical image of the hydrogel formed by 1T and the transmission electron micrograph (TEM) of the hydrogel of 1T, (D) optical image of the hydrogel formed by 2T and the transmission electron micrograph (TEM) of the hydrogel of 2T. Scale bar = 100 nm
Nucleopeptides
When z = 0, the molecular trinity reduces to a type of nucleopeptides (e.g., 4, 5, and 6 in Scheme 1a),2 and 4T, 4A, 4C, and 4G are hydrogelators.2 The addition of a tyrosine phosphate residue to the C-terminal of 4 yields another group of nucleopeptides 5, which, in the presence of an alkaline phosphatase (ALP), lose the phosphate groups to generate hydrogelators 6T, 6A, and 6G that result in supramolecular nanofibers and hydrogels at pH 7.4 and concentration of 2.0 wt%. Unlike hydrogelators 1, 2, and 3, nucleopeptide hydrogelators (4 or 5) self-assemble to form mainly thin nanofibers that adopt typical β-sheet as the secondary structures. As there is little work to use nucleopeptides for making materials, the hydrogels made of nucleopeptides represent the first kind of soft materials based on nucleopeptides.
3. Biological properties and applications
Interbase interactions
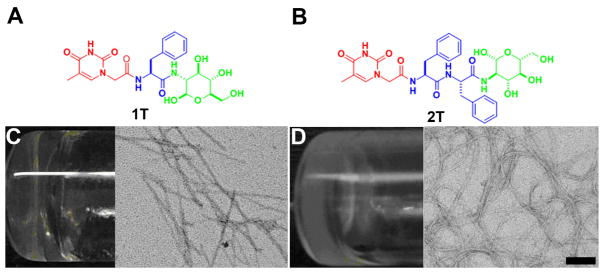
The presence of nucleobase(s) in the hydrogelators shown in Scheme 1a offers the interbase interactions (Waston-Crick and/or Hoogsteen) among the hydrogelators or between the hydrogelators and nucleic acids. While Hoogsteen interactions likely promote the self-assembly of the hydrogelators with same structure for hydrogelation, Waston-Crick interactions clearly contribute to the formation of hydrogels of the mixture of a pair of hydrogelators that have complementary structures (e.g., 1T+1A or 3T+3A).2 Moreover, the addition of an single strand oligomeric deoxyadenosine (A10) to the viscous solution of 1T (2.1 wt%, pH 7.0, Figure 2A) affords a stable gel (Figure 2B), accompanying by the increase of storage modulus (G′) from 4.4 × 102 Pa (of the solution of 1T) to 9.5 × 102 Pa (of the hydrogel of 1T plus A10) (Figure 2C). This result suggests the interbase pairing and phosphate-sugar interactions favor the formation of the supramolecular assembly of 1T and A10. Furthermore, the addition of A10 to the hydrogel of 1T results in distinctive changes in the CD spectra (Figure 2D), suggesting that the formation of A10-1T complex enhances the base stacking of A10 and affects the superstructure of the self-assembly of 1T. The preservation of the function of the nucleobase in these hydrogelators promises a new class of neutral molecules for binding with ssDNA or RNA.
Figure 2.
Optical images of (A) the highly viscous solution of 1T (2.1 wt%, pH=7.0); (B) 1T+deoxyadenosine (A10) mixed hydrogel after the addition of deoxyadenosine (A10) in 1:1 molecular ratio; (C) dynamic storage moduli (G′) of the hydrogels of 1T and 1T+deoxyadenosine (A10) mixed gel; (D) CD spectra of the hydrogels of 1T, the mixed hydrogel of 1T with deoxyadenosine (A10) in 1:1 molecular ratio, and the deoxyadenosine (A10) solution.
