Abstract
Background
Cardiotoxicity is a leading cause for drug attrition during pharmaceutical development and has resulted in numerous preventable patient deaths. Incidents of adverse cardiac drug reactions are more common in patients with pre-existing heart disease than the general population. Here we generated a library of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) from patients with various hereditary cardiac disorders to model differences in cardiac drug toxicity susceptibility for patients of different genetic backgrounds.
Methods and Results
Action potential duration (APD) and drug-induced arrhythmia were measured at the single cell level in hiPSC-CMs derived from healthy subjects and patients with hereditary long QT syndrome (LQT), familial hypertrophic cardiomyopathy (HCM), and familial dilated cardiomyopathy (DCM). Disease phenotypes were verified in LQT, HCM, and DCM iPSC-CMs by immunostaining and single cell patch clamp. Human embryonic stem cell-derived cardiomyocytes (hESC-CMs) and the human ether-a-go-go-related gene (hERG) expressing human embryonic kidney (HEK293) cells were used as controls. Single cell PCR confirmed expression of all cardiac ion channels in patient-specific hiPSC-CMs as well as hESC-CMs, but not in HEK293 cells. Disease-specific hiPSC-CMs demonstrated increased susceptibility to known cardiotoxic drugs as measured by APD and quantification of drug-induced arrhythmias such as early after depolarizations (EADs) and delayed after depolarizations (DADs).
Conclusions
We have recapitulated drug-induced cardiotoxicity profiles for healthy subjects, LQT, HCM, and DCM patients at the single cell level for the first time. Our data indicate that healthy and diseased individuals exhibit different susceptibilities to cardiotoxic drugs and that use of disease-specific hiPSC-CMs may predict adverse drug responses more accurately than standard hERG test or healthy control hiPSC-CM/hESC-CM screening assays.
Keywords: Induced pluripotent stem cells, stem cells, cardiotoxicity, arrhythmia
INTRODUCTION
Cardiac toxicity is a leading cause for drug attrition during the clinical development of pharmaceutical products 1. Of particular concern is the potential of new chemical entities (NCEs) to induce prolongation of the QT interval, a condition known to cause torsades de pointes (TdP), a rapid polymorphic ventricular tachyarrhythmia that can lead to sudden cardiac death 2–4. To minimize cardiac risks associated with drug development, both the pharmaceutical industry and U.S. Food and Drug Administration (FDA) have mandated in vitro cardiotoxicity screening early in NCE development 5, 6. In spite of such guidelines, drug-induced TdP has resulted in numerous preventable patient deaths and the costly withdrawal of associated pharmacological products from the market 7–9.
A central reason for the high rates of adverse cardiac drug reactions observed in patients is the limited capacity of preclinical screening assays to detect cardiotoxicity. Current in vitro toxicity screens rely on the artificial expression of single cardiac ion channels in genetically transformed cell lines such as Chinese hamster ovary (CHO) or human embryonic kidney (HEK) cells, which do not accurately model pertinent genetic, cellular, or biochemical characteristics of the human heart. The use of CHO and HEK cells to assess cardiotoxicity is impaired by genetic aberrations accumulated in these cells and the failure of ectopically expressed channels to accurately model the same channels found in human cardiomyocytes (CMs) 10, 11. In addition, blockade of single ion channels alone has proven to be an imperfect measure of QT prolongation as CM electrophysiology is regulated by the concurrent activity of multiple ion channels. Consequently, screening of drugs that block single ion channels alone as with CHO or HEK cells can produce false negatives (i.e., Alfuzosin) and false positives (i.e., Verapamil), leading to the market release of potentially lethal drugs and the attrition of valuable drugs, respectively 12–15.
To improve the accuracy of toxicity screening, preclinical drug tests ideally would be conducted on adult human CMs. Unfortunately, this has not been feasible in early stage drug discovery due to the difficulties in obtaining cardiac tissue from patients and the inability to propagate these cells in culture. The recent derivation of human CMs from embryonic stem cells (hESC-CMs) and induced pluripotent stem cells (hiPSC-CMs) represents a possible method to circumvent these hurdles, because both hESC-CMs and hiPSC-CMs possess many of the same electrical characteristics as primary human CMs and can be generated in unlimited quantities from pluripotent cell sources 16, 17.
The use of patient-specific hiPSC-CMs offers a unique opportunity to transform drug toxicity screening because the majority of individuals who experience adverse cardiac drug responses belong to specific high-risk demographics 18. For example, episodes of TdP and sudden cardiac death related to the gastromotility agent cisapride in the mid-1990s were largely absent from the general population and limited to patients with pre-existing heart conditions such as long QT syndrome and heart failure 19. Several recent reports have detailed the derivation hiPSC-CMs from patients with long QT syndrome (LQT), as well as other hereditary cardiac disorders such as LEOPARD syndrome, catecholaminergic polymorphic ventricular tachycardia (CPVT), familial hypertrophic cardiomyopathy (HCM), and familial dilated cardiomyopathy (DCM) 20–26. However, these reports did not examine whether such patient-specific cells accurately model torsadogenic responses to cardiotoxic drugs. In this study, we therefore generated a disease-specific hiPSC-CM library from patients with common hereditary cardiac disorders and tested the capacity of this panel to be used as a surrogate in vitro model for prediction of cardiac drug toxicity in patient groups at high risk for drug-induced TdP.
METHODS
Culture and maintenance of undifferentiated hESCs and hiPSCs
The H9 hESC line was obtained from WiCell (Madison, WI). Characteristics of hiPSC lines including mutations are summarized in Supplemental Table 1. hESCs and hiPSCs derived from healthy controls, or patients diagnosed with LQT syndrome, HCM, or DCM were maintained in feeder-free mTeSR1 medium (STEMCELL Technologies, Vancouver, Canada) on Matrigel-coated (BD Bioscience, San Jose, CA) plates at 37°C with 5% (vol/vol) CO2.
Differentiation of hESCs and hiPSCs into cardiac lineages
hESC and hiPSC lines were differentiated via embryoid body (EB) formation using a previously described protocol 27. Briefly, on day 0 of differentiation, hESC and hiPSC colonies were dissociated with Accutase (Sigma) into small clumps of 10–20 cells. Cells were resuspended in 2 ml basic media containing StemPro34 (Invitrogen), 2 mM glutamine (Invitrogen), 0.4 mM monothioglycerol (Sigma), 50 μg/ml ascorbic acid (Sigma), and 0.5 ng/ml BMP4 (R&D Systems, Minneapolis, MN) to form EBs. For the first 1–4 days of cardiac differentiation, cells were treated with 10 ng/ml BMP4, 5 ng/ml human bFGF (R&D Systems), and 3 ng/ml activin A (R&D Systems) added to the basic media. On days 4–8, EBs were re-fed with basic media containing human 50 ng/ml DKK1 (R&D Systems) and 10 ng/ml human VEGF (R&D Systems). From day 8 onwards, cells were treated with basic media containing 5 ng/ml human bFGF and 10 ng/ml human VEGF. Beating EBs were dissociated into single beating hiPSC-CMs for patch clamping and drug treatment assays.
Assessment of disease phenotype by measurement of cell size and myofilament disarray
The in vitro disease phenotypes for diseased hiPSC-CMs was assessed by immunofluorescence staining for cardiac disease markers, quantification of disorganized saromeric content, and assessment of cell size. Beating hiPSC-CMs were plated on gelatin-coated dishes. After three days in culture, beating hiPSC-CMs were trypsinized and re-plated at low density on gelatin-coated chamber slides (Nalgene Nunc International, Rochester, NY). Cells were fixed with 4% paraformaldehyde (Sigma), permeabilized in 0.3% Triton (Sigma), blocked using 5% BSA, and stained with primary antibody overnight at 4°C. Stained cells were washed three times with PBS, and then incubated with secondary antibody for 1 h at 25°C. Sarcomeric disarray and cell size of normal and diseased hiPSC-CMs were quantified using the ImageJ software package (National Institutes of Health, Bethesda, MD). Cells with disorganized sarcomeres were defined as having greater than 25% cell area of punctate troponin T distribution without striations, suggesting disorganized myofilament structure 23.
Compound solutions used in drug screening assays
All drugs used in this study were purchased from Sigma-Aldrich. Cisapride, nicorandil, and alfuzosin were dissolved in DMSO, while verapamil was dissolved in H2O. Drug compound solution was prepared as a 10 mM stock in a glass vial. Stock solution was mixed vigorously for 10 min at room temperature. For testing, pharmaceutical compounds were diluted in a glass vial using external solution (137 mM NaCl, 4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2, and 10 mM HEPES). Dilutions were prepared within 30 min of treatment. Equal amounts of DMSO (0.1%) were present at final dilution and used as vehicle controls.
Electrophysiologcal recordings of HEK293 cells and hESC-CMs/hiPSC-CMs
hERG tail currents were recorded from hERG expressing HEK293 cells with the use of standard patch-clamp techniques, as previously reported 6. To record action potentials, contracting embryoid bodies (EBs) were mechanically isolated, enzymatically dispersed into single cells, and attached to 0.1% gelatin-coated glass coverslips. Single beating cardiomyocytes were subjected to whole-cell patch-clamp at 36–37°C using an EPC-10 patch-clamp amplifier (HEKA, Lambrecht, Germany) attached to a RC-26C recording chamber (Warner) and mounted on to the stage of an inverted microscope (Nikon, Tokyo, Japan). Data were acquired using PatchMaster software (HEKA, Germany) and digitized at 1.0 kHz. Current-clamp recordings were conducted in Tyrodes solution. Significant APD90 prolongation is defined in this study as >10% change in APD90 at the indicated concentration 11.
