Abstract
BACKGROUND
Extracellular microRNAs (miRNAs) have been proposed as potentially robust and stable biomarkers of various disease conditions. The primary objective of this study was to identify miRNAs differentially occurring in the urine that could serve as potential biomarkers of acute kidney injury (AKI), because traditional AKI markers have limitations with respect to sensitivity, specificity, and timeliness of diagnosis.
METHODS
We profiled 1809 miRNAs in pooled urine samples from 6 patients with AKI and from 6 healthy controls. We measured the 378 stably detectable miRNAs in the 12 samples individually and selected the top 7 miRNAs that were most different in the urine of patients with AKI compared with the non-AKI control individuals. These miRNAs were assessed in a larger cohort of patients with AKI (n = 98:71 AKI patients in the intensive care unit (ICU) and 27 kidney transplantation patients with biopsy-proven tubular injury) and patients without AKI (n = 97: 74 healthy volunteers and 23 ICU patients without AKI).
RESULTS
We identified 4 miRNAs capable of significantly differentiating patients with AKI from individuals without AKI: miR-21 (P = 0.0005), miR-200c (P < 0.0001), miR-423 (P = 0.001), and miR-4640 (P = 0.0355). The combined cross-validated area under the ROC curve for these 4 miRNAs was 0.91. The imprecision with respect to miRNA isolation and reverse transcription efficiency was <9% across 224 samples.
CONCLUSIONS
In this study we determined the entire miRNome of human urine and identified a panel of miRNAs that are both detectable noninvasively and diagnostically sensitive indicators of kidney damage.
Acute kidney injury (AKI)7 is a devastating problem with a high incidence and in-hospital mortality rates of 40%–80% in the intensive care unit (ICU) (1). The detection of AKI relies on measuring the biomarkers blood urea nitrogen and serum creatinine (SCr), the use of which has remained largely unchanged for >50 years, despite their limitations regarding sensitivity, specificity, and capability for early diagnosis (2). A number of recent studies have examined protein biomarkers for early detection of AKI in humans (3). Advanced proteomic and metabolomic technologies are also being applied in discovery efforts (4); however, none of these investigations have advanced sufficiently for any of the biomarkers to be adopted into clinical practice. This situation can be attributed to the heterogeneity and multifactorial nature of AKI, as well as to the inherent limitations of protein biomarkers, such as the greater complexity of the proteome due to posttranslational modifications (5). These factors have prevented the emergence of biomarkers with early-detection capabilities and sufficient predictive power for use across the spectrum of AKI (6), thereby slowing the development of new therapeutics and interventions and their testing in clinical trials (7).
Small noncoding RNAs, such as microRNAs (miRNAs), have recently been identified as key regulators of diverse cellular processes, the dysregulation of which has been associated with disease pathogenesis. These findings have led to consideration of miRNAs as putative biomarkers (8, 9). The most striking advantage for using miRNAs is their stability in extracellular spaces (including such biological fluids as serum, urine, and saliva), which allows noninvasive analysis of their potential as biomarkers (10). Other characteristics that have driven the investigations into miRNAs as biomarkers of pathophysiological conditions are the high expression of genes encoding miRNAs, their low complexity, and an amenability to detection by amplification methods, such as quantitative real-time PCR (qPCR). Investigations have demonstrated that exosomal and circulating miRNAs have great potential as biomarkers for detecting cancers [such as prostate cancer (11), colorectal cancer (12), and non–small-cell lung cancer (13)], myocardial injury (14), and liver damage (15).
The majority of the research on miRNA biomarkers of kidney disease has centered on miRNAs present in the urine, but the previous studies were limited by small sample sizes (16) and poor normalizers (17). We previously conducted an analysis of miRNA production in rat kidneys subjected to ischemia reperfusion injury and found that miR-21, miR-155, and miR-18a were among the miRNAs with the highest upregulation in the kidneys after such injury. Furthermore, we have shown that miR-21 and miR-155 concentrations in the urine of patients with AKI were modestly but significantly different from those of healthy volunteers (25 participants in each group) (18). One drawback of extrapolating data from rats to humans, however, is that the number of known human miRNAs (2042 mature miRNAs) is nearly 3 times greater than that known for rats (723 mature miRNAs). The primary objective of this study was to identify miRNAs differentially present in the urine of AKI patients that could serve as sensitive, stable, and robust diagnostic biomarkers of kidney injury.