Biocompability
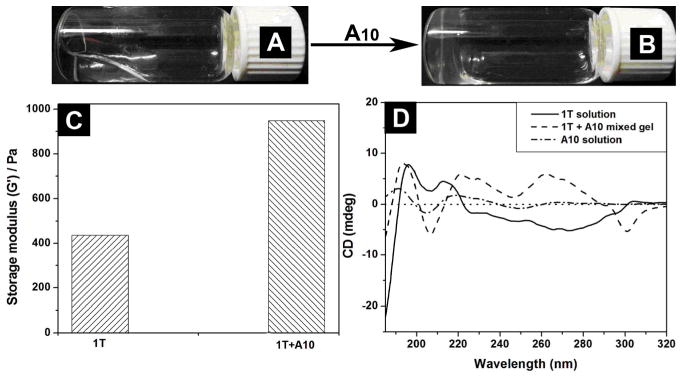
As evidenced by the cell viability tests and wound healing studies,1, 2 these hydrogelators, of molecular trinity or nucleopeptides, all exhibit excellent biocompatibility. As shown in Figure 3A, the cell viability remains above 90% after the cells are incubated with the 500 μM of the hydrogelator (1T or 4T) for 72 h. Although the cell viability decreases slightly when the cells are incubated with 500 μM of 1C, 1A, 4G or 5G for 72 h, the value of IC50 is still > 500 μM.1, 2 These results confirm that these chimeric molecules are biocompatible. In addition, in vitro wound-healing assay1, 2 shows that hydrogelator 2T or 6T exhibits little inhibitory effect on the migration of cells, further proving biocompatibility of these hydrogelators.
Figure 3.
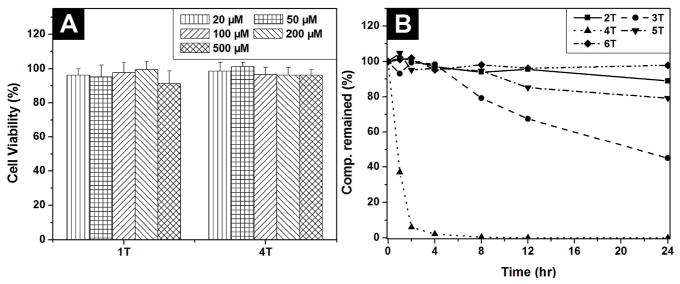
72 hr cell viability test of (A) hydrogelator 1T and 4T; (B) the time-dependent course of the digestion of hydrogelators of 2T, 3T, 4T, 5T and 6T by proteinase K.
Biostability
Besides biocompatibility, biostability is an essential requisite for the application of biomaterials. Being incubated with proteinase K,14 a powerful protease for hydrolysis of a wide range of peptidic substrates, 2T or 3T, which contains a glycoside at its C-terminal, exhibits excellent biostability; more than 90 % of 2T or 3T remains intact after being incubated with proteinase K for 24 h (Figure 3B). Less than 10% of 4T, however, remains after 4 h of the incubation with proteinase K. These results not only agree with that glycosylation is a strategy, widely used in biological systems, for enhancing the stability of proteins without comprising functions,15 but also indicate that the incorporation of glycoside to the C-terminal of amino acid/peptide would be an advantageous approach for developing biostable and multifunctional hydrogels for the applications that require long-term biostability. Interestingly, more than 80% of 5T and more than 97 % of 6T remain after 24 h of incubation with proteinase K (Figure 3B),2 suggesting that the incorporation of nucleobase may also enhance the biostability of these peptidic hydrogelators.