Statistical analysis
Statistical significance was determined by paired or unpaired Student’s t test (two-tail) for comparison between two groups. For statistical comparison between more than two groups, one-way ANOVA followed by the Dunnett test was employed. *P < 0.05 was considered statistically significant, ** indicates P < 0.01, and *** indicates P < 0.001.
A more detailed description of the methods used in this study can be found in the Supplemental Methods section of the manuscript.
RESULTS
Patient recruitment and confirmation of disease-specific mutations
Protocols for this study were approved by the Stanford University Human Subjects Research Institutional Review Board, and written consent was obtained from all study participants. Dermal fibroblasts were isolated from three distinct family cohorts of patients diagnosed at Stanford Hospital with hereditary mutations for long QT syndrome (LQT), hypertrophic cardiomyopathy (HCM), and dilated cardiomyopathy (DCM) (n=3 patients per cohort; Supplemental Figure 1a, Supplemental Table 1) as previously reported 23, 26. Nucleotide sequence analysis confirmed the presence of known causal disease mutations for each respective patient cohort, specifically the G269S missense mutation in KCNQ1 for the LQT cohort, R663H in MYH7 for the HCM cohort, and R173W in TNNT2 for the DCM cohort (Supplemental Figure 1b–d). Screening of hESCs (WA09 line) and family-matched control hiPSCs did not reveal any known mutations related to LQT, HCM, or DCM (Supplemental Table 2). To ensure genetic diversity among the control group, skin biopsies were obtained from at least one healthy individual from each of the LQT, HCM, and DCM family cohorts to serve as a disease-free control.
Generation of patient-specific iPSCs from dermal fibroblasts
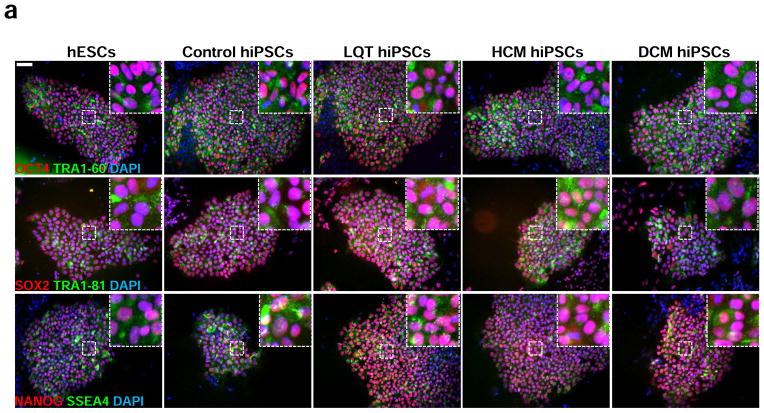
Disease-specific hiPSCs were generated from fibroblasts derived from LQT, HCM, and DCM patient cohorts through lentiviral infection with the reprogramming factors OCT4, SOX2, MYC, and KLF4. Control hiPSCs were derived from age-matched healthy subjects from the same diseased family cohorts. A minimum of 3 distinct lines was generated per patient. Pluripotency was confirmed through immunofluorescence staining for the ESC markers TRA-1-60, TRA-1-81, and SSEA-4 (Figure 1a). Silencing of reprogramming transgenes was determined by qPCR for total and endogenous expression of the pluripotency transcription factors OCT4, SOX2, MYC, and Klf4 (Supplemental Figure 2). Embryoid body (EB) and teratoma formation assays demonstrated capacity of all lines to generate cellular derivatives of all three germ layers in vitro and in vivo (Supplemental Figure 3).
Figure 1.
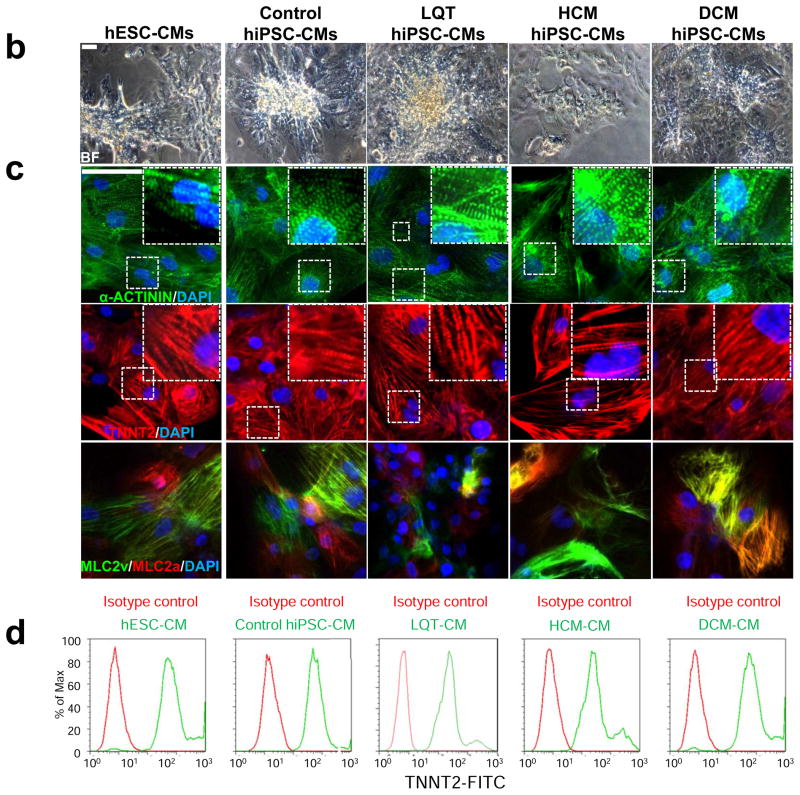
Characterization of disease-specific hiPSC lines and hiPSC-CMs from control, LQT, HCM, and DCM patients. (a) Representative immunostaining for a panel of pluripotent stem cell markers, including OCT4, SOX2, NANOG, TRA1-60, TRA1-81, and SSEA4 in H9 hESCs, control hiPSCs, LQT hiPSCs, HCM hiPSCs, and DCM hiPSCs. Scale bar, 50 μM. (b) Brightfield (BF) images of hiPSC-CMs generated from disease-specific cohorts and from H9 hESCs. Scale bar, 50 μM. (c) Immunostaining for the cardiac sarcomere markers α-ACTININ and TNNT2. Scale bar, 50 μM. (d) Flow cytometry analysis for the percentage of cells exhibiting positive TNNT2 staining. Isotype controls are shown in red, whereas stained hiPSC-CMs are shown in green.
Differentiation of patient-specific hiPSCs into cardiomyocytes
Patient-specific hiPSCs were differentiated into cardiomyocytes using standard 3D EB differentiation protocols as previously described 28. Differentiated EBs exhibited spontaneously contracting foci as early as ten days following initiation of cardiac differentiation (Supplemental Video 1). Dissociation of beating colonies into single cardiomyocytes demonstrated that single hESC-CMs and hiPSC-CMs maintained spontaneous contraction and positive staining for distinct myocyte markers (Figure 1b–c) and spontaneous contraction (Supplemental Video 2). FACS analysis for Troponin T at 30 days post-differentiation revealed beating cultures contained cardiomyocyte purities of greater than 90% (Figure 1d). No significant discrepancies in efficiency of cardiac differentiation were observed among disease-specific hiPSC lines, control hiPSC lines, and control hESC lines (Supplemental Figure 4).
Comparison of in vitro disease phenotypes for patient-specific hiPSC-CMs
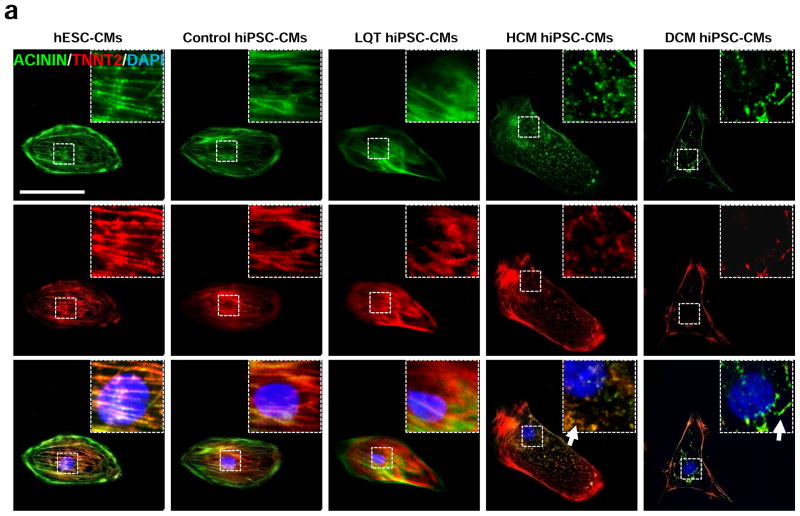
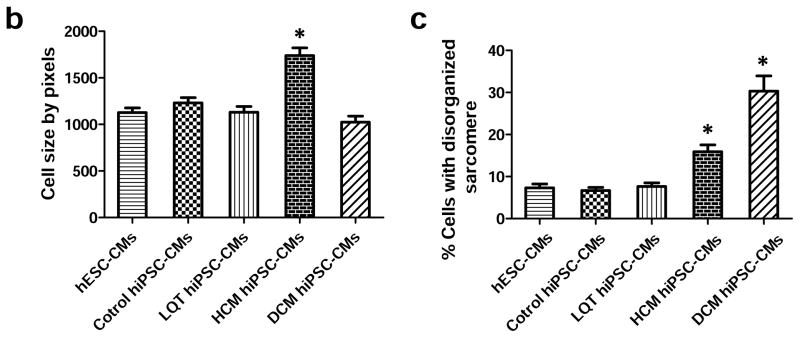
Following cardiac differentiation, patient-specific hiPSC-CMs from LQT, HCM, and DCM family cohorts were dissociated into single beating cells and characterized in vitro for expression of cardiac disease phenotypes. To ensure genetic diversity among disease-free hiPSC-CMs, patient-specific control hiPSC-CM lines were derived from separate healthy individuals belonging to the LQT, HCM, and DCM family cohorts. hESC-CMs (WA09 line) were also used as a disease-free control. Immunofluorescence staining of patient-specific hiPSC-CMs revealed immediate phenotypic differences in cells derived from patients with hereditary HCM and DCM mutations as compared to control hESC-CMs and control hiPSC-CMs. Specifically, hypertrophic hiPSC-CMs exhibited several features of HCM including cellular enlargement and sarcomeric disorganization (Figure 2a–c, Supplemental Figure 5). DCM hiPSC-CMs also exhibited disorganized sarcomeric staining patterns suggestive of myofibril disarray as previously reported 23. By comparison, control hESC-CMs, control hiPSC-CMs, and LQT hiPSC-CMs exhibited normal cell sizes and minimal sarcomeric disorganization (Figure 2a–c).