Materials and Methods
HUMAN STUDIES
All participants were patients or healthy volunteers recruited at Brigham and Women’s Hospital, Boston, or the University of Chicago Medical Center, Chicago. The Institutional Review Boards of the 2 hospitals approved the protocols for recruitment and sample collection, which were performed with informed consent of the participants.
Cross-sectional cohort
Urine samples from individuals with AKI were obtained from critically ill patients admitted to the ICU or from patients after they undergone cardiac surgery with cardiopulmonary bypass. AKI was defined by an SCr increase of at least 50% over baseline values. ICU patients with no evidence of AKI were used as hospital controls. Anuric patients were not included, because anuria reflects severe kidney injury for which biomarkers may not be of much value.
Kidney biopsy cohort
Urine samples were obtained at Brigham and Women’s Hospital from patients with biopsy-proven tubular injury to the transplanted kidney. All included biopsy samples showed only isolated acute tubular injury with degenerative changes of the tubular epithelium; they showed no clinically relevant evidence of allograft rejection or other pathologic changes.
Pheno Genetics cohort
Urine samples were obtained from healthy volunteers participating in the Pheno Genetics study, a large prospective observational cohort study examining the genetics of the immune system in healthy individuals and conducted at Brigham and Women’s Hospital. Inclusion criteria for healthy individuals were males or females 18–50 years of age and a willingness to provide urine and blood samples. Exclusion criteria were the presence of inflammatory diseases (e.g., asthma or psoriasis), autoimmune diseases (e.g., lupus or multiple sclerosis), chronic metabolic diseases (e.g., thyroid conditions or diabetes), and chronic infections (e.g., hepatitis B/C or HIV). Urine samples were collected from a subset of the healthy volunteers at a repeat visit within 1 year of the initial visit.
URINE COLLECTION
Urine was collected from spontaneous voids or from indwelling Foley catheters. Urine dipstick analysis (Multistix 8 SG; Bayer Corporation) was followed by centrifugation of the urine at 3000g for 10 min and microscopy examination of the urine sediment with an Olympus microscope. The urine supernatant was aliquoted into 1.8-mL Eppendorf tubes and frozen at −80 °C within 4 h of collection. We added no protease inhibitors or other reagents. Urinary creatinine concentrations were measured with a commercially available kit (Cayman Chemical). The urinary protein kidney injury molecule 1 (KIM-1) was measured in 30 µL urine with previously established Luminex-based assays (19, 20); the concentration was normalized to that of urinary creatinine.
RNA ISOLATION AND PROCESSING
RNA isolation
To obtain cell-free nucleic acids from urine, we centrifuged samples at 5000g for 10 min and used only the supernatant for RNA extraction. Soluble miRNAs from urine were isolated with the miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacturer’s protocol. RNA was eluted in 14 µL of RNase-free water. Full methologic details for RNA isolation are provided in the Supplemental Methods file in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol59/issue12.
Reverse transcription and preamplification
We then used the miScript RTII kit (Qiagen) to prepare cDNA from 1.5 µL of the eluted RNA, in accord with the manufacturer’s recommendations. We diluted the prepared cDNA with 4 parts of RNase free water, preamplified 5 µL of the diluted cDNA with the Qiagen miScript preAMP PCR Kit, and then diluted the preamplified cDNA 5-fold before qPCR detection (see the Supplemental Methods file in the online Data Supplement for full methodologic details of the reverse-transcription and preamplification steps).
MEASUREMENT OF miRNAs
miRNA profiling was performed with miScript miRNA PCR array human miRNome (384-well plate) from Qiagen. For subsequent evaluations of candidate miRNAs, we used custom Qiagen 384-well plates with specific primer probes. A SYBR Green–based qPCR was performed with 2 µL cDNA in a 10-µL reaction volume in the Applied Biosystems 7900HT Fast Real-Time PCR System instrument, as follows: 15 min at 95 °C and 40 cycles of 15 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C).
All qPCR experiments were performed in 384-well plates, with a set of technical controls included on each plate [Caenorhabditis elagans miR-39 (C.el-miR-39), which was used as the spike-in control, and miRTC, which is a Qiagen proprietary synthetic oligonucleotide]. To assess recoveries after RNA isolation, we spiked C.el-miR-39 into the sample before the extraction process. We assessed the efficiency of reverse transcription with miRTC, which was present in the nucleic acid mixture that was reverse transcribed, preamplified, and detected by qPCR.
