Abstract
Introduction
There are significant rates of attrition in drug development. A number of compounds fail to progress past preclinical development due to limited tools that accurately monitor toxicity in preclinical studies and in the clinic. Research has focused on improving tools for the detection of organ-specific toxicity through the identification and characterization of biomarkers of toxicity.
Areas covered
This article reviews what we know about emerging biomarkers in toxicology, with a focus on the 2012 Northeast Society of Toxicology meeting titled ‘Translational Biomarkers in Toxicology.’ The areas covered in this meeting are summarized and include biomarkers of testicular injury and dysfunction, emerging biomarkers of kidney injury and translation of emerging biomarkers from preclinical species to human populations. The authors also provide a discussion about the biomarker qualification process and possible improvements to this process.
Expert opinion
There is currently a gap between the scientific work in the development and qualification of novel biomarkers for nonclinical drug safety assessment and how these biomarkers are actually used in drug safety assessment. A clear and efficient path to regulatory acceptance is needed so that breakthroughs in the biomarker toolkit for nonclinical drug safety assessment can be utilized to aid in the drug development process.
Keywords: kidney injury, liver, predictive, qualification, testis toxicity
1. Introduction
The Northeast Regional Chapter of the Society of Toxicology held a one-day meeting titled ‘Translational Biomarkers in Toxicology’ in October 2012. The goal of this meeting was to bring together scientists from industry and academia to discuss the most recent advances in biomarker identification and development to aid in the evaluation of the safety of pharmaceuticals and environmental chemicals. Speakers at this meeting covered diverse topics, including the regulatory review and qualification of biomarkers, identification of biomarkers of testicular injury and dysfunction, translation of preclinical biomarkers to humans in the context of drug development and biomarkers of kidney injury. Issues that were discussed included identifying novel biomarkers with better sensitivity and specificity, ways to best approach demonstrating cross species translation and the challenges of biomarker qualification. All of the speakers and scientists in attendance held the common goal of streamlining the process of identifying and qualifying biomarkers for assessing potential health risks, and this meeting was a strong step forward in achieving this goal. Summaries of the presentations and issues that were discussed are shown below.
2. The current status of biomarkers for predicting toxicity
2.1 Summary
Any drug can produce an adverse response at high exposures, and it is the task of the preclinical safety expert to identify not only the response but also the exposure at which the effect is observed. The clinical risk of the identified hazard must be assessed and is dependent upon a host of factors, including exposure, indication, patient health, metabolic stability and pharmacodynamic response. Hazard identification is relatively easy: risk assessment is difficult and challenging. Fortunately, a vast historical database of animal toxicity data is available to assist in the prediction of human effect based upon the judicious use of animal toxicology studies. In general, the current preclinical in vivo models have done quite well in correlating human effect, providing ~ 70% concordance between adverse findings in clinical studies and data generated in preclinical toxicology models [1,2]. In these retrospective analyses, the best concordance was observed in hematological, gastrointestinal and cardiovascular toxicities. This conclusion has been debated over the past decade [3], since the only drugs studied were those in clinical studies, and by necessity those that failed in preclinical studies due to unacceptable toxicity were not evaluated in the clinical setting. However, the clear consensus is that these preclinical data provide translational evidence for a number of common human toxicities. Importantly, the two areas with the poorest correlation were hepatic effects and hypersensitivity/cutaneous reactions. These toxicities were not observed in the preclinical models and are often responsible for termination of development.
Although animals are the best model we have at the moment for predicting adverse human effects, there is still significant attrition in drug development, which can be attributed, in part, to adverse findings in preclinical toxicology studies, with no way of safely monitoring this toxicity in the clinic, resulting in an absence of clinical safety data [4]. Over the past decade, much of the research in this area has focused on the detection of organ-specific toxicity, both for improved preclinical:clinical translatability and for better predictivity of specific organ toxicity at early stages of development. Much of this success has occurred through the development of and consequent use of specific markers of organ toxicity, that is, biomarkers. However, the early prediction of specific organ function, such as hepatic, dermal or immunologic, is not well established. With the possible exception of cardiac function, very few novel biomarkers have been identified and accepted over the past decade. Liver enzymes such as ALT and AST are markers of tissue injury, but are not predictive of overall liver function and often do not correlate with overt hepatic pathology as assessed by light microscopy.
The National Institutes of Health defined a biomarker as follows: ‘a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.’ [5] The National Cancer Institute definition added that this ‘characteristic’ is a biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease and may be used to see how well the body responds to a treatment for a disease or condition [6]. This definition for molecular or signature biomarkers may be applied to both safety biomarkers and efficacy biomarkers. Disease-specific or efficacy biomarkers have been recognized to be useful in clinical development [7–11]. In cancer research especially, biomarker assays can be used to identify the presence of a tumor, as well as the genetic subtype and ability to respond to therapy. Efficacy biomarkers are therefore invaluable tools for cancer detection and treatments.
An expansion of the definition of biomarkers by Muller and Dieterle [12] included a number of different categories, such as pharmacogenomic, proteomic, metabolomic, pharmacological, toxicological and imaging biomarker platforms, all of which may provide different perspectives on the safety or efficacy of a particular drug in development. New biomarkers for specific organ toxicity, that is, safety biomarkers, have been recently identified. Early, predictive, noninvasive biomarkers that may be used in vitro, in vivo and in translation to the clinic have the most value in drug development.
In 2005, the European Commission started development of the Innovative Medicines Initiative (IMI) and as part of this effort, it defined desirable characteristics for a biomarker of drug safety assessment: i) specific for certain types of injury; ii) indicates injury in a variety of experimental species as well as humans; iii) can be used to bridge across nonclinical/ preclinical studies to clinical and surveillance types of studies; iv) more effective at indicating injury than any other biomarker currently used; v) used instead of classic biomarkers, not in addition; vi) can be easily measured (in real time) even at a later stage (measurement is not strongly time dependent); vii) more reproducible and sensitive than the original toxicity endpoint it would replace; and viii) reduces the number of individuals tested (animals or humans).
These IMI definitions are closely reflected in the work of the Predictive Safety Testing Consortium (PSTC) of the Critical Path Institute. The PSTC has led an extensive collaboration between a number of pharmaceutical companies and worldwide health authorities including the US Food and Drug Administration (FDA), the European Medicines Agency and the Japanese Pharmaceuticals and Medical Devices Agency (PMDA). This consortium characterized and submitted several biomarkers for detection of kidney injury for qualification by regulatory authorities for use in preclinical studies: albumin, KIM-1, cystatin, total protein, β 2-microglubulin, clusterin and trefoil factor 3 (CHMP/EMEA, 2008). Biomarkers for other organs such as the liver, skeletal muscle, cardiac toxicity and vascular injury are also being developed and are described below.
2.1.1 Liver
Standard biomarkers of drug-induced liver injury (DILI) include alanine aminotransferase and aspartate aminotransferase (AST); however, both the specificity and sensitivity of these markers are limited as stated earlier since there is lack of correlation of liver enzyme changes and observable histopathological damage. Several biomarkers being evaluated that may improve upon this lack of concordance include glutamate dehydrogenase (GLDH), malate dehydrogenase (MDH), paraoxonase/arylesterase 1 (PON-1), purine nucleoside phosphorylase (PNP), arginase (ARG-1), sorbitol dehydrogenase (SDH) and glutathione S-transferase (GST-α).
2.1.2 Muscle
Muscle toxicity is becoming more and more prevalent as an issue in drug development and efforts to predict skeletal muscle injury are ongoing. As with the liver, the traditional markers of AST and creatinine kinase (CK) lack both specificity and sensitivity. Skeletal troponin I (Tnni1, Tnni2), skeletal troponin T (Tnnt1, Tnnt3), creatinine kinase protein M, parvalbumin (Pvalb), myosin light chain 3 (Myl3), fatty acid-binding protein 3 (Fabp3), aldolase A (Aldoa) and myoglobin show promise as more sensitive and more specific biomarkers of drug-induced skeletal muscle injury.
2.1.3 Cardiac hypertrophy
Along with DILI cardiac injury is a leading cause of the termination of promising therapies in clinical development and is commonly monitored in patients utilizing echocardiography (imaging), and more recently, natriuretic peptides (NPs), which are cardiac hormones synthesized and secreted in response to myofiber stretch. Evaluations are underway to investigate whether NT-proANP (the cleaved form of atrial NP) is an appropriate marker for changes in heart weight and decreased ejection fraction.
2.1.4 Vascular injury
DIVI is common in preclinical toxicology models; however, observations of histopathology are dissimilar among preclinical species and the relevance to humans is questionable. Efforts are ongoing to evaluate sensitive and specific markers of vascular injury of relevance to man and include VEGF, GRO/CINC-1, TIMP-1, vWGpp, NGAL, TSP-1, smooth muscle alpha actin, calponin and transgelin.
Muller and Dieterle [12] provide a very good overview of many of these efforts and they also discuss the work aimed at detecting gonadotoxicity with the use of inhibins, heterodimeric polypeptide hormones secreted by the testes or ovaries, as well as the potential use of trypsinogen activation peptide as an early and specific marker for pancreatic toxicity.
Selection of markers to assess cardiotoxicity includes those for structural change as well as functional, and it is important to note that these effects may be disassociated from one another. Although natriuretic peptides are being investigated as new biomarkers for structural cardiotoxicity, the most effective translational safety biomarker remains cardiac troponin [13,14]. Troponins have been used for many years in the clinical setting and have recently been incorporated into preclinical safety assessments. However, a number of factors need to be considered when using cardiac troponin in animal studies: including cross-species assay specificity, the effect of spontaneous rodent cardiomyopathy in the troponin assessment, and likely most importantly the rapid clearance of cardiac troponin in rats compared to man. In addition to the evaluation of natriuretic peptides, other new biomarkers being considered for cardiac toxicity include interleukin 6, myeloperoxidase and soluble CD 40 ligand [14].
Functional cardiac changes include electrophysiological changes and no classical biomarkers as defined here have been developed to detect these functional changes. The International Conference of Harmonization (ICH) has mandated preclinical assessment of QT prolongation, which is most often assessed with the in vitro human ether-à-go-go related gene (hERG) product assay. Alterations of hERG channel activity often result in QT prolongation which may be associated with potentially fatal arrhythmia. Since prolonged QT in preclinical animal species is not always correlated with a clinical risk of QT prolongation, additional surrogate markers of QT-mediated proarrhythmia are being developed such as triangulation, reverse use dependence and dispersion [14].
Blood cytokines are a recent area of interest as biomarkers of toxicity. Tarrant [15] provides a comprehensive review of the use of cytokines, chemokines and growth factors in preclinical safety assessment which typically show a robust response in the inflammatory process and immune response. However the use of cytokines as biomarkers is challenging because of their short serum half-life, low basal levels, lack of tissue-specific expression and differential species and strain expression. As noted by Tarrant, cytokines could be useful as mechanistic markers and in modeling pharmacodynamic relationships and a combinatorial analysis could be useful in an integrated ‘whole’ animal approach.