Figure 2.
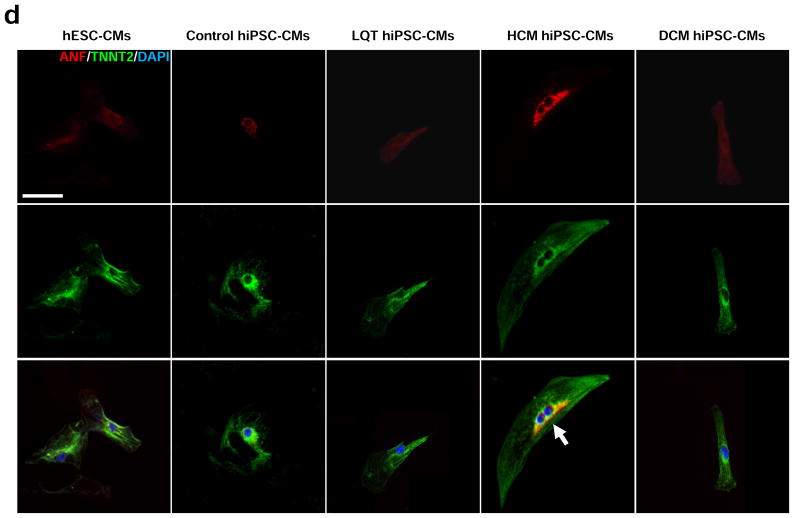
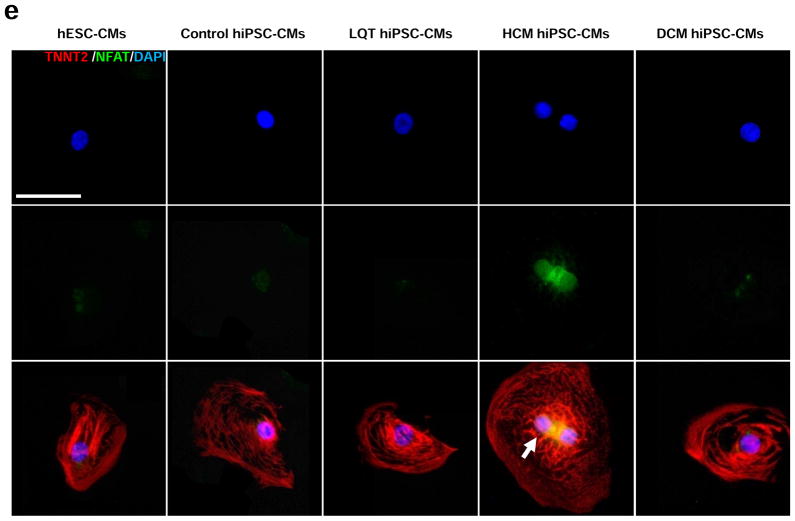
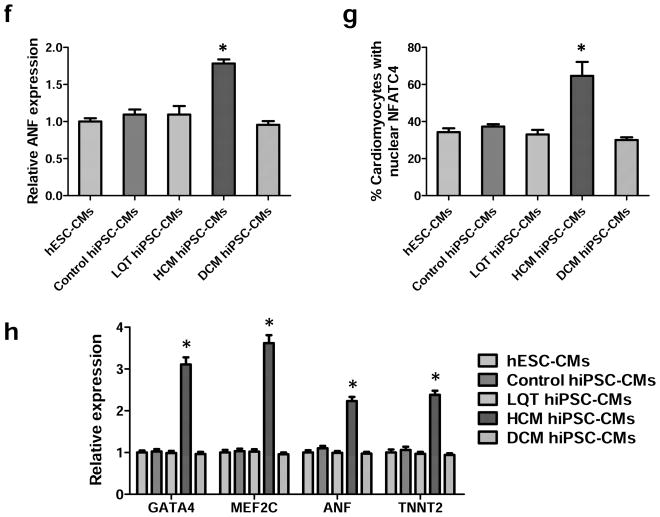
In vitro assessment of disease phenotype recapitulation for control, LQT, HCM, and DCM hiPSC-CMs. (a) Representative immunostaining of single hiPSC-CMs for cardiac troponin T and F-actin demonstrating increased cellular size in HCM hiPSC-CMs, and increased fraction of cells with disorganized sarcomeres in HCM and DCM hiPSC-CMs, compared with hESC-CMs, control hiPSC-CMs, and LQT hiPSC-CMs. White arrows indicate the disorganized sarcomeres in HCM and DCM hiPSC-CMs. Scale bar, 50 μM. (b) Quantification of cell size for hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (n=135–256, 3 lines per cohort). (c) Quantification of % cells with disorganized sarcomeres for hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (n=85–147, 3 lines per cohort). (d) Representative immunostaining of hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs reveals elevated ANF expression in HCM hiPSC-CMs as compared to other groups. White arrow indicates elevated ANF staining in HCM hiPSC-CMs. Scale bar, 50 μM. (e) Representative immunostaining of hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs and DCM hiPSC-CMs reveals nuclear translocation of NFATC4 in HCM hiPSC-CMs. White arrow indicates elevated staining for nuclear translocation of NFATC4 in HCM hiPSC-CMs. Scale bar, 50 μM. (f) Quantification of ANF expression for hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (n=43–75, 3 lines per cohort). (g) Percentage of cardiomyocytes exhibiting positive NFATC4 staining in hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (n=76–115, 3 lines per cohort). (h) HCM-related gene expressions (GATA4, MEF2C, ANF, and TNNT2) in hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs by quantitative PCR (qPCR) (n=3). * indicates P<0.05, as compared to control hiPSC-CMs.
In addition to cellular hypertrophy and disorganization of sarcomeric content, HCM hiPSC-CMs also demonstrated disease characteristics associated with HCM such as elevated expression of atrial natriuretic factor (ANF) and nuclear translocation of nuclear factor of activated T-cells (NFAT) (Figure 2d–e). By comparison, control hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, and DCM hiPSC-CMs exhibited lower levels of ANF expression and nuclear NFAT translocation (Figure 2f–g). Hypertrophic transcription factors activated by nuclear translocation of NFAT such as GATA4 and MEF2C were also found to be elevated in HCM hiPSC-CMs as compared to other patient-specific lines (Figure 2h). Taken together, these results indicate that cohorts of disease-specific hiPSC-CMs recapitulate distinct pathologic phenotypes associated with clinical presentations of LQT, HCM, and DCM, respectively.
Gene expression profiles of cardiac ion channels in hESC- and hiPSC-CMs
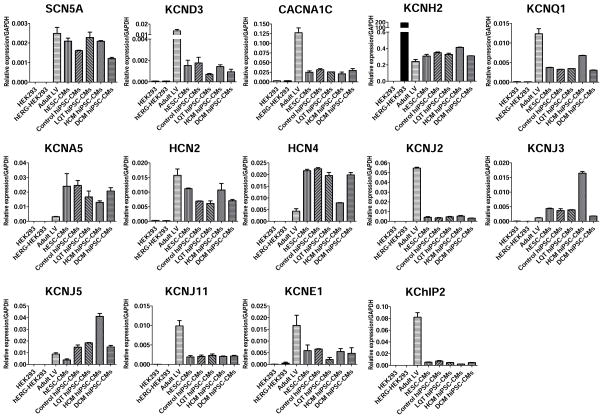
The use of pluripotent stem cell-derived cardiomyocytes to accurately model drug-induced cardiotoxicity in vitro is predicated on the expression of the full range of adult cardiac ion channels naturally found in the human heart 29. However, to date only a fraction of the ion channels endogenously expressed in adult cardiomyocytes have been verified to be present in hESC-CMs and hiPSC-CMs 30, 31. To evaluate the potential of control and disease-specific hiPSC-CMs as well as hESC-CMs to be used as surrogate models for adult cardiomyocytes, total RNA was isolated from beating cultures of hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs. RNA isolated from adult cardiac left ventricular (LV) tissue and HEK293 cells was used as positive and negative controls, respectively. Quantitative PCR demonstrated that hESC-CMs, control hiPSC-CMs, and disease-specific hiPSC-CMs expressed all cardiac ion channels found in adult LV cardiac tissue using a panel of 14 ion channel-related transcripts, including SCN5A, KCND3, CACNA1C, KCNH2, KCNQ1, KCNA5, HCN2, HCN4, KCNJ2, KCNJ3, KCNJ5, KCNJ11, KCNE1, and KChIP2 (Supplemental Table 3). By comparison, HEK293 cells overexpressing the hERG channel (hERG-HEK293) used in the hERG assay were only found to express KCNH2 (Figure 3). These findings indicate that unlike the hERG assay, hESC-CMs and hiPSC-CMs have the potential to model electrophysiological responses to pharmaceutical compounds that affect multiple cardiac ion channels.