ANALYSIS OF miRNAs in URINE AND STATISTICAL ANALYSIS
The results from the qPCR reaction with SDS 2.4 software (Applied Biosystems) were obtained as threshold cycle (Cq) values with automatic threshold and baseline values. All Cq values >32 were excluded. The Cq of a reference miRNA was subtracted from the Cq of the target to get the ∆Cq value for each sample. To derive a relative concentration for an miRNA, we linearized the value by computing 2–∆q. Data are expressed as the mean (SE), and statistical significance was evaluated with the Student t-test.
For KIM-1 and each validated miRNA biomarker, we constructed univariate, bivariate, and multivariate logistic regression models to assess their predictive capabilities. We used a 5-fold cross-validated area under the ROC curve (AUC) to evaluate the prediction performance of different models (see the Supplemental Methods file in the online Data Supplement for full details of the statistical analysis).
Results
IDENTIFYING CANDIDATE miRNAs DIFFERENTIALLY PRESENT IN PATIENTS WITH AKI COMPARED WITH HEALTHY CONTROLS
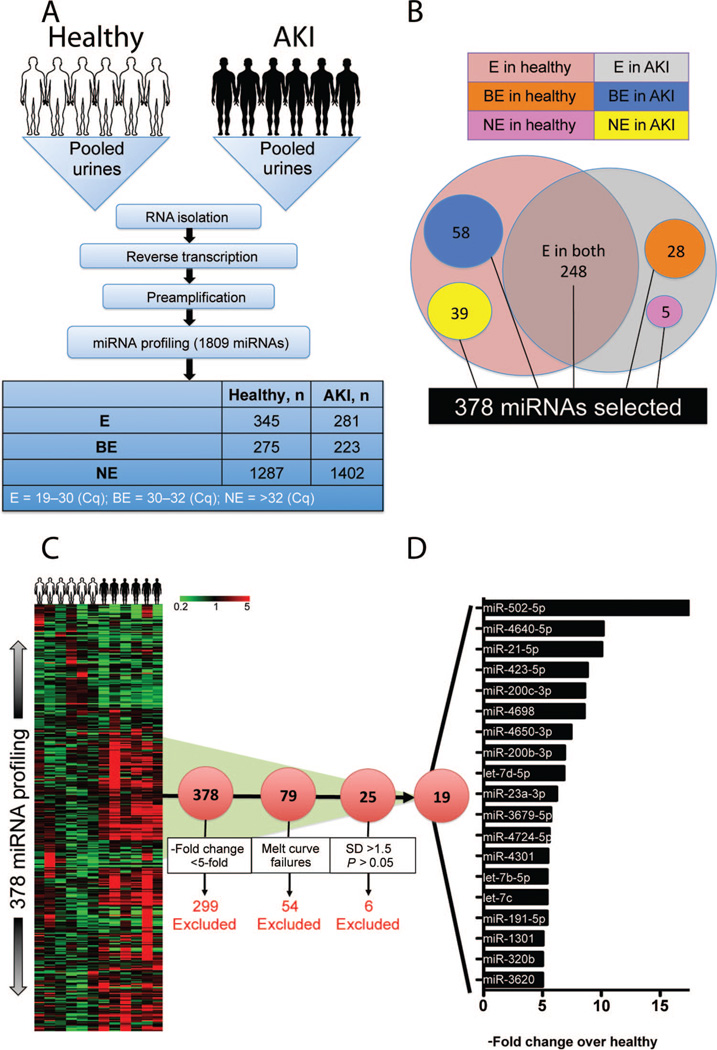
To evaluate the fraction of miRNAs detectable in human urine, we profiled urine samples pooled from 6 ICU patients with AKI (defined as a 100% increase in SCr above baseline) and from 6 healthy controls (Fig. 1A) with an miRNA PCR array (miRBase version 18, containing 1809 miRNAs; Qiagen). Each miRNA was placed into one of 3 categories according to the state of its gene expression in urine, as indicated by the Cq value obtained after qPCR (see Table 1 in the online Data Supplement): 19–30, expressed (E); 30–32, borderline expressed (BE); >32, not expressed (NE). More than 1200 miRNAs were not present in either the healthy pool or the AKI pool, 345 miRNAs were present in the pool from healthy control individuals, 281 miRNAs were present in the AKI patients, and 248 miRNAs were present in the urine of both groups (Fig. 1B). From the results of this pilot experiment, we chose 378 miRNAs for the next phase of our study. They fell into the following categories: E in both healthy individuals and AKI patients; E in healthy individuals and BE in AKI patients; E in healthy individuals and NE in AKI patients; BE in healthy individuals and E in AKI patients; NE in healthy individuals and E in AKI patients (Fig. 1B). We used a custom PCR array to analyze the relative concentrations of the 378 candidate miRNAs in the urine of the 6 AKI patients and the 6 healthy volunteers individually (Fig. 1C). The mean of the 9 miRNAs that remained unchanged across all samples was used as a “reference” for normalization (see Table 2 in the online Data Supplement). Of the 378 miRNAs investigated, 79 were upregulated >5-fold in patients with AKI; however, we excluded 54 miRNAs for melting-curve failures and 6 miRNAs that had SD values >1.5 within the group and P values >0.05 (see Table 3 in the online Data Supplement). Thus, of 19 miRNAs that passed the criteria (Fig. 1D), we chose the top 7 miRNA candidates (miR-502, miR-4640, miR-21, miR-423, miR-200c, miR-4698, and miR-4650) that were differentially present in the urine samples of AKI patients and healthy individuals to confirm the results with a larger cohort. We also chose 3 “reference” miRNAs (miR-307, miR-1287, and miR-489) that showed no difference between AKI patients and healthy individuals (see Table 2 in the online Data Supplement). We included miR-502—despite its SD of 1.76—because this miRNA showed the highest degree of upregulation (fold change, 17.42; see Table 3 in the online Data Supplement).