One of the more recent developments in the quest for identification and characterization of biomarkers is the measurement of plasma microRNAs (miRNAs) and messenger RNAs (mRNAs) [16]. miRNAs are 22 nucleotide endogenous RNA molecules that are transcribed in the nucleus, and processed in the cytoplasm before being recruited to RNAinduced silencing complexes to inhibit translation of specific target mRNA transcripts [17]. These genomic markers often display tissue-specific expression, may be released from the tissues into the plasma during toxic events, change early and with high magnitude in tissues and in the blood during specific organ toxicities, and can be measured using multiplex formats. Although validation as biomarkers has been challenged by technical difficulties, considerable progress has recently been made in assessing the potential value of miRNAs in the clinic, especially in cancer patients and cardiovascular diseases. The future of miRNAs and mRNAs as biomarkers of disease and organ toxicity depends on our ability to characterize their kinetics and to establish robust collection and measurement methods.
In addition to the work described earlier to understand serum markers of tissue injury, genomics, metabolomics and proteomics are powerful tools that have provided a considerable amount of data to the development of specific and predictive biomarkers. The detection and quantitation of biomarkers can be quite difficult since these are typically endogenous macromolecules in biological fluids which have differing immunoreactivity across species resulting in challenges in bioanalysis. Therefore, the success of these efforts is greatly enhanced by recent advances in two closely linked technologies, toxicoproteomics and metabolomics, and targeted, quantitative mass spectrometry. Mass spectrometry has emerged as the premier analytical tool compared to specific immunoassays or functional assays (Amacher, 2010) for the practical assessment of combinations of conventional and/or novel toxicity biomarkers in rodent and large animal preclinical species.
Preclinical scientists across the pharmaceutical industry have developed gene arrays consisting of genes believed to be involved in specific organ toxicity such as hepatotoxicity [18]. Fielden et al. [19] provided evidence that hepatic gene expression data from rats treated with nongenotoxic hepatocarcinogens produced a characteristic gene signature with an assay sensitivity of 86% and assay specificity of 81%. Goodsaid [20] proposed that rodent genomic biomarkers be validated across species, and Davis et al. [21] were the first to confirm rodent gene expression alterations in a nonhuman primate model associated with nephrotoxicity induced by the antibiotics gentamicin and everninomicin.
Collings and Vaidya [22] discuss advances in toxicogenomics and gene expression analysis in relation to the development of biomarkers of toxicity. Representative difference analysis of tissue specific microarrays resulted in the ability to identify up-regulation of KIM-1 in response to toxic kidney injury. Proteomics driven biomarker discovery technologies such as surface-enhanced laser desorption ionization (SELDI) and matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry have been used to investigate the response of a proteome derived from various biological matrices to toxicological insult. Better suited to the discovery of new biomarkers is the field of metabolomics since the metabolome is much smaller than that of the proteome [23]. Metabolite expression profiles in biological matrices can be evaluated with NMR and more recently with gas chromatography- and liquid chromatography–hydrophilic interaction chromatography-mass spectrometry.
The question ‘Are we ready for Novel Preclinical Safety Biomarkers?’ has been proposed [24]. The amount of effort and wealth of data over the past decade has proven that not only are we ready but that there has been success in developing, quantitating and qualifying novel biomarkers, such as those for the kidney, that are in use today. Although much progress has been made, there is much still to be done and worldwide health authorities, especially the FDA, have encouraged continuing efforts to find and integrate biomarkers into drug development and their appropriate use in clinical practice [8]. This paper will review what we know about emerging biomarkers in toxicology. It will focus on biomarkers of testicular injury and dysfunction, emerging biomarkers of kidney injury and translation of emerging biomarkers from preclinical species to human populations.
2.2 Biomarkers of testicular injury and dysfunction
2.2.1 Introduction
The assessment of human testicular function and male gamete quality currently relies on hormone measurements and semen parameters (sperm counts, motility and morphology). Semen analyses are highly variable both within and between individuals, which demonstrates their lack of sensitivity. Meanwhile, hormone measurements are generally unreliable at detecting mild testicular injury, so that only severe, potentially irreversible, injuries can be detected. A more sensitive approach is needed that allows for translation of findings in preclinical species to humans, and for monitoring of occupationally and environmentally induced testicular dysfunction at an early, reversible stage of injury.
2.2.2 Previous work/history
The human testis is very sensitive to toxicant-induced injury, yet the available tools for detecting the effects of exposures are quite limited [25,26]. The absence of symptoms associated with toxicant-induced testicular injury argues for a monitoring strategy in clinical trials and occupational and environmental exposure settings when the potential for irreversible testicular effects is a concern. The need for monitoring is illustrated by the 1977 exposure of workers to the nematocide 1,2-dibromo-3-chloropropane (DBCP) in a pesticide manufacturing plant [27]. A problem with testicular function came to attention because few male workers had recently fathered children [4]. An investigation revealed that a high proportion of workers exposed to DBCP were either azoospermic (13%) or oligospermic (33%); duration of exposure was related to the severity of the testicular injury [28]. A followup study of these workers 7 years later showed that permanent destruction of the germinal epithelium had occurred in most of the highly exposed men [29].
Monitoring the effects of exposure to a testicular toxicant using current methods is problematic, because serum hormones and semen parameters are variable and relatively insensitive [30]. Histopathology, a sensitive measure of testicular injury, can be performed in animal models, but its invasive nature makes it impractical to monitor exposures in humans. The potential value of measuring a sperm molecular biomarker to monitor testicular toxicity has been espoused by Dr. Gary Klinefleter of the US Environmental Protection Agency (EPA) who has studied SP22, a sperm protein that declines in abundance after exposure to both epididymal and testicular toxicants [31]. However, difficulty in measuring this low abundance sperm protein and the limitations of relying on a single indicator instead of a panel of indicators has slowed the development of SP22 as a biomarker.
About 40% of testicular mRNAs are detected in sperm, indicating that the sperm transcriptome can be used to monitor gene expression during spermatogenesis [32]. Various studies have investigated the association between altered testicular function and sperm mRNA transcript content, finding: i) significantly different motility-related sperm mRNA transcript abundance between normal and motility-impaired men [33], ii) altered sperm protamine mRNA levels in men with infertility [34,35] and iii) higher sperm Bcl2 mRNA in infertile men [35]. Sperm RNAs may be passively retained or play an active role in chromatin structure, imprinting, gene silencing and embryogenesis [36]. These results have led to the proposal that ‘.… As a wholly non-invasive proxy for the testis, this (sperm) RNA offers considerable potential as a marker for fertility status and the genetic and environmental influences that could make all the difference between a fertile and infertile phenotype’ [36].
Sperm DNA methylation marks offer a similar potential for insight into disrupted spermatogenesis. The germline DNA is demethylated early during embryonic development and then remethylated in a sex-specific manner later in development and during spermatogenesis [37]. Since DNA methylation remodeling is occurring during spermatogenesis, various studies have investigated the association between altered testicular function and sperm DNA methylation marks, finding aberrant DNA methylation of both imprinted [38–46] and nonimprinted genes [39,47,48]. A recent study compared genome-wide DNA methylation profiles for men with poor in vitro fertilization-related embryogenesis and abnormal sperm chromatin compaction, finding a subset of men with genome-wide DNA methylation defects with imprinted genes more prone to being abnormal than the entire genome [49]. Studies of altered sperm DNA methylation following toxicant exposure in adult animals have included extensive studies of the effects of a chemotherapeutic regimen in a rat model [50], and the effects of tamoxifen exposure of rats on sperm imprinted gene DNA methylation [44]. In summary, sperm mRNA transcripts and DNA methylation marks are acquired during spermatogenesis and reflect the integrity of that process. Measuring these sperm molecular biomarkers can provide insight into the testicular response to environmental and occupational chemical exposures.
2.2.3 Current research and new findings
Our animal-related work has made several unique contributions to this field, including i) developing a sperm mRNA transcript panel to detect responses over a time course of testicular toxicant exposure in a rat model [51] and will be extended to examine dose-dependent responses, ii) using the rat model to show that a sperm mRNA transcript panel is more sensitive than standard histopathology in detecting toxicant-induced perturbations [51] and iii) determining that sperm mRNA transcript patterns are toxicant specific [51]. We have also published a study of human sperm CpG methylation profiles and mRNA alterations associated with low sperm motility [52]. In this publication, we showed that i) low motility sperm have genomewide DNA hypomethylation that may be due to a failure of the sperm to complete chromatin compaction properly because of increased histone deacetylase 1 (HDAC1) presence; ii) low motility sperm have reduced mitochondrial NAD-dependent deacetylase sirtuin-3 (SIRT3) mRNA content which might be related to increased subcellular reactive oxygen species during spermatogenesis leading to the abnormal motility phenotype; and iii) this oxidative stress may be impeding the ability of DNA methyltransferase-3a (DNMT3A) to set the correct methylation marks which would also contribute to the hypomethylated phenotype.
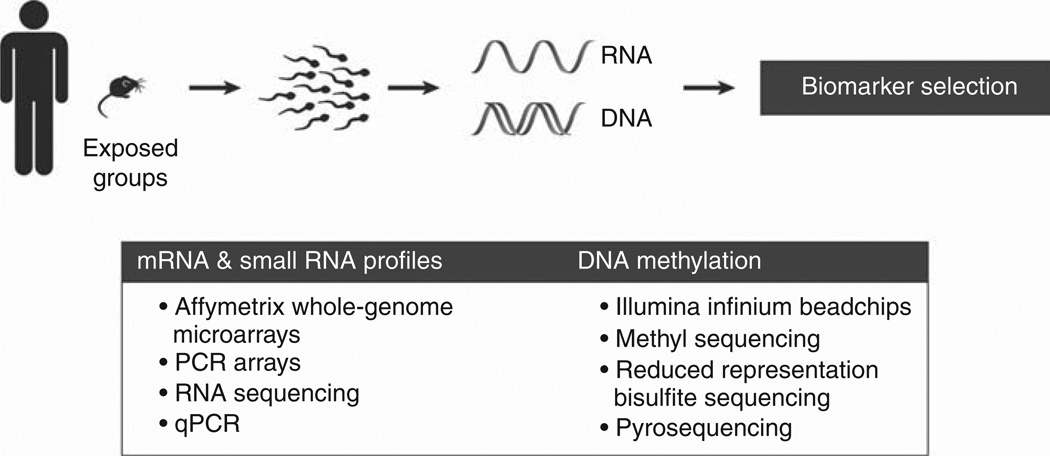
We have very strong evidence that a panel of sperm mRNA transcripts can detect and predict low level exposures to Sertoli cell toxicants in the rat [51]. To develop our current 12-transcript PCR array panel, we exposed rats to the Sertoli cell toxicant 2,5-hexanedione (2,5-HD) and selected one time point (3 months) and one dose (0.33% in the drinking water) to study. Repeatedly testing this exposure condition, we performed whole-genome Affymetrix GeneChip arrays (128 significantly altered transcripts out of ~ 27,000) followed by a 29-transcript PCR array and a 12-transcript PCR array. Transcripts chosen for the PCR arrays were driven by exposurerelated fold-change, and known expression and function within the testis. With each refinement of the PCR array panel, we improved the yield of significantly altered transcripts among those measured (Figure 1).