Figure 3.
Gene expression of cardiac ion channel transcripts in disease-specific hiPSC-CMs. Quantitative PCR results for expression of the primary 14 cardiac ion channel transcripts (SCN5A, KCND3, CACNA1C, KCNH2, KCNQ1, KCNA5, HCN2, HCN4, KCNJ2, KCNJ3, KCNJ5, KCNJ11, KCNE1, and KChIP2) responsible for the electrophysiological function in adult CMs. HEK-293, and hERG expressing HEK293 cells (hERG-HEK293) were used as negative controls, whereas adult LV tissue was used as a positive control (n=3, 2 lines per group).
Electrophysiological characterization of control and disease-specific iPSC-CMs
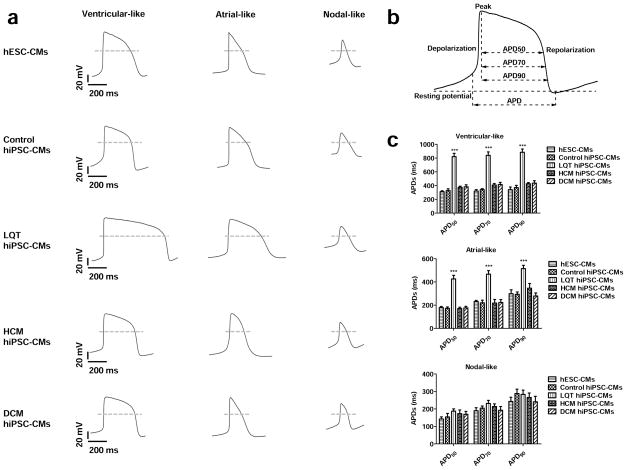
To establish electrophysiological baselines of control and disease-specific hiPSC-CMs, spontaneous action potentials were recorded from single beating cardiomyocytes derived from H9 hESCs and from control, LQT, HCM, and DCM hiPSCs. Nodal-, atrial-, and ventricular-like waveforms were detected in CMs from all lines as determined by the morphology of the action potentials generated and by the APD90/APD50 ratio (Figure 4a–b, Supplemental Table 4). No significant differences in beating rate, maximum diastolic potential (MDP), overshoot, action potential amplitude (APA), and maximal rate of depolarization (Vmax) were detected between hESC-CMs and control, LQT, HCM, and DCM hiPSC-CMs (Supplemental Figure 6). However, LQT hiPSC-CMs exhibited significantly longer action potential durations at 50%, 70%, and 90% repolarization (APD50, APD70, APD90) as compared to control lines and other patient-specific lines (Figure 4c, Supplemental Table 4). Control hiPSC-CMs derived from LQT, HCM, and DCM family cohorts did not exhibit significant differences in electrophysiological baselines, suggesting that healthy hiPSC-CMs derived from different genetic backgrounds are phenotypically similar (Supplemental Figure 7a–c).
Figure 4.
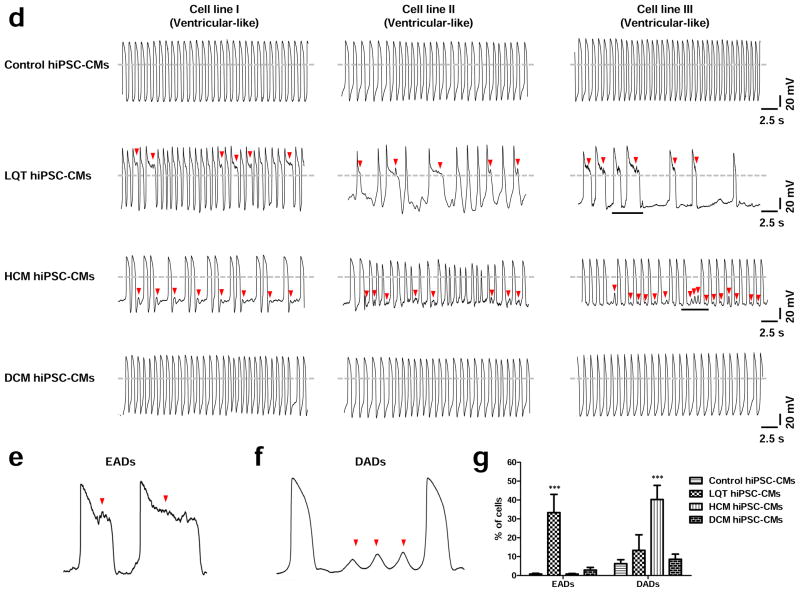
Baseline AP characteristics of control, LQT, HCM, and DCM hiPSC-CMs. (a) Typical recordings of ventricular-, atrial-, and nodal-like action potentials from hESC-CMs, control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs. (b) Schematic diagram of a patch-clamp trace, showing how results were analyzed to calculate action potential duration at 50, 70, and 90% repolarization (APD50, APD70, and APD90, respectively). (c) APD50, APD70, and APD90 for each of the three myocyte subtypes in control, LQT, HCM, and DCM patient groups (n=70, 3 lines per cohort). (d) Representative recordings of ventricular-like action potentials from control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (n=70, 3 lines per cohort). Arrowheads show putative early afterdepolarizations (EADs) or delayed afterdepolarizations (DADs). (e) Magnification of the underlined segment of the LQT AP tracing shows a representative EAD waveform at an expanded timescale. (f) Magnification of the underlined segment of the HCM AP tracing shows a representative DAD waveform at an expanded timescale. (g) Percentage of disease-specific cardiomyocytes displaying DADs and EADs, respectively (Control: n=128 cells, 3 control subject lines; LQT: n=90 cells, 3 patient lines; HCM: n=134 cells, 3 patient lines; DCM: n=70 cells, 3 lines). Dashed lines show 0 mV. *** indicates P<0.001.
Studies modeling disease progression of LQT using patient-specific hiPSC-CMs have demonstrated recapitulation of arrhythmic phenotypes in vitro 20, 21, 32. We therefore profiled incidence of arrhythmia in control and disease-specific hiPSC-CMs to determine normal electrophysiological baselines for our platform. To ensure analyses were standardized between different cell lines, we only profiled cell subpopulations with ventricular-like waveforms. Significantly, both LQT and HCM hiPSC-CMs exhibited a high incidence of arrhythmia including early afterdepolarizations (EADs) in LQT hiPSC-CMs (33.3 ± 9.7%) and delayed afterdepolarizations (DADs) in HCM hiPSC-CMs (40.3 ± 7.5%) (Figure 4d–g). By contrast, hESC-CMs, control hiPSC-CMs, and DCM hiPSC-CMs exhibited very low frequencies of irregular electrophysiological waveforms. Comparison of control hiPSC-CM lines revealed no significant differences between control hiPSC-CMs derived from healthy individuals from LQT. HCM, and DCM family cohorts (Supplemental Figure 7d).
Comparison of hiPSC-CM and hERG assay for detection of QT prolongation
Preclinical drug toxicity screening using hERG blockade has been shown to incorrectly predict cardiotoxicity for a number of pharmaceutical compounds 12. These include false positives such as verapamil that are not cardiotoxic but block the hERG channel, as well as false negatives such as alfuzosin, which do not bind hERG but elicit QT prolongation through interactions with other ion channels 12, 14. To determine whether our platform is superior to hERG assay for detection of cardiotoxicity, we screened both verapamil and alfuzosin on commercial HEK293 cells stably expressing hERG (hERG-HEK293) and control hiPSC-CMs from our patient library. Cardiotoxic readout of the hERG-HEK293 assay was evaluated through calculation of the half maximal inhibitory concentration (IC50) to inhibit the hERG peak tail current, a surrogate measure for QT prolongation. By comparison, cardiotoxic readout of drug-treated hiPSC-CMs was determined through assessment of the (IC50) to significantly prolong the APD90, which is a more direct and physiologically relevant measurement of the QT interval.