Fig. 1. Human urinary miRNome profiling identifies 19 miRNAs significantly upregulated in patients with kidney injury, compared with healthy volunteers.
(A), Work flow for processing pooled urines from 6 healthy volunteers and 6 AKI patients and then screening 1809 miRNAs. Each miRNA was designated according to the Cq value as expressed (E) (Cq, 19–30), borderline expressed (BE) (Cq, 30–32), or not expressed (NE) (Cq, >32). (B), Selection of 378 miRNAs with Cq values between 19 and 30 in at least one of the pooled samples. (C), Heat map showing the fold changes in values of the 378 miRNAs in 6 healthy volunteers and 6 AKI patients individually, compared with the mean of the healthy volunteers. (D), A panel of the top 19 miRNAs with proper melting curves, with miRNA values >5-fold higher (P< 0.05) in AKI patients than in healthy individuals and with SD values <1.5 within the group.
RELATIVE URINARY LEVELS OF miR-21, miR-200C, AND miR-423 CAN DISTINGUISH BETWEEN PATIENTS WITH AND WITHOUT AKI
We confirmed the relative levels of miRNAs in the urine in 4 cohorts: healthy volunteers (n = 74), ICU patients without kidney disease (n = 23), ICU patients with AKI (n = 71), and kidney transplantation patients with biopsy-proven tubular injury in the transplanted kidney (n = 27). Demographic data and clinical information for the participants are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of the human participantsa
| AKI, ICU (n = 71)b,c |
AKI, ATId on biopsy (n = 27) |
No AKI, ICU (n = 23) |
Healthy (n = 74) |
|
|---|---|---|---|---|
| Age, years | 61.1 (13.1) | 54.6 (12.9) | 55.2 (16.8) | 36.3 (10.7) |
| Caucasian, % | 75 | 70 | 35 | 77 |
| Female sex, % | 38 | 37 | 52 | 47 |
| Baseline SCr, mg/dLe,f | 1.0 (0.5) | NAg | 1.0 (0.4) | NAh |
| Peak SCr, mg/dLf | 3.6 (1.9) | 2.6 (1.7) | 1.3 (1.4) | NAh |
Data are presented as the mean (SD) as indicated.
The severity of AKI was determined according to KDIGO (Kidney Disease, Improving Global Outcomes) staging: stage I,1%; stage II, 35%; stage III, 64%. Dialysis was required for 33% of patients in KDIGO stage III.
Causes of AKI were ischemia (24%), cardiac surgery (13%), nephrotoxins (6%), sepsis (44%), volume depletion (3%), pancreatitis (1%), hepatorenal syndrome (1%), catastrophic antiphosolipid syndrome (1%), and multifactorial (7%).
ATI, acute tubular injury; NA, not applicable.
Baseline SCr values were not available for 2 patients with no AKI and for 9 patients with AKI in the ICU.
SCr concentrations are converted to the SI unit of measure as follows: 1 mg/dL = 88.4 µmol/L
Baseline SCr values were not available for patients with kidney biopsies.
SCr was not measured in healthy volunteers.