Figure 1.
Technical approaches used in identifying sperm molecular biomarker from rodent toxicity studies and chemically exposed populations in humans.
We then applied the 12-transcript PCR array panel to all 17 exposure groups tested to date, and found that each of the 12 transcripts is significantly altered in at least one exposure setting. This includes validation with a second Sertoli cell toxicant that yielded overlapping results, as well as exposure to a germ cell toxicant (DBCP) and multiple time points of both exposure and recovery from exposure. With a set of only 4 transcripts (Abi2, Clu, Ptgds and Sod3), we can detect at least one significantly altered sperm mRNA transcript in each of the 17 exposure models tested. We have begun to apply this same assay refinement strategy to DBCP, our model germ cell toxicant, to expand the PCR array panel to better include responses to germ cell injury.
In addition to mRNA transcript characterization, we are also interested in how these transcripts may correlate with decreased sperm performance. We are using in utero insemination with sperm from exposed rats (2,5-HD for 3 months) to determine i) if the sperm from exposed rats has a decreased ability to fertilize oocytes and ii) if this decreased function correlates with abundance of our candidate mRNA transcripts. This will bring together much of the work we have accomplished in our animal models by integrating molecular indicators of testicular injury with functional relevance.
2.2.4 Conclusions
Current methods for evaluating testicular injury and sperm quality are highly variable and/or are observed when the severity of injury is potentially irreversible. This raises the need for more reliable and sensitive approaches to assess injury. Over the past decade, there has been progress in understanding the underlying biology, with emerging evidence that both sperm mRNA transcripts and the epigenetic state of the sperm DNA modulate early embryonic events [36,53,54]. Furthermore, our work has demonstrated that quantifiable changes in sperm mRNA transcripts and DNA methylation marks are associated with alterations in sperm parameters and fertility [52].
The molecular signatures are established during spermatogenesis during which the germ cells are maturing for over 2 months within the seminiferous epithelium. Testicular insults sustained during this time window can affect the complex programming of both mRNA and miRNA transcripts, and DNA methylation marks that occur during germ cell proliferation and maturation. The complexity of germ cell development provides the opportunity to identify minor exposure-related perturbations of this process and determine the molecular basis for the downstream physiological responses. The development of a sperm molecular biomarker panel is an important step towards providing a reliable and sensitive monitoring strategy for chemical exposures from environmental and therapeutic settings, and has the capability to facilitate the translation of preclinical animal study findings to a monitoring strategy in clinical trials of new drug candidates.
2.2.5 Future directions
The ultimate goal of this project is to develop a predictive tool that will improve the detection of altered testicular function following exposure to a range of chemicals that include environmental contaminants and pharmaceutical compounds. We can leverage our current rat model to further develop and refine the biomarker panel by screening germ and Sertoli cell-specific toxicants.
Clinically, the biomarker panel will be used in screening human samples to uncover potential testicular dysfunction. Currently, over 450 semen samples from men presenting to the Division of Urology at Rhode Island Hospital have had traditional semen analyses performed, and the samples have been further processed for RNA and DNA. A subset of ~ 250 of these men have answered a detailed questionnaire regarding their lifestyle habits (i.e., smoking status, alcohol intake and body mass index) and pharmaceutical drug prescriptions, and have a range of exposures and semen parameters typical for a young male population. The sensitivity of our panel will be tested to see if it can identify alterations in the molecular biomarkers that are associated with these lifestyle and exposure scenarios. Of particular interest are men diagnosed with either lymphoma or leukemia who will undergo treatment with cytotoxic chemotherapeutic agents. There is evidence suggesting that chemotherapeutics can induce epigenetic changes in rodent sperm that have the potential for transgenerational impact [50], warranting further investigation in human populations.
2.3 Biomarkers of acute kidney injury
2.3.1 Introduction
Acute kidney injury (AKI) is a public concern with an incidence rate of 533 cases per million person-years in 2009, and increasing at nearly 10% annually [55]. The cases of mortality associated with dialysis-requiring AKI have more than doubled over the past decade, owing to the high risk of developing progressive chronic kidney disease (CKD) and end-stage renal disease (ESRD), apart from nonrenal complications [55,56]. Anywhere from 19 – 33% of cases of hospital associated AKI can be caused by drug induced nephrotoxicity [57]. Exposure to environmental contaminants such as occupational chemicals (arsine gas, carbon tetrachloride, trichloroethylene, etc.) [58,59], heavy metals (cadmium, lead, mercury, arsenic, uranium, etc.) [60–66], and other endemic toxins (ochratoxin A, aristolochic acid) [67–72] represent a very important etiologic factor in hospital and communityacquired kidney disease [73,74]. Avid tubular uptake of these drugs/environmental contaminants increases intracellular concentrations of toxic agents multifold, leading to alteration of renal hemodynamics, interstitial nephritis, tubular toxicity and obstruction by deposition of the metabolites [75]. The kidneys also receive nearly 25% of the cardiac output and significantly contribute to the biotransformation of drugs to toxic metabolites [76], making it highly susceptible to tubular epithelial cell necrosis and apoptosis. Tissue repair and regression of injury are dependent on the extent of the insult, and involve epithelial cell migration, proliferation and differentiation to recover lost structure and function [77]. Irreversible damage to the kidney leads to a reduction in the glomerular filtration rate (GFR), compromising kidney function and ultimately resulting in renal failure and death [77].
The traditional biomarkers of kidney function, serum creatinine (SCr) and blood urea nitrogen (BUN) are not entirely indicative of the GFR, nonspecific and insensitive in differentiating between stages of injury, reflecting the effects of cellular injury on renal function rather than the causative injury itself [57]. These considerable drawbacks are confounding factors in clinical trials for putative drug candidates because of poor diagnostic capabilities resulting in delayed commencement of therapy. Thus, there is an imperative need for the development and qualification of biomarkers for the early detection of AKI, to be used in risk assessment studies for drug development, predictive toxicological screens for chemical compounds and in evaluating therapeutic agents for stimulation of kidney regeneration. The goal of this presentation was to provide a succinct overview of some novel and emerging biomarkers for early detection of kidney damage.
2.3.2 Previous work/history
Kidney injury molecule-1 (Kim-1 in rodents and KIM-1 in humans), also known as T cell immunoglobulin and mucin-1 (TIM-1) or Hepatitis A Virus Cellular Receptor 1 (HAVCR-1) is a type I membrane glycoprotein containing a six-cysteine immunoglobulin-like domain and a mucin domain in the extracellular region. Kim-1 is the highest upregulated gene after kidney injury and is localized to the apical membrane of proximal tubular epithelial cells [78]. Kim-1 recognizes the phosphatidylserine and oxidized lipid epitopes on apoptotic cell surfaces and promotes their phagocytosis by the Kim-1 expressing epithelial cells [79]. The ectodomain of Kim-1 is cleaved by matrix metalloproteases and shed both in vitro and in vivo into the urine of rats and humans following injury, and can be detected as early as day 1 and day 2 after nephrotoxicant administration in rodent studies [80]. The Critical Path Institute’s Preventive Safety Testing Consortium (PSTC) recognized Kim-1 as an early biomarker for assessment of kidney injury induced by eleven structurally and mechanistically diverse models of tubular injury in rats. The diagnostic capability of Kim-1 far outstripped the traditional markers of kidney injury—SCr, BUN and NAG, in being able to accurately ascertain even the subtlest forms of tubular damage that could only be identified by histopathological analyses. The receiver operator characteristics–area under the curve (ROC–AUC) values were 0.91 to 0.99 for urinary Kim-1, with the threshold obtained for 95 – 96% specificity for injured animals compared to the average of controls being 1.87 [81]. On the basis of these studies, the US FDA and the EMA qualified Kim-1 for evaluating kidney toxicity in drug review processes. To determine the clinical potential of KIM-1 as a biomarker, a clinical cross sectional study was conducted and nine urinary biomarkers were tested individually and in combination for estimating their sensitivity and specificity in detecting AKI and associated mortality. Although all biomarkers individually were able to distinguish patients with and without AKI with an AUC > 0.83, a logic regression model used to combine the top 4 biomarkers (KIM-1, NGAL, HGF and total protein) yielded an AUC of 0.94 [82]. This combinatorial approach to biomarker studies not only offers a more structured and comprehensive overview of the pathological state, but is also technologically feasible to multiplexed biomarker measurements at the bedside.
In a span of 5 years, techniques for detection of Kim-1 have developed from immunoblotting, to ELISA assays, to multiplexed measurement of multiple analytes using microbeadbased Luminex xMAP technology [57]. The development of a rapid, immunochromatographic lateral flow assay for urinary Kim-1/KIM-1 is a significant step in making KIM-1 measurement a point-of-care diagnostic tool [83]. The RenaStick (a dipstick to measure urinary KIM-1) utilizes two epitopically distinct antibodies against KIM-1, to capture the protein in a colloidal gold nanoparticle complex and to colorimetrically determine the presence of the protein. The assay has a detection limit in the pg/ml range comparable to the Luminex assay, requires only 70 µl of sample, is independent of an analyzer and delivers results in 15 min while the Luminex or ELISA take up to 3 – 4 h [83]. The rapid advancements in the technology and its commercial availability have enabled many longitudinal studies to evaluate the temporal expression pattern and predictive potential of urinary KIM-1 as compared with other markers in various forms of AKI and is currently the subject of extensive ongoing clinical studies [84,85].