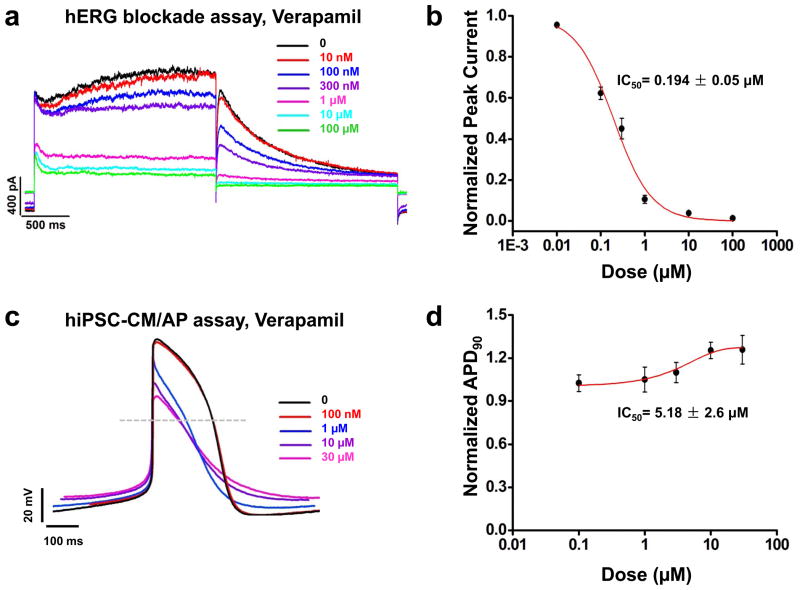
Patch clamp recordings in hERG-HEK293 cells demonstrated verapamil inhibits hERG tail current in a dose-dependent manner at a low concentration, suggesting high potential for cardiotoxicity (IC50 = 0.19 ± 0.05 μM; Figure 5a–b, Supplemental Table 5), whereas vehicle (0.1% DMSO) showed no effect on hERG tail current (Supplemental Figure 8). By comparison, treatment of hESC-CMs and hiPSC-CMs with verapamil demonstrated both dose-dependent shortening of the Phase 2 APD due to L-type Ca2+ channel blockade and dose-dependent prolongation of the Phase 3 APD due to hERG channel blockade (Figure 5c–d, Supplemental Figure 9a, Supplemental Table 6). As a result, significant APD90 prolongation in hESC-CMs and hiPSC-CMs was not observed at physiological concentrations of the drug (Figure 5c–d, Supplemental Figure 9a, Supplemental Table 6).
Figure 5.
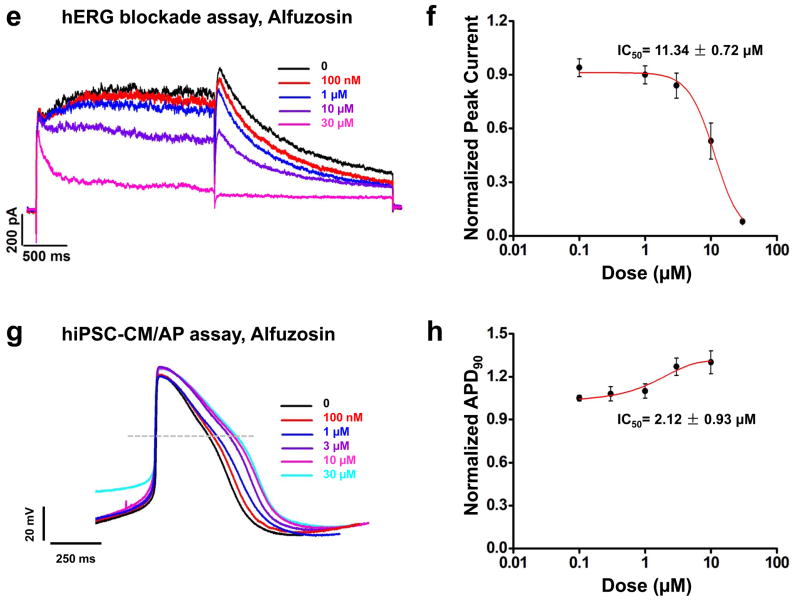
Comparison of hERG blockade assay with hiPSC-CMs for detection of cardiotoxicity using verapamil and alfuzosin. (a) Representative tracings demonstrating the effect of verapamil on HEK293 hERG current at doses of 0, 0.01, 0.1, 0.3, 1, 10, and 100 μM (black, red, blue, purple, magenta, cyan, and green traces, respectively) (n=3). Dose-dependent hERG current inhibition was observed. (b) Dose-response relationship demonstrating steady-state inhibition of hERG by verapamil. IC50 was calculated by fitting a Hill function to the data constrained to 100% inhibition. IC50 for verapamil was 0.194 ± 0.05 μM (n=3 for each dose). (c) Representative traces of the effect of verapamil, a hERG and L-type Ca2+ channel blocker, on the control hiPSC-CM AP at doses of 0, 0.1, 1, 3, 10, and 30 μM (black, red, blue, purple, and magenta traces, respectively) (n=8, 3 lines). Dose-dependent prolongation of the Phase III of the AP was observed, demonstrating blockade of the hERG channel. Dose-dependent shortening of the Phase II of the AP was also observed, indicating that AP prolongation induced by hERG blockade was offset by AP shortening caused by L-type Ca2+ channel inhibition. (d) Dose-response relationship showing the steady-state prolongation of hiPSC-CM APD90 by verapamil. IC50 was calculated by fitting a Hill function to the data, providing an IC50 value of verapamil at 5.18 ± 2.6 μM (n=8, 3 lines). (e) Representative tracings showing the effect of alfuzosin on HEK293 hERG current at doses of 0, 0.1, 1, 10, and 30 μM (black, red, blue, purple, and magenta traces, respectively) (n=3). Dose-dependent hERG current inhibition was observed. (f) Dose-response relationship showing the steady-state inhibition of hERG by alfuzosin. IC50 was calculated by fitting a Hill function to the data constrained to 100% inhibition. IC50 for alfuzosin was 11.34 ± 0.72 μM (n=3 for each dose). (g) Representative tracings showing the effect of alfuzosin on the control hiPSC-CM AP at doses of 0, 0.1, 0.3, 1, 3, and 10 μM (black, red, blue, purple, magenta, and cyan traces, respectively) (n=8, 3 lines). Dose-dependent AP prolongation was observed. (h) Dose-response relationship showing the steady-state prolongation of hiPSC-CM APD90 by alfuzosin. IC50 was calculated by fitting a Hill function to the data, providing a IC50 value of alfuzosin at 2.12 ± 0.93 μM (n=8, 3 lines). Dashed lines show 0 mV.
We next treated hERG-HEK293 cells with alfuzosin, a drug that can increase sodium channel current that is known to cause QT prolongation in patients 14. Alfuzosin treatment demonstrated little potential to block hERG (IC50 = 11.3 ± 0.7 μM; Figure 5e–f, Supplemental Table 5). However, when tested on control hESC-CMs and hiPSC-CMs, alfuzosin was found to produce a significantly dose-dependent prolongation of the APD90 from concentrations as low as 1μM (hESC-CM IC50 = 2.9 ± 1.1 μM, hiPSC-CM IC50 = 2.1 ± 0.9 μM; Figure 5g–h, Supplemental Figure 9b, Supplemental Table 6). Vehicle alone (0.1% DMSO) had no effect on AP waveforms (Supplemental Figure 10). Taken together, these findings indicate that hESC-CMs and hiPSC-CMs are more accurate predictors of drug-induced cardiotoxicity than hERG-HEK293 cells.
Evaluation of drug-induced cardiotoxicity in control and disease-specific hiPSC-CMs
Several recent studies have demonstrated proof-of-principle for hESC-CMs and hiPSC-CMs to model pharmaceutical responses of various drugs 22, 30, 33–36. However, the majority of these studies have been conducted using hESCs and hiPSCs derived from subjects without known risk for cardiac disorders. These cells therefore could not recapitulate the clinical experience of individuals at high risk for adverse cardiac drug reactions. As the majority of cardiotoxic incidents occur in patients with pre-existing heart conditions and not healthy individuals, we aimed to test whether disease-specific hiPSC-CMs have higher susceptibilities to cardiotoxicity as compared to control hESC-CMs and hiPSC-CMs. Diseased hiPSC-CMs derived from individuals with LQT, HCM, and DCM were treated in parallel with control hESC-CMs and hiPSC-CMs with a panel of drugs known to interact with different cardiac ion channels (Supplemental Table 7). Cardiotoxic readouts for each of these compounds were measured by its potential to induce arrhythmia or affect the APD in single hiPSC-CMs.
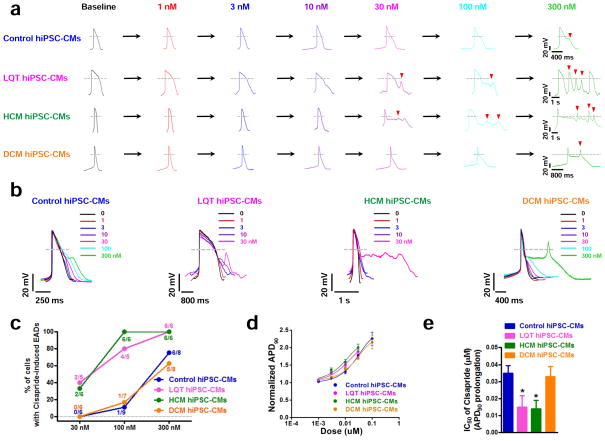
Distinct control and disease-specific hiPSC-CMs were first screened for toxicity to cisapride, a potent hERG blocking drug removed from market in 2000 (Supplemental Figure 11a–c, Supplemental Table 5) 19. Control, LQT, HCM, and DCM hiPSC-CMs were treated with a range of doses (1 nM – 300 nM), and cardiotoxic responses were evaluated by whole-cell patch clamp. Cisapride was observed to elicit EADs at different concentrations for each disease-specific cell line (Figure 6a–b). LQT and HCM hiPSC-CMs, which exhibited EADs and DADs at electrophysiological baselines, were found to have increased susceptibility to cisapride-induced arrhythmia. While control and DCM hiPSC-CMs largely demonstrated EADs at concentrations of cisapride ≥ 300 nM, LQT and HCM hiPSC-CMs exhibited cisapride-induced EADs at concentrations as low as 30 nM (Figure 6b–c, Supplemental Table 6). The arrhythmic waveforms elicited by cisapride in LQT and HCM lines were also more punctuated than in control or DCM hiPSC-CMs. In addition to arrhythmia, LQT and HCM hiPSC-CMs were also found to have more adverse reactions to cisapride-induced APD prolongation than DCM hiPSC-CMs and control hiPSC-CMs (Figure 6d–e, Supplemental Table 6). Control hiPSC-CMs derived from healthy individuals within the LQT, HCM, and DCM family cohorts exhibited similar patterns of ciaspride-induced cardiotoxicity (Supplemental Figure 12a–b). These results indicate that as compared to cells derived from control or DCM patients, hiPSC-CMs containing LQT or HCM mutations are more sensitive to cardiotoxicity caused by pharmacological blockade of hERG.