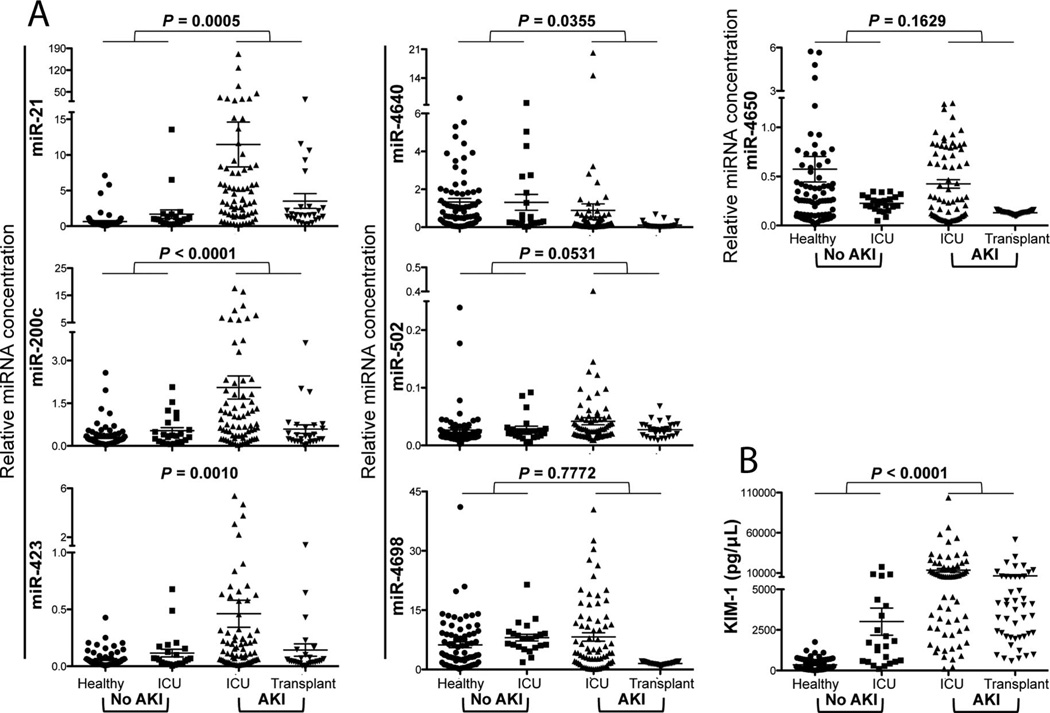
The mean relative levels of miR-21, miR-200c, and miR-423 in the urine were significantly higher in patients with AKI than in the patients without AKI, whereas miR-4640 was significantly lower in the AKI group than in the non-AKI group (Fig. 2). The degree of upregulation was approximately 10-fold for miR-21 (P = 0.0005), approximately 4.5-fold for miR-200c (P < 0.0001), and approximately 5-fold for miR-423 (P = 0.0010). miR-4640 was downregulated approximately 2-fold (P = 0.0355) (Fig. 2). Urinary KIM-1, an advanced protein biomarker for assessing kidney damage, was measured to confirm the injury. KIM-1 was increased significantly in patients with AKI compared with patients without AKI (P < 0.0001) (Fig. 2). miR-21, miR-200c, miR-423, and miR-4650 were able to distinguish between ICU patients with and patients without evidence of AKI (P < 0.01; see Table 4 in the online Data Supplement), and the data for the healthy volunteers and the ICU control group were significantly different with respect to both miR-21 (P = 0.014) and miR-4650 (P = 0.01). The ICU AKI group and transplant biopsy AKI group had significantly different (P < 0.05) relative levels of all 7 candidate miRNAs, a result indicating the different etiologies of the injuries in these 2 groups (see Table 4 in the online Data Supplement).
Fig. 2. Differences in the relative levels for the 7 candidate urinary miRNAs in the patients with and the patients without AKI.
(A), qPCR analysis for the 7 miRNAs in healthy volunteers (n = 74), patients in the ICU without AKI (n = 23), patients in the ICU with AKI (n = 71), and patients with biopsy-proven tubular injury (n = 27). The relative level of each miRNA was calculated according to the formula 2–∆Cq, and miR-1287 was used as the internal normalizer. (B), The urinary KIM-1 concentration was normalized to the urinary creatinine concentration as a biomarker for differentiating healthy and AKI patients. P values as evaluated with the Student t-test are shown, and the data in each group are summarized as the mean and SE.