2.3.3 Current research and new findings
2.3.3.1 Fibrinogen
Microvascular endothelial injury, hemodynamic alterations and inflammatory response are some of the hallmarks of AKI [77]. Given the overwhelming role of the vascular system in the regulation of inflammatory damage, components of the hemostatic cascade are potential novel markers of injury as well as targets for therapeutic intervention. Fibrinogen, which comprises of 3 distinct polypeptides, is a versatile protein being a vital member of the coagulation machinery and an acute response protein with a well-documented mechanistic role in several inflammatory diseases [86]. Fibrinogen α, β and γ chains are significantly upregulated in the kidney following rodent renal ischemia reperfusion injury (IRI), in an expression pattern similar to Kim-1 [87]. The constituent chains are encoded by 3 individual genes and are transcribed and translated in the kidney tissue after the insult [88]. In a genome-wide expression analysis we found Fgβ to be the second highest gene upregulated amongst 22,523 genes in the rat kidney following IRI [87]. Fibrinogen protein is upregulated in the kidney and is also excreted in the urine following mechanistically distinct forms of AKI in mice, rats and humans supporting its claim as a reproducible and consistent marker of kidney injury [89]. Urinary protein as well as fibrinogen immunoreactivity in the kidney can successfully distinguish between patients with and without AKI, with the excreted protein achieving it with an AUC-ROC of 0.98, comparable to the established biomarkers of AKI [87,89]. Aside from being a biomarker of the injury process, fibrinogen and fibrinderived peptides perpetuate proinflammatory signals by acting as ligands for distinct receptors on immune cells. Thus, small bioactive peptides that compete with native protein for ligand-binding sites have been shown to have therapeutic effects by reducing the availability of free-binding centers and halting the progression of injury in multiple models of inflammatory diseases [90,91]. A naturally occurring Fgβ-derived peptide, Bβ15–42 has been shown to be protective against an ischemically induced model of AKI in mice, reducing tubular injury and expediting the resolution of injury [87,92]. Genetic reduction of global fibrinogen levels by deleting the Aα gene, which prevents assembly of the total protein, has also been shown to be protective against a myriad of diseases [93–95]. Fibrinogen deficient mice (Fg−/−) have reduced transcription and translation of the Aα chain, and the total protein is practically absent from circulation, while in heterozygotes (Fg+/−) it is reduced to about 75% of the control levels [96]. In an evaluation of the response of all 3 phenotypes to an ischemic injury, Fg+/− were protected the most from tubular injury as evidenced by SCr and BUN, reduced apoptotic cell count and heightened immune cell clearance [88]. Diminished availability of fibrinogen reduces binding to ICAM-1, thereby attenuating the apoptotic response cascade and assisting in speedy recovery from the insult [88]. Thus, fibrinogen is not only emerging as a new translational biomarker for detection of AKI but can also be further investigated as a therapeutic target to mitigate AKI.
2.3.3.2 Urinary MicroRNAs
As discussed earlier, outside of the world of protein biomarkers, there exist other small molecules, most notably small RNA species (e.g., miRNAs) that are gaining repute as early, sensitive and robust indicators of disease and damage conditions. Following ischemia induced AKI in rodents, miR-21, −155 and −18a were among the top upregulated miRNAs, and were found to have differential expression profiles in the blood and urine of ischemic rats compared to control animals [97]. The results were duplicated in a gentamicininduced model of nephrotoxicity, and were not observed following hepatotoxicity, establishing the robustness and specificity of the expression profile. In a pilot experiment, the urinary profiles of miR-21 and −155 were able to differentiate between a cohort of patients with AKI and healthy volunteers [97]. Plasma profiles of miRNAs have been used to identify cancers and various organ damage conditions [98–101]. Whether small urinary RNAs can serve as reproducible, quantifiable, early diagnostic and prognostic indicators of kidney disease in humans will be investigated in the near future.
2.3.4 Conclusions
There is an urgent need for the development of early and sensitive biomarkers for AKI that will enable opportune diagnosis in clinical settings for the timely administration of therapeutics, as well as in preclinical trials and safety assessment studies for an accurate feedback of drug performance. Emerging urinary protein biomarkers such as KIM-1 are promising candidates, which perform far better than traditional markers in terms of the sensitivity and predictive capability in animal models. Molecules such as fibrinogen, that are mechanistically involved in the pathophysiology of AKI also hold promise, both as early indicators of the injury process as well as putative therapeutic targets that can be manipulated to attenuate the course of damage. Small extracellular RNA species are being increasingly introduced as the new generation of biomarkers, by virtue of their early expression, stability, ease of detection, amplifiable signal and reduced complexity. Work has already begun in characterizing and validating small RNA expression reflective of AKI, in an attempt to identify a new ‘KIM,’ a ‘Kidney Injury MicroRNA.’
2.3.5 Future directions
Future attempts will need to focus on developing technologies for convenient detection of existing biomarkers, and evolving them to point of care diagnostic tools. Techniques for quantitatively assaying a combination of biomarkers in parallel will be invaluable in obtaining the maximum amount of information before making an informed decision about treatment options in the clinical setting. Further qualification and validation of new protein and nonprotein biomarkers in large clinical cohorts and long-term prospective studies will be necessary to evaluate their efficacy in the accurate diagnosis of AKI and prognosis of risk and development of disease.
2.4 Translation of emerging biomarkers from preclinical species to human populations
2.4.1 Introduction
Insufficient therapeutic index is a major cause of failures in drug development. Throughout the drug discovery process, therapeutic and toxic exposures are determined using in vitro assays and in vivo studies with biomarkers being of crucial importance. Safety biomarkers are essential for maximizing therapeutic index in several ways. Typically, they are applied to address candidate selection and manage risk by monitoring the no-adverse-effect levels of exposure in preclinical and clinical studies. Safety biomarkers are also useful for assessing human relevance of preclinical safety findings and enabling the development of safe or safer dosing paradigms. In addition, due to the recent advances in genome sequencing and system biology, there is a potential for safety biomarkers to influence patient selection. Although the premise of safety biomarkers is clear, there are challenges that need to be addressed. The most important aspects in biomarker research are the development of more sensitive and specific safety biomarkers for injury and their translation across preclinical species to the clinic. The challenge is multiplied by a lack of investigative models and limitations of clinical study designs. Further, biomarker qualification and regulatory acceptance provide additional hurdles in the application of safety biomarkers in drug development.
Here, we provide two examples where safety biomarkers have been applied that address the above-mentioned challenges. The first example shows the application of emerging biomarkers of acute kidney injury in the development of kidney sparing polypeptide-based antibiotics [102]. The second example will summarize our recently published retrospective study design for clinical evaluation of emerging biomarkers of liver injury in human subjects [103].
2.4.2 Current research
Polypeptide-based antibiotics such as polymyxins are essential in the treatment of serious, life threatening, gram-negative bacterial infections often caused by multiresistant bacterial strains. Since the utilization of polymyxins in the clinic has been severely limited due to drug-induced AKI, the development of safer alternatives is essential. Unfortunately, the serum creatinine and blood urea nitrogen are not suitable biomarkers in preclinical rat studies due to their low sensitivity. Thus, development of polymyxin analogues with improved kidney safety profiles necessitates the identification of more sensitive renal biomarkers to detect AKI earlier in the drug development process. This is also needed in the clinic for providing optimal clinical care and management of druginduced kidney injury. In addition to KIM-1, as discussed earlier, additional biomarkers of AKI such as clusterin, microalbumin, trefoil factor 3 (TFF3), alpha-glutathione S-transferase (GSTα), N acetyl-β-D-glucosaminidase (NAG), neutrophil gelatinase-associated lipocalin (NGAL), and osteopontin have recently been identified and evaluated [82,104–107]. In some cases these markers are more sensitive and selective than classical markers [105,107]. In addition, these biomarkers are capable of distinguishing among damage to the proximal tubules, distal tubules or the glomeruli.
To facilitate drug development of kidney sparing polymyxin analogues, we have developed a biomarker driven strategy across our preclinical species (rat, dog, and monkey). The rat is considered a species of choice for initial in vivo toxicologic evaluation of compounds due to practical and ethical reasons. Thus, the biomarker strategy consisted of identifying the most suitable biomarkers in short term rat toxicity studies and then evaluating translation to dogs and monkeys, and potentially humans in clinical studies. We first studied responses of a biomarker panel consisting of classical biomarkers such as serum creatinine, serum blood urea nitrogen, urinary protein and emerging biomarkers urinary NGAL, Kim1 and NAG after treatment with a model agent polymyxin B. Although all tested biomarkers were sensitive to polymyxin B-induced AKI in dogs and monkeys, in rats the classical biomarkers sCRE, BUN and urinary protein as well as emerging biomarkers NAG and Kim1 were insensitive. Only NGAL showed sufficient sensitivity for monitoring AKI in polymyxin B treated rats. Since it is not possible to utilize dogs and monkeys for screening of a large number of drug candidates due to ethical and practical aspects, the identification of NGAL as a sensitive biomarker in rats was crucial for further progress of the project. Maximum NGAL levels were reached 48 h after polymyxin treatment enabling us to use a 2-day toxicology study for effectively screening dozens of drug candidates in a short period of time. The benefits of this biomarker driven compound selection strategy in the early drug discovery stage are several fold: i) fast evaluation of dozens of leads while reducing the need for histopathologic examination of tissues and ii) reduced numbers of animals and amount of test compound needed compared to conventional toxicity studies. In summary, this work demonstrates that a preclinical biomarker-based strategy consisting of selection of biomarkers for appropriate species is essential. Its application for selection of drug candidates with improved kidney safety profiles can serve as a case study for addressing injury of other tissues.
Clinical translation of safety biomarkers provides a major challenge for application of emerging safety biomarkers in drug development. Since it is impossible to treat humans with toxicants for the sake of biomarker development, assessing performance of biomarkers of toxicity in the human clinic is difficult. The benchmarking against histopathology is challenging due to a lack of biopsies. Because of the sporadic nature and complicating factors in identification of drug induced organ damage, the access to human samples from cases of acute drug induced organ failure is very limited. In addition, the funding for clinical translational studies is not sufficient. Several international consortia such as HESI, PSTC and IMI SAFE-T are pursuing biomarker translation activities in the areas of kidney, liver and cardiovascular safety [104,108,109]. Several factors need to be addressed when evaluating clinical translation of emerging biomarkers. It includes determining the baseline biomarker value across gender, age and ethnic groups and establishing threshold values for biomarker response. Since the utility of safety biomarkers is its application in drug development, the biomarker performance should be studied in human diseases or conditions that approximate drug-induced injury including monitoring biomarker performance in standard treatments that are known to carry a risk of injury, for example, acetaminophen injury. Although prospective clinical biomarker studies are preferred, the ethical considerations, large cost and time needed for their conduct call for the exploration of alternative strategies.
In our recently published study [103] we explored an alternative biomarker study design utilizing discarded samples from subjects undergoing routine clinical evaluation by their physicians. Using this paradigm, we characterized the background levels of emerging biomarkers of liver injury, glutamate dehydrogenase (GLDH), malate dehyrogenase (MDH), purine nucleoside phosphorylase (PNP) and paraoxonase-1 (PON1) and effects of age and gender in the healthy subject population. Healthy was defined by the absence of clinical manifestation of liver disease. By carefully selecting patients with a variety of liver impairments, we were able to assess the performance of biomarkers in human subjects. The major obstacle for assessing the performance of emerging biomarkers in human subjects is the absence of a histologic correlate of the damage, that is, liver biopsies. Since liver biopsy is an invasive procedure, the evaluation of biomarker performance is measured by correlation with the biochemical benchmark of liver damage, serum alanine aminotransferase (ALT) activity. ALT is a widely used clinical biomarker [110] of liver damage but not all increases necessarily progress to severe liver injury. The increased ALT activity could be in response to transient hepatocellular injury that will not progress to severe DILI or might be a due to hepatic induction of the enzyme in response to certain drugs or metabolic states [111]. To get a better assessment of biomarker performance and limit the influence of ALT associated issues, we have used a biochemical criterion for defining drug induced liver injury that utilizes ALP and BIL in addition to ALT [112]. The ROC analysis of 843 subjects revealed that GLDH had the highest diagnostic power with an AUC of 0.98, followed by MDH (AUC = 0.91), PON1 (AUC = 0.70) and PNP (AUC = 0.62) [103]. Although the added value of GLDH needs to be further refined, the data suggests that GLDH might become an alternative biomarker of liver injury in the clinic. Furthermore, the analysis of the biomarker response in several cases of acetaminophen induced liver injury showed an interesting pattern. In some cases, the levels of GLDH decreased more rapidly than ALT [103]. Whether the faster rate of decline observed with GLDH better reflects the liver’s recovery from injury and thus shows added value in comparison to ALT needs to be further studied.