DNA delivery
The proof of interbase interaction between 1T and A10 in CD and rheology studies (Figure 2) leads to the use of such interactions for the delivery of DNA or RNA into cells because it could offer a new non-viral vector for gene therapy.16 The incubation of HeLa cells with 1T and fluorescein-labeled single strand oligonucleotide (FAM-A10) result in the green fluorescence in both the cytosols and the nuclei of the HeLa cells (Figure 4); in the control experiment (i.e., without using 1T), green fluorescence is absent from the cytosols and nuclei of the HeLa cells. This result indicates that 1T interacts with and delivers the oligonucleotide FAM-A10 into live cells. Although intensive research activities have focused on the development of various non-viral vectors for gene delivery in recent years, including cationic lipids and polymers, those synthetic vectors have suffered from low gene-transfer efficiency, toxicity, or in vivo instability. Thus, 1T, as the first example of the use of the molecular trinity hydrogelators for the delivery of nucleic acids into cells, represents a new type of neutral, biocompatible and biostable agent for DNA delivery, which could lead to the development of new non-viral vectors for both in vitro and in vivo applications.
Figure 4.
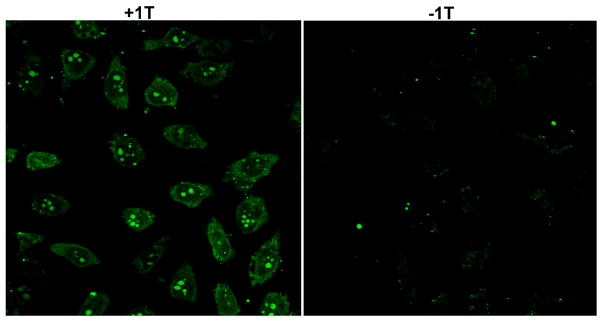
Fluorescence microscopy images show subcellular distribution of A10, which is labeled with fluorescein dye (green). (Left) 500 μM 1T and 1 μM FAM-A10 incubated with HeLa cells for 24 h. (Right) 1 μM FAM-A10 incubated with HeLa cells for 24 h.
4. Conclusion and outlook
In summary, the simple conjugation of nucleobases, amino acids, and glycosides turns out to be an effective way to generate useful, functional small molecules that self-assemble in water. This discovery not only introduces a facile way to expand the current repertoire of building blocks for generating supramolecular nanofibers and hydrogels from small, biologically important molecules, but also promises novel, functional soft nanomaterials for a variety of potential applications in biomedicines. The further development of these chimeric molecules would generate more significant results if the following aspects are explored.
Molecular design
Besides the change of x, y, z in molecules shown in Scheme 1a, what are the other structures or geometry suitable for generating molecular trinities for self-assembly? For example, must the molecules have the order of nucleobase-amino acid-glycoside for self-assembling in water? What are the options for making molecular trinities that have non-linear geometry? Of course, when one designs the molecules, the functions of the resulting supramolecular architectures should be a key factor. But the production and exploration of the unknown structures, in this case, might lead to surprise discovery and exciting opportunities as well.
Functions and applications
Clearly, the judicious selection of the peptide or glycoside segments to be introduced into the molecular trinities promises the delivery of DNA or RNA17 to specifically targeted cells, which deserves further exploration. In addition, the incorporation of other functional peptides or glycoside into the molecular trinities provides a facile way to explore many other functions, including nucleic acids condensation,18 endosomal escape,19 blocking metabolism,20 cell receptor targeting10, 19 and nuclear localization.21 The recent work on aminoglycosyl nucleosides and sugar-amino acid-nucleosides as the potential ligands targeting RNA or the inhibitors for glycosyltransferase,22 in fact, supports that the integration of sugar, amino acid, nucleoside/nucleotide together will lead to multifunctional hydrogelators as potential molecular therapeutics.
Other fundamental issues
It remains a mystery that why all cells use the similar collection of small molecules (nucleotide, amino acid, sugar, and lipid) for living. We hope the exploration of the self-assembly of these small molecules to provide insights on some questions that relate to the origin of life. For example, the hypothesis of “RNA world”23 has gained the popularity for recent years. Can the self-assemblies (or hydrogels)24 of the molecular trinity act like catalysts in water, like RNAs do? Can the supramolecular assemblies of the molecular trinities regulate biological processes, like proteins do? Of course, the answers may not come easily, but the problems certainly are intriguing and worth the exploration.