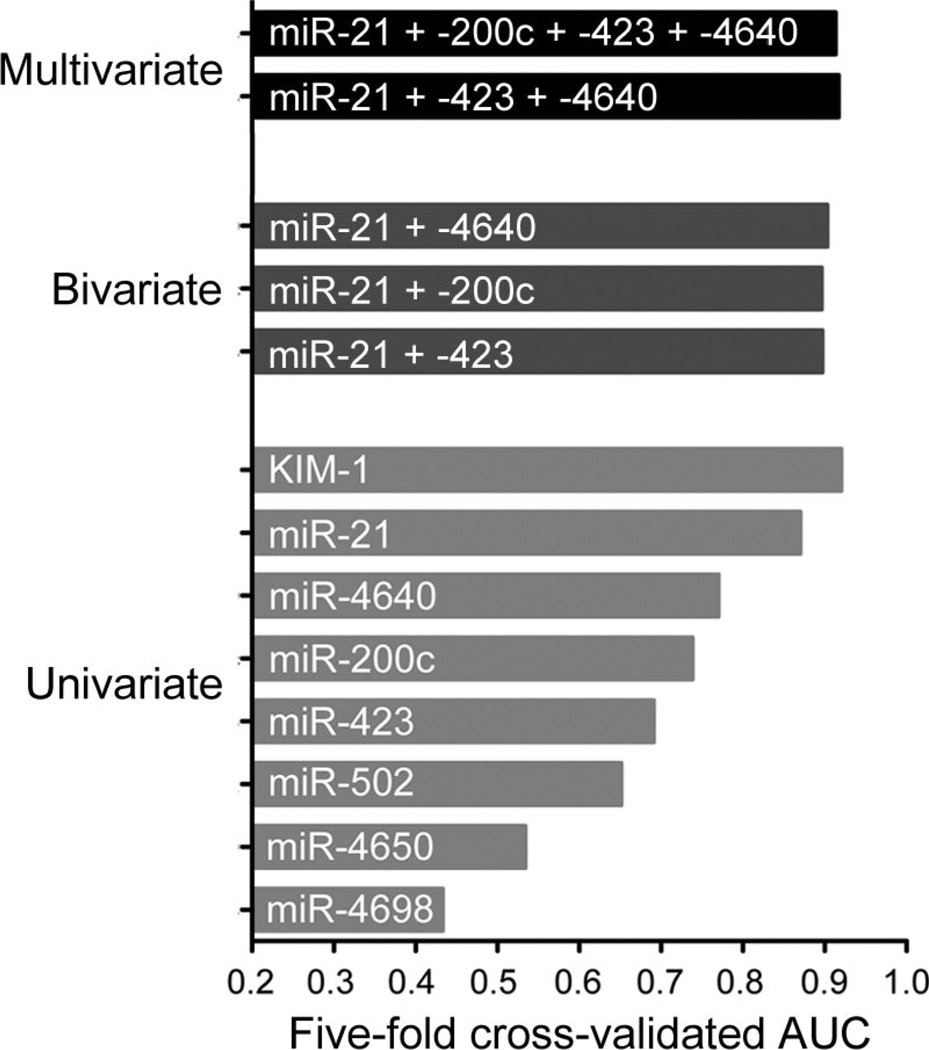
The diagnostic performance calculated with the 5-fold cross-validated AUC for KIM-1 and the individual miRNAs in the univariate model were as follows: miR-21, 0.87 (95% CI, 0.82–0.92); miR-200c, 0.74 (95% CI, 0.66–0.81); miR-423, 0.69 (95% CI, 0.61– 0.77); miR-4640, 0.77 (95% CI, 0.43–0.84); miR-502, 0.65 (95% CI, 0.45–0.72); miR-4650, 0.53 (95% CI, 0.43–0.63); miR-4698, 0.43 (95% CI, 0.38–0.68); KIM-1, 0.92 (95% CI, 0.89–0.97); see Fig. 3. In the bivariate analysis, the 3 other miRNAs that were capable of significantly (P < 0.05) distinguishing between AKI and non-AKI samples with the Student t-test were combined one at a time with the miRNA with the highest AUC in the univariate model (miR-21). The results obtained with this bivariate model increased the crossvalidated AUC values marginally: to 0.90 (95% CI, 0.80–0.93) for miR-21 plus miR-200c, to 0.90 (95% CI, 0.83–0.95) for miR-21 plus miR-423, and to 0.90 (95% CI, 0.84–0.94) for miR-21 plus miR-4640 (Fig. 3). Using the stepwise variable selection method for multivariate analysis for the full data set, we selected miR-21, miR-423, and miR-4640 and calculated their combined cross-validated AUC (0.92; 95% CI, 0.81– 0.95). We selected the panel of 4 biomarkers (miR-21, miR-200c, miR-423, and miR-4640) that were significantly different in AKI patients compared with non-AKI controls (P < 0.05) according to the Student t-test and obtained an AUC of 0.91 (95% CI, 0.86–0.92) for these 4 miRNAs (Fig. 3). No two-way interaction terms were selected, in accordance with the results of lasso logistic regression.