2.4.3 Conclusions and future directions
Application of translational safety biomarkers throughout the drug development process to aid compound selection, contribute to understanding of risk and address human relevance of preclinical findings by monitoring drug induced tissue injury in clinical studies, is becoming increasingly recognized. Although several biomarkers of tissue injury exist, there is an unmet need for more sensitive and predictive biomarkers of drug induced injury applicable across preclinical species and clinic. Therefore, scientific endeavors to develop and qualify translational safety biomarkers by international organizations and consortia in the United States and Europe such as HESI, PSTC and IMI SAFE-T have been initiated [104,108,109]. These efforts are enabled by a biomarker qualification process put first in place at the USFDA [104]. Although this approach has proven successful in cases of preclinical biomarker development for kidney and cardiovascular injury, the assessment of biomarker performance in the clinical setting to support biomarker qualification provides a great challenge. This has been addressed by designing prospective clinical evaluation strategies consisting of carefully designed clinical trials for detection of drug induced kidney and liver injury by SAFE-T consortium [113]. However, the complexity, feasibility and ethical issues surrounding prospective clinical studies require exploring alternative strategies. Therefore we have designed a retrospective strategy that exploited discarded samples from patients undergoing hospital visits [103]. The study design builds on collaboration with clinicians and provides a blueprint for an approach that could be replicated with other biomarkers.
2.5 Regulatory qualification of translational biomarkers
2.5.1 Introduction
How can a biomarker qualification process be developed by regulatory agencies? A process has now been used by the pharmaceutical industry over the past few years. We can look back now at its impact on pharmaceutical product development: i) What are the strengths and weaknesses of the different versions of a biomarker qualification process developed in each ICH region? ii) How can we develop metrics with which we can measure the impact of these processes on drug development and their acceptance by the pharmaceutical industry?
The information publicly available at this time is insufficient to answer these questions comprehensively. However, a healthy discussion about the opportunities and challenges of biomarker qualification may be started with what we know thus far.
2.5.2 Definitions for biomarker qualification
At the FDA, the immediate regulatory documentation leading to the biomarker qualification process may be traced back to the Pharmacogenomics Guidance [114] and the Critical Path Opportunities list [115]. The Pharmacogenomics Guidance defined:
Valid biomarker: A biomarker that is measured in an analytical test system with well-established performance characteristics and for which there is an established scientific framework or body of evidence that elucidates the physiologic, toxicologic, pharmacologic, or clinical significance of the test results. The classification of biomarkers is context specific.
There are two main challenges with this definition: the evidentiary standards that would need to be drafted to support them, and the process which would determine the validity of a biomarker. Technical specifications for a valid biomarker to be measured in an analytical test system with well-established performance characteristics can be developed, but how do we know that a biomarker has an established scientific framework or body of evidence? What makes a scientific framework or body of evidence ‘established’? This definition for the standards of evidence is difficult to translate into viable metrics and to translate these viable metrics into a regulatory decision about the validity of a biomarker. A well-trodden path in the past has been to have biomarkers accepted as ‘valid’ not necessarily on the basis of specific standards of evidence, but on the basis of long and exhausting periods of public debate about their validity so that a final decision was often made decades later by the exhaustion of scientists and clinicians in these debates. Even after this exhausting process, critical questions about the application of accepted biomarkers may still remain incompletely addressed. A long process for acceptance through exhaustion also generates along the way a steady stream of biases which carry over to the final context of use for the biomarker.
The Guidance introduced a concept in this definition which allowed for a different discussion about how to accept biomarkers. The definition added that: The classification of biomarkers is context specific. While the Guidance itself referred to ‘valid biomarkers’ with additional subclassifications of ‘known valid’ and ‘probable valid’, the introduction of the concept of context of use opened a different discussion about how to accept biomarkers. If the determination of the validity of a biomarker is contingent on its context of use, then a biomarker need not be accepted as a ‘valid’, but rather, as ‘qualified for a specific context of use.’
The context of use in biomarker qualification is a concept antithetical to the notion that a single, definitive validation is possible for a biomarker. The context of use of a biomarker aligns with the available data on the basis of how the biomarker is to be used, and not on the perceived quality of the data, which some authors have proposed as a potential classification [116] for ‘validated’ biomarkers. Biomarker qualification in this case is driven by the data available to support a specific context of use for the biomarker, rather than by a definitive assessment about the size or quality of the database available to evaluate a biomarker for all plausible contexts of use. This is an incremental concept, where the context of use for a biomarker expands as additional data are available: there is no definitive and universal context of use. A biomarker qualification driven by its context of use is also inconsistent with the Institute of Medicine Report [117] proposal for a single, definitive validation for biomarkers.
The introduction of the concept of ‘qualification,’ and its contrast with the concept of ‘validity’ transformed the discussion about regulatory acceptance of biomarkers. A regulatory process for biomarker acceptance is viable if its goal has a finite set of evidentiary standards and a finite review cycle span. Biomarker validation is, by definition, an open-ended process with open-ended evidentiary standards, where every potential application of the biomarker needs to be supported by independent studies and datasets. Biomarker qualification, however, is a finite process with finite evidentiary standards, because for this concept a finite context of use needs to be justified by finite datasets.
Context of use is a critical concept in biomarker qualification, because it is the basic metric against which evidentiary standards need to be defined. This is clear from the perspective of the clinical or nonclinical data needed to support the qualification of a biomarker. The decades-long processes for biomarker validation often examined every conceivable scenario for the application of a biomarker, requiring also every conceivable dataset to support its use. However, if the context of use of a biomarker is circumscribed to a specific application, the datasets needed to support this application will also be circumscribed to specific, well-designed studies from which these datasets may be generated. A broad context of use for a biomarker requires i) a clear question, ii) a scientific body of evidence and iii) a concurrent scientific consensus, which often prevent a swift and effective application of biomarkers in drug development. Narrowing the context in the application of a biomarker facilitates the acceptance and application of this biomarker. For example, the application of biomarkers to better understand mechanisms of safety or efficacy is likely to require straightforward scientific evidence for a number of examples of compounds or studies justifying the mechanistic conclusions. At the other extreme, the acceptance of the application of the same biomarkers for surrogate testing may be expected to include a comprehensive evidence database across multiple compounds and studies, with thorough analytical and biological support for the proposed replacement of a preexisting clinical endpoint.
While analytical validity may be measured through its own independent objective metrics, it is also subject to the context of use of a biomarker. For example, a specific context of use for a biomarker may require an analytical specification range for its measurement which is only a subset of the global analytical specification range available for this measurement. This prescribed range can facilitate the design of measurement platforms for this biomarker.
The process that would determine the validity of a biomarker is a separate challenge: who would determine that a biomarker is valid? As long as the decision was made through a drawn-out, exhausting public debate process, it is likely that most stakeholders in this process would also have an opportunity to participate in it. However, if a process is to be developed to review the scientific and clinical evidence supporting the validity of a biomarker, an owner for this process would need to be identified. While we have discussed here evidentiary standards and the process itself as two independent challenges, the Guidance also implied that a replacement for a decades-long process would require a consensus reached through precompetitive collaborations by interested stakeholders organized into consortia which would supply datasets to support a qualification. The Pharmacogenomics Guidance itself didn’t dwell on this, but the experience gained at the FDA [118–120] through the Pilot Process for Biomarker Qualification in the period of 2005 – 2008 led to formal regulatory biomarker qualification processes at the FDA, EMA and PMDA.
2.5.3 Biomarker qualification process
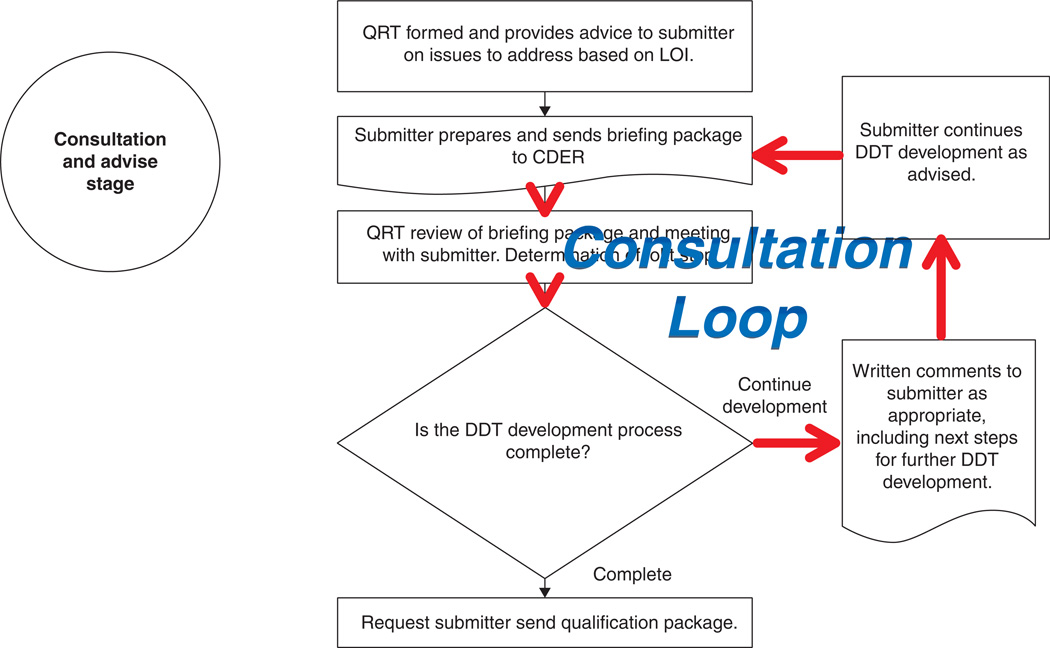
The Guidance on the Qualification of Drug Development Tools [121] summarized the experience gained from each of these concepts and activities associated with the Pilot Process for Biomarker Qualification. This Guidance also supported a long-term concept for a qualification process which has been under development since the draft version of this Guidance was published. The resulting Biomarker Qualification Process captured the need shown in the Pilot Process for a Consultation and Advice period previous to a qualification submission review. The Process includes a Pre-Submission Stage where a ‘prereview’ review requires an assessment by CDER of the scientific readiness of the submission. This is followed by a Consultation and Advice stage throughout which a submitter would be able to reach agreement with CDER reviewers on the context of use and evidentiary standards needed for qualification. In the absence of guidance on context of use determination and evidentiary standards assessment, this Consultation and Advice stage is essential for a viable Biomarker Qualification Process. Feedback in the Consultation and Advice stage requires a total of 16 steps. This Consultation and Advice stage is followed by a data review process which closely follows what would be expected in an NDA review, and adds another 5 steps to the Process. The Pre-Submission and Consultation and Advice stages together overshadow the length and complexity of the Review stage (Figure 2) [122]. A Consultation and Advice stage will not be needed once guidance is issued by CDER on Evidentiary Standards in Biomarker Qualification.