Acknowledgments
This work was partially supported by a NIH grant (R01CA142746), a HFSP grant (RGP0056/2008), and start-up fund from Brandeis University.
Notes and references
- 1.Li X, Kuang Y, Shi J, Gao Y, Lin HC, Xu B. J Am Chem Soc. 2011;133:17513–17518. doi: 10.1021/ja208456k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Kuang Y, Lin HC, Gao Y, Shi J, Xu B. Angew Chem Int Ed. 2011;50:9365–9369. doi: 10.1002/anie.201103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science Publishing; 2002. [Google Scholar]
- 4.Nelson DL, Cox MM, Lehninger . Principles of Biochemistry. 4. New York: W.H. Freeman; 2005. [Google Scholar]; Youngson RM. Collins Dictionary of Human Biology. 7. Glasgow: HarperCollins; 2006. [Google Scholar]
- 5.Snip E, Koumoto K, Shinkai S. Tetrahedron. 2002;58:8863–8873. [Google Scholar]
- 6.Iwaura R, Yoshida K, Masuda M, Yase K, Shimizu T. Chem Mater. 2002;14:3047–3053. [Google Scholar]
- 7.Moreau L, Barthelemy P, El Maataoui M, Grinstaff MW. J Am Chem Soc. 2004;126:7533–7539. doi: 10.1021/ja039597j. [DOI] [PubMed] [Google Scholar]; Latxague L, Ziane S, Chassande O, Patwa A, Dalila MJ, Barthelemy P. Chem Commun. 2011;47:12598–12600. doi: 10.1039/c1cc13948g. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SG, Marini DM, Hwang W, Santoso S. Curr Opin Chem Biol. 2002;6:865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]; Zhang Y, Yang ZM, Yuan F, Gu HW, Gao P, Xu B. J Am Chem Soc. 2004;126:15028–15029. doi: 10.1021/ja044401g. [DOI] [PubMed] [Google Scholar]; Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Soft Matter. 2006;2:822–835. doi: 10.1039/b607706d. [DOI] [PubMed] [Google Scholar]; Pal A, Ghosh YK, Bhattacharya S. Tetrahedron. 2007;63:7334–7348. [Google Scholar]; Adams DJ, Butler MF, Frith WJ, Kirkland M, Mullen L, Sanderson P. Soft Matter. 2009;5:1856–1862. [Google Scholar]; Branco MC, Schneider JP. Acta Biomater. 2009;5:817–831. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adams DJ, Morris K, Chen L, Serpell LC, Bacsa J, Day GM. Soft Matter. 2010;6:4144–4156. [Google Scholar]; Adams DJ, Topham PD. Soft Matter. 2010;6:3707–3721. [Google Scholar]
- 9.Yang Z, Liang G, Xu B. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Acharya SNG. Chem Mater. 1999;11:3504–3511. [Google Scholar]; Wang G, Cheuk S, Yang H, Goyal N, Reddy PVN, Hopkinson B. Langmuir. 2009;25:8696–8705. doi: 10.1021/la804337g. [DOI] [PubMed] [Google Scholar]; Goyal N, Cheuk S, Wang G. Tetrahedron. 2010;66:5962–5971. [Google Scholar]
- 11.Estroff LA, Hamilton AD. Chem Rev. 2004;104:1201–1217. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]; Banerjee S, Das RK, Maitra U. J Mater Chem. 2009;19:6649–6687. [Google Scholar]
- 12.Mann S. Angew Chem Int Ed. 2008;47:5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 13.Hart SL, Harbottle RP, Cooper R, Miller A, Williamson R, Coutelle C. Gene Ther. 1995;2:552–554. [PubMed] [Google Scholar]; Ruoslahti E. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]; Hart SL, Collins L, Gustafsson K, Fabre JW. Gene Ther. 1997;4:1225–1230. doi: 10.1038/sj.gt.3300513. [DOI] [PubMed] [Google Scholar]; Temming K, Schiffelers RM, Molema G, Kok RJ. Drug Resist Update. 2005;8:381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]; Chen K, Chen X. Theranostics. 2011;1:189–200. doi: 10.7150/thno/v01p0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bromme D, Peters K, Fink S, Fittkau S. Arch Biochem Biophys. 1986;244:439–446. doi: 10.1016/0003-9861(86)90611-9. [DOI] [PubMed] [Google Scholar]; Liang GL, Yang ZM, Zhang RJ, Li LH, Fan YJ, Kuang Y, Gao Y, Wang T, Lu WW, Xu B. Langmuir. 2009;25:8419–8422. doi: 10.1021/la804271d. [DOI] [PubMed] [Google Scholar]