Fig. 3. AUC values for potential biomarkers of AKI.
AUC values were computed with a univariate logistic regression model for the 7 candidate miRNAs and KIM-1 individually, with a bivariate logistic regression model combining miR-21 with another miRNA (miR-423, miR-200c, and miR-4640, one at a time), and by multivariate logistic regression with a 2 different miRNA panels (miR-21 plus miR-423 plus miR-4640, and miR-21 plus miR-200c plus miR-423 plus miR-4640).
MINIMAL INTERASSAY AND INTRAASSAY IMPRECISION
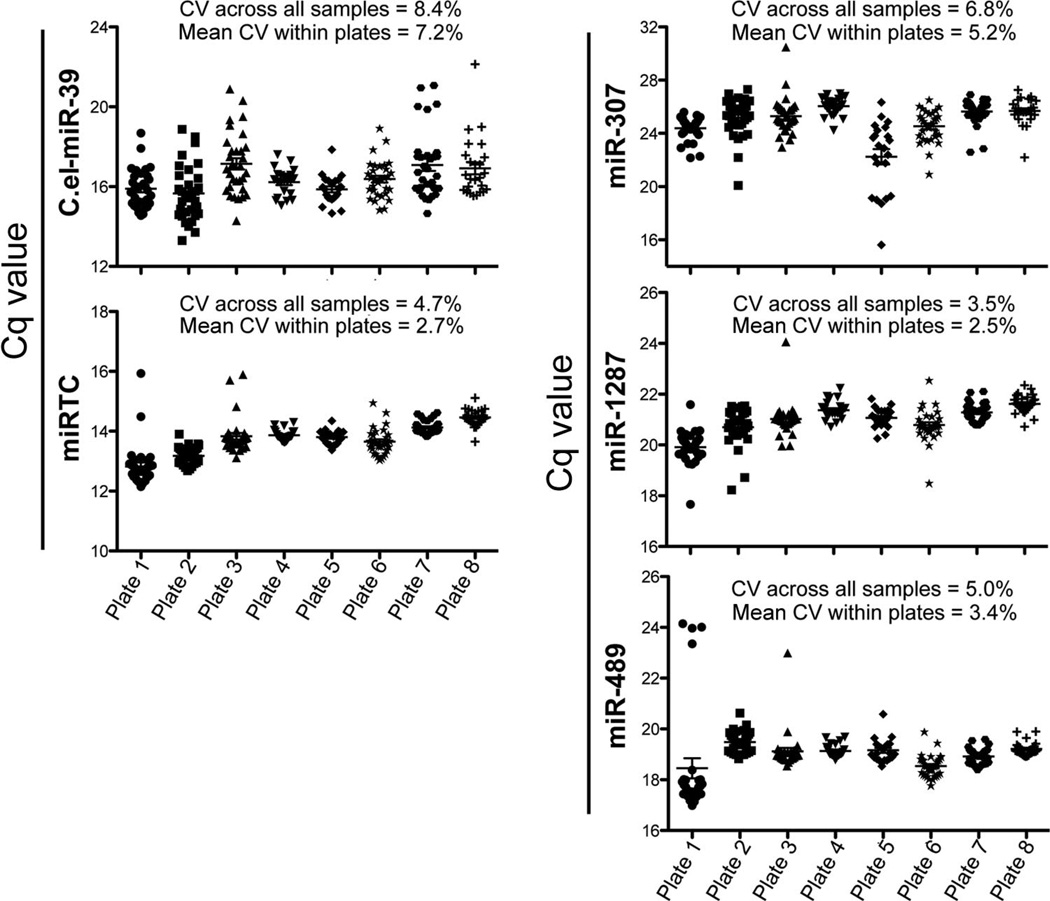
The entire experiment was performed with eight, 384-well custom plates, each of which contained probes for the 7 candidate miRNAs, C.el-miR-39, the miRTC oligonucleotide, and 3 normalizers (miR-307, miR-1287, and miR-489). All samples demonstrated good RNA recoveries (as measured by C.el-miR-39), with a mean intraassay CV of 7.2% and a mean interassay CV of 8.4% across all 224 samples and 8 plates (Fig. 4A). We used miRTC to measure the efficiency of reverse transcription. The mean intraplate CV of 2.7% and the mean interplate CV of 4.7% indicated that the reverse-transcription reactions occurred at equivalent efficiencies across all samples (Fig. 4A). No template-control reactions and reactions without the reverse transcriptase were performed to determine whether the reverse-transcription and PCR reactions worked properly. The Cq values for these reactions were either undetermined or 5–10 cycles higher than the observed Cq for a urine sample from a healthy individual. These results indicate the robustness of the results (see Table 5 in the online Data Supplement). The melting temperatures of each miRNA were checked for all 244 samples to ensure the specificity of the probe and the mean (SD) melting temperature across all samples (see Table 5 in the online Data Supplement).