Figure 2. Consultation loop in the Consultation and Advice stage of a biomarker qualification process.
Several stages in current biomarker qualification processes throughout regulatory agencies are vulnerable to prolonged consultation cycles where applicants and reviewers go through multiple cycles of discussion about context of use and evidentiary standards for biomarker qualification.
2.5.4 Future of biomarker qualification
How well has this Biomarker Qualification Process fulfilled what the Pharmacogenomics Guidance and the Critical Path Opportunities List proposed? The development of the concept behind the Process itself has transformed the perspective about what is possible within a regulatory framework to introduce novel biomarkers in drug development. It has changed the popular view of a heterogeneous and unpredictable review landscape for the acceptance of new biomarkers as experienced through NDA reviews into a process and evidentiary standards which could be integrated into the critical path for drug development.
The rendering of this concept and this Process is yet to deliver on what the Guidance and the Critical Path Opportunities List promised. As of May 2013, the Biomarker Qualification Program Web page [122] showed three completed biomarker qualifications. The table in this webpage shows that the process yield is of about one qualification review completed every other year over the course of the six years since the Pilot Process for Biomarker Qualification was started in 2006. The Biomarker Qualification Program Web page does not provide information about the cycle time for each qualification, and it also does not provide information about the number of ongoing qualifications in the Program. However, the Web page for the Biomarker Qualification Process does outline the Process as reflected in the draft Qualification Guidance for Drug Development Tools [121], and as described in the previous section, this is a slow process. The complexity and multiple steps in this process represent difficult burdens to overcome for a precompetitive collaboration leading to a biomarker qualification. While this process accounts for critical checkpoints for qualification data, it seems counterintuitive, and contrary to the original goal of biomarker qualification, that drug approval would take a shorter time than a biomarker qualification. This process is difficult to integrate with critical path product development planning and to sell within pharmaceutical companies.
The experience available thus far in this process could be used to accelerate its performance by drafting a guidance outlining evidentiary standards expected for a biomarker qualification submission. Knowledge by a submitter about the evidentiary standards expected would obviate the need for the Consultation and Advice stage in the process. A direct access to regulatory review of biomarker qualification submissions could reduce the total number of steps and time needed to reach a decision.
3. Conclusions
This article summarizes the topics discussed at the 2012 meeting of the Northeast Society of Toxicology entitled ‘Translational Biomarkers in Toxicology,’ which focused on the most recent advances in biomarker development to aid in the assessment of organ-specific toxicity. As detailed here, there have been significant advances in the identification and characterization of novel biomarkers of testis, kidney, and liver injury that show improved sensitivity and specificity over the more traditional biomarkers of injury for these tissues. These biomarkers will aid in improved preclinical: clinical translatability and for better predictivity of specific organ toxicity at early stages of drug development if they can make it through the qualification process, which is currently a slow and complex process. The experienced gained thus far in the biomarker qualification process can be exploited to improve and streamline this process so that novel, improved biomarkers can more rapidly be used to evaluate the safety of pharmaceuticals and environmental chemicals.
4. Expert opinion
One of the most striking observations from this review is the gap between the scientific work in the development and qualification of novel biomarkers for nonclinical drug safety assessment and how these biomarkers are actually used in drug safety assessment. Without a clear and efficient path to regulatory acceptance, even the most impressive breakthroughs in the biomarker toolkit for nonclinical drug safety assessment will remain underutilized. The perennial acknowledged level of utility for these biomarkers associated with internal decision-making in the prioritization of development candidates is unsustainable for the future development of novel biomarkers, not only because of the lack of impact on regulatory review, but also because their public invisibility hinders development and qualification of clinical applications for these biomarkers.
An excellent case study for the value of regulatory acceptance is found in the nonclinical safety assessment qualification of biomarkers of nephrotoxicity. Even with the modest context of use for which these biomarkers were qualified by the FDA, EMA and PMDA, these biomarkers have already had a major impact, not only on internal decision-making by pharmaceutical companies, but also on nonclinical safety assessment data submitted for review to regulatory agencies. The most impressive impact of this qualification has been on the willingness of pharmaceutical companies in the PSTC and the FNIH Biomarkers Consortium to fund prospective clinical studies to generate data for the clinical qualification of these biomarkers. This ongoing effort will eventually show what it takes for a qualified nonclinical safety assessment context of use to evolve into a clinical safety biomarker.
Over the past decade, changes which lead to the development of the Biomarker Qualification Process were lead from within regulatory agencies, with the support of pharmaceutical industry consortia such as the PSTC. These changes need to gain relevance and permanence through appropriate regulatory guidance, reflecting the knowledge gained thus far about what works and what does not work in biomarker qualification. The leadership which regulatory agencies can provide to turn these guidance documents into reality is overdue.
Article highlights.
Recent research has identified sperm mRNA transcripts and DNA methylation marks in sperm that show promise as predictive biomarkers of altered testicular function following chemical exposure.
In addition to the use of KIM-1 as an improved biomarker for acute kidney injury, fibrinogen and urinary miRNAs have recently been identified as potential biomarkers that show promise as being more sensitive and selective than traditional biomarkers of kidney function.
A biomarker-driven compound selection strategy across preclinical species helped to facilitate the identification of polymyxin analogues with improved kidney safety profiles, which can be used as a strategy for addressing toxicity in other tissues.
A retrospective strategy that utilized discarded samples from patients undergoing routine clinical evaluation allowed for the characterization of background levels of emerging biomarkers of liver injury in relation to the widely used clinical biomarker of ALT activity, and revealed GLDH as a potential alternative biomarker of liver injury.
Much progress has been made in the development and qualification of novel biomarkers, but there is still a great deal of work to be done to identify novel biomarkers and integrate these into the risk assessment process for drug candidates and environmental chemicals and to determine their appropriate use in clinical practice.
Acknowledgments
Declaration of interest
S Campion, J Aubrecht, D Burt and S Schomaker are all employees of Pfizer, Inc. F Goodsaid and DW Brewster are employees of Vertex Pharmaceuticals. The research was funded in part by the Superfund Research Program (NIH/NIEHS) with K Boekelheide receiving grant P42ES013660 and L Anderson and S Pacheco being awarded grant T32ES007272-17 for ‘Training in Environmental Pathology.’ Research in Vaidya laboratory is supported by National Institutes of Health—Outstanding New Environmental Scientist Award (ES017543). K Boekelheide does occasional expert consulting with pharmaceutical and chemical manufacturing companies (Akros, Dow, Pfizer, Zafgen). He also owns stock in Exxon-Mobil and Pfizer, and in a small start-up biotechnology company (CytoSolv) developing a wound healing therapeutic.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Olson H, Betton G, Robinson D, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32(1):56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 2.Sistare FD, DeGeorge JJ. Preclinical predictors of clinical safety: opportunities for improvement. Clin Pharmacol Ther. 2007;82(2):210–214. doi: 10.1038/sj.clpt.6100243. [DOI] [PubMed] [Google Scholar]
- 3.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson AJ, Colburn WA, DeGruttola VG, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 6.NCI dictionary of terms. Available from: http://www.cancer.gov/dictionary?cdrid=45618.
- 7.Taube SE. Biomarkers in oncology. Ann NY Acad Sci. 2009;1180:111–118. doi: 10.1111/j.1749-6632.2009.05019.x. [DOI] [PubMed] [Google Scholar]
- 8. Amur S, Frueh FW, Lesko LJ, Huang SM. Integration and use of biomarkers in drug development, regulation and clinical practice: a US regulatory perspective. Biomarkers Med. 2008;2(3):305–311. doi: 10.2217/17520363.2.3.305. • Regulatory perspective on characterization and qualification of new biomarkers.
- 9.Jones R. Biomarkers: casting the net wide. Nature. 2010;466(7310):S11–S12. doi: 10.1038/466S11a. [DOI] [PubMed] [Google Scholar]
- 10.Dienstmann R, Rodon J, Tabernero J. Biomarker-driven patient selection for early clinical trials. Curr Opin Oncol. 2013;25(3):305–312. doi: 10.1097/CCO.0b013e32835ff3cb. [DOI] [PubMed] [Google Scholar]
- 11.George S, Reichardt P, Lechner T, et al. Hypertension as a potential biomarker of efficacy in patients with gastrointestinal stromal tumor treated with sunitinib. Ann Oncol. 2012;23(12):3180–3187. doi: 10.1093/annonc/mds179. [DOI] [PubMed] [Google Scholar]
- 12. Muller PY, Dieterle F. Tissue-specific, non-invasive toxicity biomarkers: translation from preclinical safety assessment to clinical safety monitoring. Expert Opin Drug Metab Toxicol. 2009;5(9):1023–1038. doi: 10.1517/17425250903114174. ••Definition and comprehensive review of biomarkers for several different types of tissues.
- 13.O’Brien PJ. Cardiac troponin is the most effective translational safety biomarker for myocardial injury in cardiotoxicity. Toxicology. 2008;245(3):206–218. doi: 10.1016/j.tox.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Piccini JP, Whellan DJ, Berridge BR, et al. Current challenges in the evaluation of cardiac safety during drug development: translational medicine meets the Critical Path Initiative. Am Heart J. 2009;158(3):317–326. doi: 10.1016/j.ahj.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 15. Tarrant JM. Blood cytokines as biomarkers of in vivo toxicity in preclinical safety assessment: considerations for their use. Toxicol Sci. 2010;117(1):4–16. doi: 10.1093/toxsci/kfq134. • Review of the use of cytokines as biomarkers.