- 15.Sola RJ, Griebenow K. J Pharm Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]; Culyba EK, Price JL, Hanson SR, Dhar A, Wong CH, Gruebele M, Powers ET, Kelly JW. Science. 2011;331:571–575. doi: 10.1126/science.1198461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulligan RC. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]; Anderson WF. Nature. 1998;392:25–30. doi: 10.1038/32058. [DOI] [PubMed] [Google Scholar]; Niidome T, Huang L. Gene Ther. 2002;9:1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]; El-Aneed A. J Controlled Release. 2004;94:1–14. doi: 10.1016/j.jconrel.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R, Ramsey P, Martin M, Felgner PL. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]; Koltover I, Salditt T, Radler JO, Safinya CR. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]; Luo D, Saltzman WM. Nature Biotechnology. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]; Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Gottschalk S, Sparrow JT, Hauer J, Mims MP, Leland FE, Woo SLC, Smith LC. Gene Ther. 1996;3:448–457. [PubMed] [Google Scholar]; Plank C, Tang MX, Wolfe AR, Szoka FC. Hum Gene Ther. 1999;10:2272–2272. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 19.Midoux P, Kichler A, Boutin V, Maurizot JC, Monsigny M. Bioconjugate Chem. 1998;9:260–267. doi: 10.1021/bc9701611. [DOI] [PubMed] [Google Scholar]; Deshayes S, Morris MC, Divita G, Heitz F. Cell Mol Life Sci. 2005;62:1839–1849. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu GY, Wu CH. J Biol Chem. 1987;262:4429–4432. [PubMed] [Google Scholar]; Wu GY, Wu CH. J Biol Chem. 1988;263:14621–14624. [PubMed] [Google Scholar]; Wu GY, Wu CH. Biochemistry. 1988;27:887–892. doi: 10.1021/bi00403a008. [DOI] [PubMed] [Google Scholar]; Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. J Pharm Sci. 1998;87:678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 21.Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD. Nature. 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]; Lanford RE, Kanda P, Kennedy RC. Cell. 1986;46:575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhang GS, Guan Z, Zhang LR, Min JM, Zhang LH. Bioorg Med Chem. 2003;11:3273–3278. doi: 10.1016/s0968-0896(03)00278-5. [DOI] [PubMed] [Google Scholar]; Vembaiyan K, Pearcey JA, Bhasin M, Lowary TL, Zou W. Bioorg Med Chem. 2011;19:58–66. doi: 10.1016/j.bmc.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert W. Nature. 1986;319:618–618. [Google Scholar]; Szostak JW. Nature. 2009;459:171–172. doi: 10.1038/459171a. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Yang Z, Zhang X, Xiao X, Chang CK, Xu B. Angew Chem Int Ed. 2007;46:4285–4289. doi: 10.1002/anie.200700404. [DOI] [PubMed] [Google Scholar]; Wang Q, Yang Z, Gao Y, Ge W, Wang L, Xu B. Soft Matter. 2008;4:550–553. doi: 10.1039/b715439a. [DOI] [PubMed] [Google Scholar]