Figure 6.
Differences in susceptibilities of disease-specific hiPSC-CMs to cisapride-induced cardiotoxicity. (a) Representative tracings showing dose-dependent effect of cisapride on the AP of control hiPSC-CMs, LQT hiPSC-CMs, HCM hiPSC-CMs, and DCM hiPSC-CMs (Baseline: black, 1 nM: red, 3 nM: blue, 10 nM: purple, 30 nM: magenta, 100 nM: cyan, 300 nM: green). Arrowheads indicate cisapride-induced EADs. LQT hiPSC-CMs and HCM hiPSC-CMs were observed to exhibit cisapride-induced EADs at lower concentrations (30 nM) as compared to family matched control hiPSC-CMs and DCM hiPSC-CMs (300 nM) (n=10, 3 patient or control subject lines per cohort). (b) Aligned APs (in Panel a) showing the effects of cisapride on control, LQT, HCM, and DCM hiPSC-CMs (n=10, 3 patient or control subject lines per cohort). (c) Percentage of cardiomyocytes exhibiting cisapride-induced EADs in control, LQT, HCM, and DCM hiPSC-CMs at 30 nM, 100 nM, and 300 nM, respectively (n=6–9, 3 patient or control subject lines per cohort). Dashed lines show 0 mV. (d) Dose-response relationship showing the steady-state prolongation of APD90 by cisapride for control, LQT, HCM, and DCM hiPSC-CMs. Data were fit by the Hill function (n=10, 3 patient or control subject lines per cohort). (e) IC50 values of prolongation of APD90 of cisapride for control, LQT, HCM, and DCM hiPSC-CMs (n=10, 3 patient or control subject lines per cohort). Dashed lines show 0 mV. * indicates P<0.05, ** indicates P<0.01.
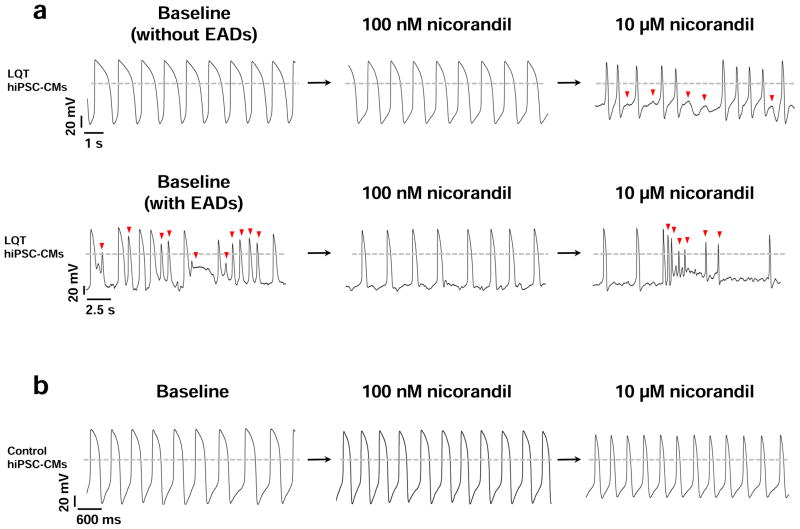
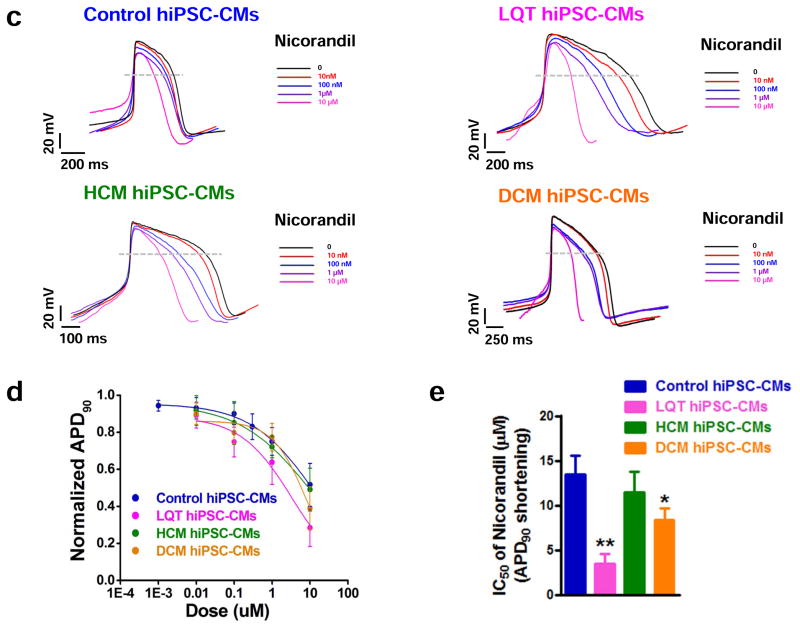
We next evaluated the potential of the K+ ATP channel opener nicorandil to elicit differential drug responses between control, LQT, HCM, and DCM hiPSC-CMs. Previous studies have shown nicorandil shortens the QT interval through enhancement of K+ outflux and can ameliorate LQT-induced arrhythmias at moderate doses 20, 32, 37. Treatment of our LQT hiPSC-CMs at low doses of nicorandil supported these results, as 100 nM was found to both normalize APD prolongation (18 out of 18 cells, 3 patient lines) and abolish spontaneously occurring EADs (7 out of 10 cells, 3 patient lines) (Figure 7a). Several recent studies have also reported induction of arrhythmia and ventricular fibrillation in patients receiving high doses of nicorandil due to excessive shortening of the QT interval 11,38, 39. We therefore aimed to assess whether patient-specific hiPSC-CMs could recapitulate short QT cardiotoxicity. Interestingly, LQT hiPSC-CMs exhibited heightened sensitivity to nicorandil-induced cardiotoxicity at high doses, including exhibition of proarrhythmias (7 out of 9 cells, 3 patient lines) (Figure 7a). These abnormalities were not observed in control cells (Figure 7b). Moreover, LQT and DCM hiPSC-CMs demonstrated extreme QT shortening induced by nicorandil (IC50 of APD90 shortening for LQT = 3.44 ± 1.11 μM, IC50 of APD90 shortening for DCM = 8.32 ± 1.35 μM; Figure 7c–e, Supplemental Table 6). By comparison, control and HCM hiPSC-CMs were not found to exhibit excessive QT shortening as induced by nicorandil (IC50 of APD90 shortening for control = 13.44 ± 2.1 μM, IC50 of APD90 shortening for HCM = 11.45 ± 2.30 μM; Figure 7c–e, Supplemental Table 6). Control hiPSC-CMs derived from healthy individuals within the LQT, HCM, and DCM family cohorts exhibited similar response patterns to nicorandil-induced cardiotoxicity (Supplemental Figure 12c–d). Taken together, these findings suggest disease-specific hiPSC-CMs are characterized by distinct pharmacological responses to drug-induced cardiotoxicities, and that these differences may underlie the sensitivities of specific patient populations to adverse cardiac drug reactions.
Figure 7.
Differences in susceptibilities of disease-specific hiPSC-CMs to short-QT arrhythmia induced by nicorandil. (a) Treatment of LQT hiPSC-CMs with 100 nM nicorandil normalizes the QT interval and abolishes baseline EADs. High doses (>10 μM) of nicorandil induce proarrhythmias. Upper panel: LQT hiPSC-CM AP baseline without EADs, 100 nM nicorandil is able to normalize the APD values. Treatment with 10 μM nicorandil induces arrhythmias (n=18, 3 patient lines per cohort). Bottom panel: LQT hiPSC-CM AP baseline with EADs (indicated by red arrows). 100 nM nicorandil can abolish baseline EADs. However, at treatment with 10 μM nicorandil, the QT interval is excessively shortened, resulting in nicorandil-induced arrhythmia as indicated by red arrows (n=9, 3 patient lines per cohort). (b) Treatment of control hiPSC-CMs with nicorandil. APD shortening is observed without arrhythmia or drug-induced toxicity at both 100 nM and 10 μM nicorandil (n=10, 3 control subject lines per cohort). (c) Representative tracings showing the dose-dependent effect of nicorandil on control, LQT, HCM, and DCM hiPSC-CM APs (Baseline: black, 10 nM: red, 100 nM: blue, 1 μM: purple, 10 μM: magenta). Dose-dependent shortening of the AP was observed in all groups (n=18, 3 patient or control subject lines per cohort). (d) Dose-response relationships showing shortening of the steady-state APD90 by nicorandil on control, LQT, HCM, and DCM hiPSC-CMs. Data were fit by the Hill function (n=18, 3 patient or control subject lines per cohort). (e) IC50 values of APD90 shortening induced by nicorandil on control, LQT, HCM, and DCM hiPSC-CMs (n=18, 3 patient or control subject lines per cohort). Dashed lines show 0 mV. * indicates P<0.05, ** indicates P<0.01.