- 16.Mikaelian I, Scicchitano M, Mendes O, et al. Frontiers in preclinical safety biomarkers: microRNAs and messenger RNAs. Toxicol Pathol. 2013;41(1):18–31. doi: 10.1177/0192623312448939. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Mendrick DL. Genomic and genetic biomarkers of toxicity. Toxicology. 2008;245(3):175–181. doi: 10.1016/j.tox.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007;99(1):90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- 20.Goodsaid FM. Identification and measurement of genomic biomarkers of nephrotoxicity. J Pharmacol Toxicol Methods. 2004;49(3):183–186. doi: 10.1016/j.vascn.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Davis JW, II, Goodsaid FM, Bral CM, et al. Quantitative gene expression analysis in a nonhuman primate model of antibiotic-induced nephrotoxicity. Toxicol Appl Pharmacol. 2004;200(1):16–26. doi: 10.1016/j.taap.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 22. Collings FB, Vaidya VS. Novel technologies for the discovery and quantitation of biomarkers of toxicity. Toxicology. 2008;245(3):167–174. doi: 10.1016/j.tox.2007.11.020. • Discussion of advances in toxicogenomics and gene expression analysis in relation to develompent of biomarkers of toxicity.
- 23.Amacher DE. The discovery and development of proteomic safety biomarkers for the detection of drug-induced liver toxicity. Toxicol Appl Pharmacol. 2010;245(1):134–142. doi: 10.1016/j.taap.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Goodsaid F. Are we ready for novel preclinical safety biomarkers? Preclinica. 2004;2(4):1–3. [Google Scholar]
- 25.Jurewicz J, Hanke W, Radwan M, Bonde JP. Environmental factors and semen quality. Int J Occup Med Environ Health. 2009;22(4):305–329. doi: 10.2478/v10001-009-0036-1. [DOI] [PubMed] [Google Scholar]
- 26.Boekelheide K. Commentary on “incidence and nature of testicular toxicity findings …”. Birth Defects Res B Dev Reprod Toxicol. 2011;92(6):501–503. doi: 10.1002/bdrb.20319. [DOI] [PubMed] [Google Scholar]
- 27.Whorton D, Krauss RM, Marshall S, Milby TH. Infertility in male pesticide workers. Lancet. 1977;2(8051):1259–1261. doi: 10.1016/s0140-6736(77)92665-4. [DOI] [PubMed] [Google Scholar]
- 28.Whorton D, Milby TH, Krauss RM, Stubbs HA. Testicular function in DBCP exposed pesticide workers. J Occup Med. 1979;21(3):161–166. [PubMed] [Google Scholar]
- 29.Eaton M, Schenker M, Whorton MD, et al. Seven-year follow-up of workers exposed to 1,2-dibromo-3-chloropropane. J Occup Med. 1986;28(11):1145–1150. [PubMed] [Google Scholar]
- 30.Bonde JP, Giwercman A, Ernst E. Identifying environmental risk to male reproductive function by occupational sperm studies: logistics and design options. Occup Environ Med. 1996;53(8):511–519. doi: 10.1136/oem.53.8.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klinefelter GR. Saga of a sperm fertility biomarker. Anim Reprod Sci. 2008;105(1–2):90–103. doi: 10.1016/j.anireprosci.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Ostermeier GC, Dix DJ, Miller D, et al. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360(9335):772–777. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Zhou Z, Xu M, et al. A spermatogenesis-related gene expression profile in human spermatozoa and its potential clinical applications. J Mol Med (Berl) 2004;82(5):317–324. doi: 10.1007/s00109-004-0526-3. [DOI] [PubMed] [Google Scholar]
- 34.Aoki VW, Liu L, Carrell DT. A novel mechanism of protamine expression deregulation highlighted by abnormal protamine transcript retention in infertile human males with sperm protamine deficiency. Mol Hum Reprod. 2006;12(1):41–50. doi: 10.1093/molehr/gah258. [DOI] [PubMed] [Google Scholar]
- 35.Steger K, Wilhelm J, Konrad L, et al. Both protamine-1 to protamine-2 mRNA ratio and Bcl2 mRNA content in testicular spermatids and ejaculated spermatozoa discriminate between fertile and infertile men. Hum Reprod. 2008;23(1):11–16. doi: 10.1093/humrep/dem363. [DOI] [PubMed] [Google Scholar]
- 36. Miller D, Ostermeier GC. Towards a better understanding of RNA carriage by ejaculate spermatozoa. Hum Reprod Update. 2006;12(6):757–767. doi: 10.1093/humupd/dml037. • Review of spermatozoal RNA and its potential as a marker for fertility status.
- 37.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18(1–2):63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 38.Hammoud SS, Purwar J, Pflueger C, et al. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94(5):1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Houshdaran S, Cortessis VK, Siegmund K, et al. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2(12):e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363(9422):1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 41.Marques CJ, Costa P, Vaz B, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14(2):67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 42.Poplinski A, Tuttelmann F, Kanber D, et al. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/ H19 ICR1. Int J Androl. 2010;33(4):642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi H, Sato A, Otsu E, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16(21):2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 44.Pathak S, Kedia-Mokashi N, Saxena M, et al. Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil Steril. 2009;91(5 Suppl):2253–2263. doi: 10.1016/j.fertnstert.2008.07.1709. [DOI] [PubMed] [Google Scholar]
- 45.Sato A, Hiura H, Okae H, et al. Assessing loss of imprint methylation in sperm from subfertile men using novel methylation polymerase chain reaction Luminex analysis. Fertil Steril. 2011;95(1):129–134. doi: 10.1016/j.fertnstert.2010.06.076. 34 e1–4. [DOI] [PubMed] [Google Scholar]
- 46.Boissonnas CC, Abdalaoui HE, Haelewyn V, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18(1):73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W, Shen O, Qin Y, et al. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS ONE. 2010;5(11):e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro-Costa P, Nogueira P, Carvalho M, et al. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010;25(10):2647–2654. doi: 10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aston KI, Punj V, Liu L, Carrell DT. Genome-wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil Steril. 2012;97(2):285–292. doi: 10.1016/j.fertnstert.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Chan D, Delbes G, Landry M, et al. Epigenetic alterations in sperm DNA associated with testicular cancer treatment. Toxicol Sci. 2012;125(2):532–543. doi: 10.1093/toxsci/kfr307. [DOI] [PubMed] [Google Scholar]
- 51. Pacheco SE, Anderson LM, Sandrof MA, et al. Sperm mRNA transcripts are indicators of sub-chronic low dose testicular injury in the Fischer 344 rat. PLoS One. 2012;7(8):e44280. doi: 10.1371/journal.pone.0044280. • Research demonstrating the utility of sperm mRNA transcripts as biomarkers of toxicity in a rat model of toxicant exposure.
- 52. Pacheco SE, Houseman EA, Christensen BC, et al. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One. 2011;6(6):e20280. doi: 10.1371/journal.pone.0020280. • Paper demonstrating the methylation profiles and mRNA alterations associated with low sperm motility in human subjects.
- 53.Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97(2):267–274. doi: 10.1016/j.fertnstert.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 54.Jenkins TG, Carrell DT. The paternal epigenome and embryogenesis: poising mechanisms for development. Asian J Androl. 2011;13(1):76–80. doi: 10.1038/aja.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu RK, McCulloch CE, Dudley RA, et al. Temporal changes in incidence of dialysis-requiring AKI. J Am Soc Nephrol. 2013;24(1):37–42. doi: 10.1681/ASN.2012080800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bucaloiu ID, Kirchner HL, Norfolk ER, et al. Increased risk of death and de novo chronic kidney disease following reversible acute kidney injury. Kidney Int. 2012;81(5):477–485. doi: 10.1038/ki.2011.405. [DOI] [PubMed] [Google Scholar]
- 57. Vaidya VS, Ferguson MA, Bonventre JV. Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48:463–493. doi: 10.1146/annurev.pharmtox.48.113006.094615. • Comprehensive review on biomarkers of kidney injury.
- 58.Hodgson S, Nieuwenhuijsen MJ, Hansell A, et al. Excess risk of kidney disease in a population living near industrial plants. Occup Environ Med. 2004;61(8):717–719. doi: 10.1136/oem.2003.010629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Vleet TR, Schnellmann RG. Toxic nephropathy: environmental chemicals. Semin Nephrol. 2003;23(5):500–508. doi: 10.1016/s0270-9295(03)00094-9. [DOI] [PubMed] [Google Scholar]
- 60.Wu X, Liang Y, Jin T, et al. Renal effects evolution in a Chinese population after reduction of cadmium exposure in rice. Environ Res. 2008;108(2):233–238. doi: 10.1016/j.envres.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Nordberg GF, Jin T, Wu X, et al. Prevalence of kidney dysfunction in humans - relationship to cadmium dose, metallothionein, immunological and metabolic factors. Biochimie. 2009;91(10):1282–1285. doi: 10.1016/j.biochi.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 62.Gonick HC. Nephrotoxicity of cadmium & lead. Indian J Med Res. 2008;128(4):335–352. [PubMed] [Google Scholar]
- 63.Roels HA, Hoet P, Lison D. Usefulness of biomarkers of exposure to inorganic mercury, lead, or cadmium in controlling occupational and environmental risks of nephrotoxicity. Ren Fail. 1999;21(3–4):251–262. doi: 10.3109/08860229909085087. [DOI] [PubMed] [Google Scholar]
- 64.Roels H, Lauwerys R, Konings J, et al. Renal function and hyperfiltration capacity in lead smelter workers with high bone lead. Occup Environ Med. 1994;51(8):505–512. doi: 10.1136/oem.51.8.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Burbure C, Buchet JP, Leroyer A, et al. Renal and neurologic effects of cadmium, lead, mercury, and arsenic in children: evidence of early effects and multiple interactions at environmental exposure levels. Environ Health Perspect. 2006;114(4):584–590. doi: 10.1289/ehp.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarup L. Hazards of heavy metal contamination. Br Med Bull. 2003;68:167–182. doi: 10.1093/bmb/ldg032. [DOI] [PubMed] [Google Scholar]
- 67.Hassan AM, Sheashaa HA, Fattah MF, et al. Study of ochratoxin A as an environmental risk that causes renal injury in breast-fed Egyptian infants. Pediatr Nephrol. 2006;21(1):102–105. doi: 10.1007/s00467-005-2033-3. [DOI] [PubMed] [Google Scholar]
- 68.Petrik J, Zanic-Grubisic T, Barisic K, et al. Apoptosis and oxidative stress induced by ochratoxin A in rat kidney. Arch Toxicol. 2003;77(12):685–693. doi: 10.1007/s00204-003-0501-8. [DOI] [PubMed] [Google Scholar]
- 69.Pozdzik AA, Salmon IJ, Debelle FD, et al. Aristolochic acid induces proximal tubule apoptosis and epithelial to mesenchymal transformation. Kidney Int. 2008;73(5):595–607. doi: 10.1038/sj.ki.5002714. [DOI] [PubMed] [Google Scholar]
- 70.Isnard Bagnis C, Deray G, Baumelou A, et al. Herbs and the kidney. Am J Kidney Dis. 2004;44(1):1–11. doi: 10.1053/j.ajkd.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Grollman AP, Jelakovic B. Role of environmental toxins in endemic(Balkan) nephropathy 2006, Zagreb Croatia. J Am Soc Nephrol. 2007 Oct;18(11):2817–2823. doi: 10.1681/ASN.2007050537. [DOI] [PubMed] [Google Scholar]
- 72.Grollman AP, Shibutani S, Moriya M, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA. 2007;104(29):12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soderland P, Lovekar S, Weiner DE, et al. Chronic kidney disease associated with environmental toxins and exposures. Adv Chronic Kidney Dis. 2010;17(3):254–264. doi: 10.1053/j.ackd.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Bacchetta J, Dubourg L, Juillard L, Cochat P. Non-drug-induced nephrotoxicity. Pediatr Nephrol (Berlin, Germany) 2009;24(12):2291–2300. doi: 10.1007/s00467-009-1180-3. [DOI] [PubMed] [Google Scholar]
- 75.Thadhani R, Pascual M, Bonventre JV. Acute renal failure. N Engl J Med. 1996;334(22):1448–1460. doi: 10.1056/NEJM199605303342207. [DOI] [PubMed] [Google Scholar]
- 76.Anders MW. Metabolism of drugs by the kidney. Kidney Int. 1980;18(5):636–647. doi: 10.1038/ki.1980.181. [DOI] [PubMed] [Google Scholar]
- 77.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121(11):4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ichimura T, Bonventre JV, Bailly V, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273(7):4135–4142. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 79.Ichimura T, Asseldonk EJ, Humphreys BD, et al. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vaidya VS, Ramirez V, Ichimura T, et al. Urinary kidney injury molecule-1: a sensitive quantitative biomarker for early detection of kidney tubular injury. Am J Physiol Renal Physiol. 2006;290(2):F517–F529. doi: 10.1152/ajprenal.00291.2005. [DOI] [PubMed] [Google Scholar]
- 81. Vaidya VS, Ozer JS, Dieterle F, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28(5):478–485. doi: 10.1038/nbt.1623. • Research demonstrating the utility of KIM-1 as a sensitive biomarker of kidney injury.
- 82.Vaidya VS, Waikar SS, Ferguson MA, et al. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin Transl Sci. 2008;1(3):200–208. doi: 10.1111/j.1752-8062.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vaidya VS, Ford GM, Waikar SS, et al. A rapid urine test for early detection of kidney injury. Kidney Int. 2009;76(1):108–114. doi: 10.1038/ki.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonventre JV, Yang L. Kidney injury molecule-1. Curr Opin Crit Care. 2010;16(6):556–561. doi: 10.1097/MCC.0b013e32834008d3. [DOI] [PubMed] [Google Scholar]
- 85.Peralta CA, Katz R, Bonventre JV, et al. Associations of urinary levels of kidney injury molecule 1 (KIM-1) and neutrophil gelatinase-associated lipocalin (NGAL) with kidney function decline in the Multi-Ethnic Study of Atherosclerosis (MESA) Am J Kidney Dis. 2012;60(6):904–911. doi: 10.1053/j.ajkd.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davalos D, Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin Immunopathol. 2012;34(1):43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 87.Krishnamoorthy A, Ajay AK, Hoffmann D, et al. Fibrinogen {beta}-derived B{beta}15–42 peptide protects against kidney ischemia/reperfusion injury. Blood. 2011;118(7):1934–1942. doi: 10.1182/blood-2011-02-338061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ajay AK, Saikumar J, Bijol V, Vaidya VS. Heterozygosity for fibrinogen results in efficient resolution of kidney ischemia reperfusion injury. PLoS One. 2012;7(9):e45628. doi: 10.1371/journal.pone.0045628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hoffmann D, Bijol V, Krishnamoorthy A, et al. Fibrinogen excretion in the urine and immunoreactivity in the kidney serves as a translational biomarker for acute kidney injury. Am J Pathol. 2012;181(3):818–828. doi: 10.1016/j.ajpath.2012.06.004. • Research evaluating the utility of fibronigen as as biomarker for early detection of acute kidney injury.
- 90.Adams RA, Bauer J, Flick MJ, et al. The fibrin-derived gamma377–395 peptide inhibits microglia activation and suppresses relapsing paralysis in central nervous system autoimmune disease. J Exp Med. 2007;204(3):571–582. doi: 10.1084/jem.20061931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petzelbauer P, Zacharowski PA, Miyazaki Y, et al. The fibrin-derived peptide Bbeta15–42 protects the myocardium against ischemia-reperfusion injury. Nat Med. 2005;11(3):298–304. doi: 10.1038/nm1198. [DOI] [PubMed] [Google Scholar]
- 92.Sorensen I, Rong S, Susnik N, et al. Bbeta(15–42) attenuates the effect of ischemia-reperfusion injury in renal transplantation. J Am Soc Nephrol. 2011;22(10):1887–1896. doi: 10.1681/ASN.2011010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cruz-Topete D, Iwaki T, Ploplis VA, Castellino FJ. Delayed inflammatory responses to endotoxin in fibrinogen-deficient mice. J Pathol. 2006;210(3):325–333. doi: 10.1002/path.2060. [DOI] [PubMed] [Google Scholar]
- 94.Drew AF, Tucker HL, Liu H, et al. Crescentic glomerulonephritis is diminished in fibrinogen-deficient mice. Am J Physiol Renal Physiol. 2001;281(6):F1157–F1163. doi: 10.1152/ajprenal.2001.281.6.F1157. [DOI] [PubMed] [Google Scholar]
- 95.Akassoglou K, Adams RA, Bauer J, et al. Fibrin depletion decreases inflammation and delays the onset of demyelination in a tumor necrosis factor transgenic mouse model for multiple sclerosis. Proc Natl Acad Sci USA. 2004;101(17):6698–6703. doi: 10.1073/pnas.0303859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suh TT, Holmback K, Jensen NJ, et al. Resolution of spontaneous bleeding events but failure of pregnancy in fibrinogen-deficient mice. Genes Dev. 1995;9(16):2020–2033. doi: 10.1101/gad.9.16.2020. [DOI] [PubMed] [Google Scholar]
- 97. Saikumar J, Hoffmann D, Kim TM, et al. Expression, circulation, and excretion profile of microRNA-21, −155, and −18a following acute kidney injury. Toxicol Sci. 2012;129(2):256–267. doi: 10.1093/toxsci/kfs210. • Work conducted to characterize miRNAs as potential biomarkers of kidney injury.
- 98.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 99.Asaga S, Kuo C, Nguyen T, et al. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 100.Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119(2):87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Burt D, Crowell SJ, Ackley DC, et al. Application of emerging biomarkers of acute kidney injury in development of kidney-sparing polypeptide-based antibiotics. Drug Chem Toxicol. doi: 10.3109/01480545.2013.834360. In press. • Excellent example of a preclinical biomarker-based strategy for identifying drug candidates with improved safety profiles.
- 103. Schomaker S, Warner R, Bock J, et al. Assessment of emerging biomarkers of liver injury in human subjects. Toxicol Sci. 2013;132(2):276–283. doi: 10.1093/toxsci/kft009. • Describes a study in which the performance of emerging biomarkers of liver injury was evaluated in human subjects.
- 104. Goodsaid FM, Frueh FW, Mattes W. The predictive safety testing consortium: a synthesis of the goals, challenges and accomplishments of the critical path. Drug Discov Today Technol. 2007;4(2):47–50. doi: 10.1016/j.ddtec.2007.10.010. • Introduction to the PSTC as collaborative effort and novel approach to biomarker qualification.
- 105.Goodsaid FM, Blank M, Dieterle F, et al. Novel biomarkers of acute kidney toxicity. Clin Pharmacol Ther. 2009;86(5):490–496. doi: 10.1038/clpt.2009.149. [DOI] [PubMed] [Google Scholar]
- 106.Bonventre JV, Vaidya VS, Schmouder R, et al. Next-generation biomarkers for detecting kidney toxicity. Nat Biotechnol. 2010;28(5):436–440. doi: 10.1038/nbt0510-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lock EA. Sensitive and early markers of renal injury: where are we and what is the way forward? Toxicol Sci. 2010;116(1):1–4. doi: 10.1093/toxsci/kfq128. [DOI] [PubMed] [Google Scholar]
- 108.Sistare FD, Dieterle F, Troth S, et al. Towards consensus practices to qualify safety biomarkers for use in early drug development. Nat Biotechnol. 2010;28(5):446–454. doi: 10.1038/nbt.1634. [DOI] [PubMed] [Google Scholar]
- 109. Vaidya VS, Bonventre JV. Biomarkers: in medicine, drug discovery, and environmental health. Hoboken, NJ: John Wiley & SOns, Inc.; 2010. • Comprehensive review of biomarkers.
- 110.Ozer J, Ratner M, Shaw M, et al. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245(3):194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 111.FDA. Guidance for Industry. Drug-Induced Liver Injury: premarketing Clinical Evaluation. 2007 Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM174090.pdf.
- 112.Aithal GP, Watkins PB, Andrade RJ, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther. 2011;89(6):806–815. doi: 10.1038/clpt.2011.58. [DOI] [PubMed] [Google Scholar]
- 113.Dieterle F. The European IMI SAFE-T consortium: qualification of translation safety biomarkers. Toxicol Lett. 2009;189:S157. [Google Scholar]
- 114.FDA. Guidance for Industry. Pharmacogenomic Data Submissions. 2005 Available from: www.fda.gov/downloads/RegulatoryInformation/Guidances/UCM126957.pdf.
- 115.FDA. Critical Path Opportunities List. 2006 Available from: www.fda.gov/downloads/scienceresearch/specialtopics/CriticalPathinitiative/CriticalPathOpportunitiesreports/UCM077258.pdf.
- 116.Altar CA, Amakye D, Bounos D, et al. A prototypical process for creating evidentiary standards for biomarkers and diagnostics. Clin Pharmacol Ther. 2008;83(2):368–371. doi: 10.1038/sj.clpt.6100451. [DOI] [PubMed] [Google Scholar]
- 117.Institute of Medicine of the National Academy of Sciences. Consensus Report. Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease. 2010 [Google Scholar]
- 118.Goodsaid FM, Freuh F. Regulatory guidance and validation of toxicologic biomarkers. In: DeCaprio AP, editor. Toxicologic biomarkers. New York: Taylor & Francis Group; 2006. pp. 187–220. [Google Scholar]
- 119.Goodsaid F, Frueh F. Process map proposal for the validation of genomic biomarkers. Pharmacogenomics. 2006;7(5):773–782. doi: 10.2217/14622416.7.5.773. [DOI] [PubMed] [Google Scholar]
- 120. Goodsaid FM, Frueh FW, Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245(3):219–223. doi: 10.1016/j.tox.2007.12.023. •• Discussion of approaches for qualification of biomarkers.
- 121.FDA. Guidance for Industry. Qualification Process for Drug Development Tools. 2010 Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM230597.pdf.
- 122.FDA. Biomarker Qualification Program. Available from: http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugDevelopmentToolsQualificationProgram/ucm284076.htm.