DISCUSSION
Drug-induced cardiac toxicity is a major risk in pharmaceutical development. The US Department of Health and Human Services estimates that nearly 1 million patients suffer from adverse drug reactions each year, of which drug-induced arrhythmia is the leading cause 40. As a result, both the FDA and pharmaceutical industry have instituted mandatory preclinical drug screening guidelines to detect potential cardiac toxicity before market release. Current preclinical cardiotoxicity assays are based upon the International Conference on Harmonization (ICH) Supplement S7B, and focus on screening for compounds that prolong the QT interval through blockade of the hERG potassium channel. The hERG assay is known to be an inaccurate predictor of cardiac toxicity, however, and regularly results in false negative and false positive readouts 11, 12. Use of animal hearts as surrogate models for the human heart are also limited because murine, canine, and rabbit cardiomyocytes are characterized by different electrophysiological properties and fail to respond to certain drugs in a similar manner to humans 29, 41. The generation of cardiomyocytes from hESCs and hiPSCs offers a potential method to circumvent the inaccuracies of hERG and animal models by allowing for drug screening to be performed directly on human heart cells in vitro for the first time36.
Derivation of patient-specific hiPSCs presents the unprecedented opportunity to improve our understanding of hereditary cardiac disorders through “disease in a dish” modeling (Supplemental Figure 13). Recent studies have demonstrated potential of hiPSCs to recapitulate disease phenotypes of genetic cardiac diseases such as dilated cardiomyopathy, long QT, CPVT, and LEOPARD syndrome 20–25, 32. The capacity for hiPSC-based disease modeling has important ramifications for drug toxicity screening as the majority of adverse cardiac drug reactions occur in patients with hereditary cardiac disease. Patients with LQT, HCM, and DCM, are particularly sensitive to cardiotropic drugs and are vulnerable to fatal arrhythmias 42. To test whether hiPSC-CMs could model the increased susceptibility of genetic cardiac disorders to drug-induced cardiotoxicity, we characterized and screened a library of hiPSC-CMs derived from patients with LQT, HCM, and DCM for a panel of drugs known to affect cardiac ion channels.
Our findings have several important implications for the use of patient specific hiPSC-CMs to detect drug-induced cardiotoxicity. First, we demonstrate the patient-specific hiPSC-CMs in our cell library exhibit distinct in vitro phenotypes associated with the control, LQT, HCM, and DCM patients from which they were derived. Second, we profiled control and disease-specific hiPSC-CMs for expression of a full panel of cardiac ion channels known to be associated with drug-induced cardiotoxicity. While previous reports have demonstrated proof-of-principle for hESC-CMs and hiPSC-CMs to model Na+, Ca2+, and K+ ionic currents, these studies have only assessed gene expression for a subset of cardiac ion channels 30, 31, 36, 43. Furthermore, the analyses in these reports are largely limited to 1–2 cell lines, which are not representative of the genetic diversity embodied by the general population. In our study, we have profiled gene expression for all major cardiac ion channels for a library of hiPSC-CMs derived from 12 individuals of different genetic backgrounds. We demonstrate that regardless of presence or absence of in vitro disease phenotype, the patient-specific hiPSC-CMs assessed in our study express all major cardiac ion channels at similar levels to hESC-CMs. As a result, in head-to-head comparisons conducted within our study, hiPSC-CMs modeled cardiotoxicities more accurately than commercial hERG cell lines. To date, this is the first report we know of to directly compare hiPSC-CM and hERG assays for detection of drug-induced cardiotoxicity at the single cell level.
Screening of our patient cell library also demonstrated disease-specific hiPSC-CMs are more susceptible to drug-induced cardiac toxicity than control hiPSC-CMs as assessed by measurement of the QT interval prolongation and induction of arrhythmia. Diseases characterized by an elevated baseline of arrhythmia such as LQT and HCM exhibited a particular propensity for exhibition of drug-induced EADs by hERG blockade with cisapride. The high frequency of baseline arrhythmias and electrophysiologcal abnormalities observed in LQT and HCM hiPSC-CMs may be explained by imbalances in ion channel homeostasis caused by mutations in KCNQ1 and MYH7, respectively. Previous LQT hiPSC-CM modeling studies have observed baseline EADs and attributed these to sensitization of late-membrane inward currents 20. For HCM, several studies have demonstrated that increased intracellular calcium retention by MYH7 mutations can lead to presentation of DADs 44.
Interestingly, we observed a cut-off within the therapeutic window of cisapride (3 nM) at which LQT and HCM hiPSC-CMs exhibited significant QT prolongation, but control hiPSC-CMs did not. In addition, we also observed a second cut-off (30 nM) at which cisapride was found to induce EADs in LQT and HCM hiPSC-CMs, but not in control or DCM cells. The heightened sensitivity of LQT and HCM hiPSC-CMs to cisapride toxicity are likely caused by drug-induced exacerbation of the baseline arrhythmias observed in both of these cell cohorts as shown in previous studies 24. Control and DCM hiPSC-CMs, which did not exhibit baseline arrhythmia or APD prolongation, required higher doses of cisapride for exhibition of drug-induced cardiotoxicity at the single cell level. These results replicate epidemiological findings in which the majority of patients found to undergo adverse drug reactions to cisapride exhibited pre-existing QT prolongation and arrhythmia 19.
Our findings further indicate that hiPSC-CMs recapitulate patient-specific susceptibilities to drug-induced toxicities other than blockade of hERG. Nicorandil is a K+ ATP channel opener that shortens APD by increasing in K+ outflux. While experimental models have shown K+ ATP channel openers can ameliorate LQT at low to moderate doses, the use of nicorandil has also been associated with induction of arrhythmia and ventricular fibrillation in patients with ischemic dilated cardiomyopathy 38, 39. High doses of nicorandil have been shown to induce arrhythmia in hiPSC-CM and animal models of LQT through excessive shortening of the QT interval 38, 45. In DCM, several studies have demonstrated that depletion of cellular ATP reserves impedes proper regulation of KATP channel opening, resulting in arrhythmias following potassium outflow 39. Our findings support these results as both DCM and LQT hiPSC-CMs were found to exhibit drug-induced proarrhythmias when treated with doses of nicorandil > 10 μM. By comparison, treatment of HCM or control hiPSC-CMs with the same dosage of nicorandil was not observed to induce arrhythmia.
In summary, our results indicate that disease-specific hiPSC-CMs can recapitulate clinical susceptibilities of high-risk populations to drug-induced cardiotoxicities, and that the use of a patient-specific hiPSC-CM library to detect cardiotoxicity may be superior to utilization of hESC-CMs or hERG assay alone. However, it is important to note there are several limitations to our approach. These include the developmental immaturity of iPSC-CMs as compared to adult cardiomyocytes, and the inability of single cells to model arrhythmic triggers at the tissue level such as interstitial fibrosis and re-entry 20, 22, 26. Furthermore it is critical to recognize the iPSC-CM libraries used in this study are not equivalent substitutes for adult cardiomyocytes found in our patient cohorts. Rather, our approach is an in vitro method to model the effects of disease-specific differences in adverse drug response between different patient populations. We believe the future use of a diverse patient hiPSC-CM library in future drug toxicity screens will contribute toward the stratification of clinical trials by refinement of thresholds for cardiotoxicty as well as identification of patient populations at high-risk for cardiac toxicity.
Supplementary Material
Clinical Perspective.
Cardiac toxicity is a side effect of many pharmaceutical compounds, and is a leading cause for drug withdrawal from market due to safety concerns. Current preclinical methods to measure cardiotoxicity are inefficient and rely on genetically altered cell lines, such as human embryonic kidney (HEK) cells, or Chinese hamster ovary (CHO) cells, which do not accurately resemble human heart cells (cardiomyocytes). Recent technological advancement has enabled the generation of human induced pluripotent stem cells (hiPSCs) from skin, which can be used to generate patient-specific cardiomyocytes (hiPSC-CMs)in vitro. The hiPSC-CMs generated in this fashion carry all the genetic information from the individuals from whom they are derived. In this report, we validate the capacity of a hiPSC-CM library to be used as a “clinical trial in a dish” model for accurate detection of patient-specific drug responses. Our results demonstrate that hiPSC-CMs detect drug-induced cardiac toxicity more accurately than the preclinical tests currently in use by pharmaceutical companies and mandated by the FDA (hERG assay). We further demonstrate that hiPSC-CMs can be used in personalized medical assays to assess the genetic susceptibilities of distinct individuals to drug-induced cardiac toxicities. This is important given that the majority of cardiotoxic drugs have a low incidence of harmful effects among the general population, and are largely only toxic to specific patient populations with determined genetic traits. We believe these findings strongly support the addition of hiPSC-CM based assays to pre-clinical drug toxicity screening protocols and drug development for the acceleration of safe pharmaceutical compounds to the market.
Acknowledgments
We thank Ziyan Zhang for preparing high-resolution figures. We gratefully acknowledge funding support from NIH New Innovator Award DP2OD004437, R01 HL113006, CIRM RB3-05129, Leducq Foundation 11CDV02, Burroughs Wellcome Foundation (JCW), Bio-X (ASL), and U01 HL099776 (RCR).
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Dykens JA, Marroquin LD, Will Y. Strategies to reduce late-stage drug attrition due to mitochondrial toxicity. Expert Rev Mol Diagn. 2007;7:161–175. doi: 10.1586/14737159.7.2.161. [DOI] [PubMed] [Google Scholar]
- 2.Kannankeril PJ, Roden DM. Drug-induced long qt and torsade de pointes: Recent advances. Curr Opin Cardiol. 2007;22:39–43. doi: 10.1097/HCO.0b013e32801129eb. [DOI] [PubMed] [Google Scholar]
- 3.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 4.Roden DM. Mechanisms and management of proarrhythmia. Am J Cardiol. 1998;82:49I–57I. doi: 10.1016/s0002-9149(98)00472-x. [DOI] [PubMed] [Google Scholar]
- 5.International conference on harmonisation; guidance on s7a safety pharmacology studies for human pharmaceuticals; availability. Notice. Federal register. 2001;66:36791–36792. [PubMed] [Google Scholar]
- 6.Fermini B, Fossa AA. The impact of drug-induced qt interval prolongation on drug discovery and development. Nature reviews Drug discovery. 2003;2:439–447. doi: 10.1038/nrd1108. [DOI] [PubMed] [Google Scholar]
- 7.Roden DM. Pharmacogenetics and drug-induced arrhythmias. Cardiovasc Res. 2001;50:224–231. doi: 10.1016/s0008-6363(00)00302-3. [DOI] [PubMed] [Google Scholar]
- 8.De Bruin ML, Pettersson M, Meyboom RH, Hoes AW, Leufkens HG. Anti-herg activity and the risk of drug-induced arrhythmias and sudden death. Eur Heart J. 2005;26:590–597. doi: 10.1093/eurheartj/ehi092. [DOI] [PubMed] [Google Scholar]
- 9.van Noord C, Sturkenboom MC, Straus SM, Witteman JC, Stricker BH. Non-cardiovascular drugs that inhibit herg-encoded potassium channels and risk of sudden cardiac death. Heart. 2011;97:215–220. doi: 10.1136/hrt.2009.188367. [DOI] [PubMed] [Google Scholar]
- 10.McNeish J. Embryonic stem cells in drug discovery. Nature reviews Drug discovery. 2004;3:70–80. doi: 10.1038/nrd1281. [DOI] [PubMed] [Google Scholar]
- 11.Lu HR, Vlaminckx E, Hermans AN, Rohrbacher J, Van Ammel K, Towart R, Pugsley M, Gallacher DJ. Predicting drug-induced changes in qt interval and arrhythmias: Qt-shortening drugs point to gaps in the ichs7b guidelines. British J Pharmacol. 2008;154:1427–1438. doi: 10.1038/bjp.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redfern WS, Carlsson L, Davis AS, Lynch WG, MacKenzie I, Palethorpe S, Siegl PK, Strang I, Sullivan AT, Wallis R, Camm AJ, Hammond TG. Relationships between preclinical cardiac electrophysiology, clinical qt interval prolongation and torsade de pointes for a broad range of drugs: Evidence for a provisional safety margin in drug development. Cardiovasc Res. 2003;58:32–45. doi: 10.1016/s0008-6363(02)00846-5. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann P, Warner B. Are herg channel inhibition and qt interval prolongation all there is in drug-induced torsadogenesis? A review of emerging trends. J Pharmacol Toxicol Methods. 2006;53:87–105. doi: 10.1016/j.vascn.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Lacerda AE, Kuryshev YA, Chen Y, Renganathan M, Eng H, Danthi SJ, Kramer JW, Yang T, Brown AM. Alfuzosin delays cardiac repolarization by a novel mechanism. J Pharmacol Exp Ther. 2008;324:427–433. doi: 10.1124/jpet.107.128405. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Menchaca AA, Navarro-Polanco RA, Ferrer-Villada T, Rupp J, Sachse FB, Tristani-Firouzi M, Sanchez-Chapula JA. The molecular basis of chloroquine block of the inward rectifier kir2.1 channel. Proc Natl Acad Sci U S A. 2008;105:1364–1368. doi: 10.1073/pnas.0708153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, Gepstein L. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 17.Otsuji TG, Minami I, Kurose Y, Yamauchi K, Tada M, Nakatsuji N. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: Qualitative effects on electrophysiological responses to drugs. Stem Cell Res. 2010;4:201–213. doi: 10.1016/j.scr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Ray WA. Population-based studies of adverse drug effects. N Engl J Med. 2003;349:1592–1594. doi: 10.1056/NEJMp038145. [DOI] [PubMed] [Google Scholar]
- 19.Smalley W, Shatin D, Wysowski DK, Gurwitz J, Andrade SE, Goodman M, Chan KA, Platt R, Schech SD, Ray WA. Contraindicated use of cisapride: Impact of food and drug administration regulatory action. JAMA. 2000;284:3036–3039. doi: 10.1001/jama.284.23.3036. [DOI] [PubMed] [Google Scholar]
- 20.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long qt syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 21.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-qt syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 22.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of leopard syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra147. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012 Sep 11;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. Epub 2012 Jun 27. [DOI] [PubMed] [Google Scholar]
- 25.Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz KL. Dantrolene rescues arrhythmogenic ryr2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human ips cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 29.Davis RP, van den Berg CW, Casini S, Braam SR, Mummery CL. Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends Mol Med. 2011;17:475–484. doi: 10.1016/j.molmed.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Honda M, Kiyokawa J, Tabo M, Inoue T. Electrophysiological characterization of cardiomyocytes derived from human induced pluripotent stem cells. J Pharmacol Sci. 2011;117:149–159. doi: 10.1254/jphs.11038fp. [DOI] [PubMed] [Google Scholar]
- 31.Yokoo N, Baba S, Kaichi S, Niwa A, Mima T, Doi H, Yamanaka S, Nakahata T, Heike T. The effects of cardioactive drugs on cardiomyocytes derived from human induced pluripotent stem cells. Biochem Biophys Res Commun. 2009;387:482–488. doi: 10.1016/j.bbrc.2009.07.052. [DOI] [PubMed] [Google Scholar]
- 32.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng S, Lacerda AE, Kirsch GE, Brown AM, Bruening-Wright A. The action potential and comparative pharmacology of stem cell-derived human cardiomyocytes. J Pharmacol Toxicol Methods. 61:277–286. doi: 10.1016/j.vascn.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Otsuji TG, Minami I, Kurose Y, Yamauchi K, Tada M, Nakatsuji N. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: Qualitative effects on electrophysiological responses to drugs. Stem Cell Res. 4:201–213. doi: 10.1016/j.scr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson MK, Duker G, Tropp C, Andersson B, Sartipy P, Vos MA, van Veen TA. Quantified proarrhythmic potential of selected human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 4:189–200. doi: 10.1016/j.scr.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu W, Antzelevitch C. Effects of a k(+) channel opener to reduce transmural dispersion of repolarization and prevent torsade de pointes in lqt1, lqt2, and lqt3 models of the long-qt syndrome. Circulation. 2000;102:706–712. doi: 10.1161/01.cir.102.6.706. [DOI] [PubMed] [Google Scholar]
- 38.Shah RR. Drug-induced qt interval shortening: Potential harbinger of proarrhythmia and regulatory perspectives. Br J Pharmacol. 2010;159:58–69. doi: 10.1111/j.1476-5381.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Black SC, Lucchesi BR. Potassium channel openers are likely to be proarrhythmic in the diseased human heart. Cardiovasc Res. 1994;28:923–924. doi: 10.1093/cvr/28.6.923. discussion 926–929. [DOI] [PubMed] [Google Scholar]
- 40.Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277:301–306. [PubMed] [Google Scholar]
- 41.Zicha S, Moss I, Allen B, Varro A, Papp J, Dumaine R, Antzelevich C, Nattel S. Molecular basis of species-specific expression of repolarizing k+ currents in the heart. Am J Physiol Heart Circ Physiol. 2003;285:H1641–1649. doi: 10.1152/ajpheart.00346.2003. [DOI] [PubMed] [Google Scholar]
- 42.Saumarez RC, Chojnowska L, Derksen R, Pytkowski M, Sterlinski M, Huang CL, Sadoul N, Hauer RN, Ruzyllo W, Grace AA. Sudden death in noncoronary heart disease is associated with delayed paced ventricular activation. Circulation. 2003;107:2595–2600. doi: 10.1161/01.CIR.0000068342.96569.A1. [DOI] [PubMed] [Google Scholar]
- 43.Caspi O, Itzhaki I, Kehat I, Gepstein A, Arbel G, Huber I, Satin J, Gepstein L. In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev. 2009;18:161–172. doi: 10.1089/scd.2007.0280. [DOI] [PubMed] [Google Scholar]
- 44.Semsarian C, Ahmad I, Giewat M, Georgakopoulos D, Schmitt JP, McConnell BK, Reiken S, Mende U, Marks AR, Kass DA, Seidman CE, Seidman JG. The l-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J Clin Invest. 2002;109:1013–1020. doi: 10.1172/JCI14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long qt syndrome type 2 mutation. Eur Heart J. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.