Fig. 4. Inter- and intraplate variation for technical controls and normalizers.
(A), Cq values of the 2 technical controls plotted for all 8 plates (244 samples) used in the confirmation experiments. C.el-miR-39 was used to evaluate RNA recoveries, and miRTC values were used to indicate reverse-transcription efficiency. (B), Cq values for the 3 normalizers (miR-307, miR-1287, and miR-489) tested for all 244 samples. Plate 1, 30 samples; plate 2, 32 samples; plate 3, 31 samples; plate 4, 22 samples; plate 5, 22 samples; plate 6, 31 samples; plate 7, 31 samples; plate 8, 25 samples. The interplate efficiency is represented by the CV across all samples, and the mean CV within each plate represents the intraplate efficiency. The data on the plots are summarized by the mean and SE.
Of the 9 invariant miRNAs used to normalize the 378 miRNAs in the 6 healthy controls and the 6 patients with AKI, we tested 3 miRNAs that had low SDs (see Table 2 in the online Data Supplement) as potential normalizers. miR-1287 was the most consistent miRNA (mean intraplate CV, 2.5%; mean interplate CV, 3.5%), compared with miR-307 and miR-489 (Fig. 4B). Hence, we used miR-1287 as the endogenous normalizer to calculate the respective relative concentrations for the candidate miRNAs in Fig. 2.
CHARACTERIZING TEMPORAL VARIATION IN THE CANDIDATE URINARY miRNAs
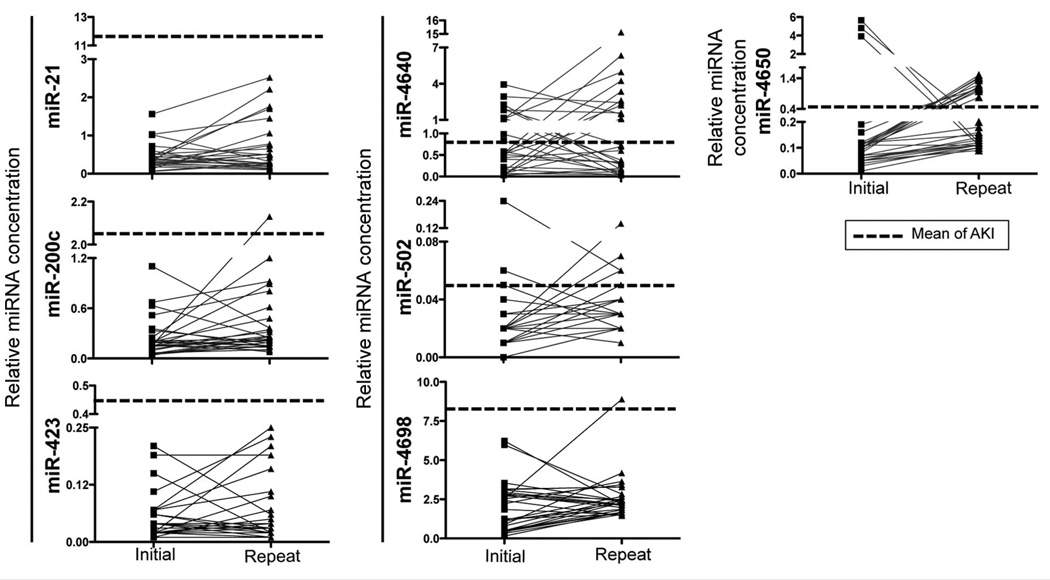
To evaluate the endogenous variation of the 7 candidate miRNAs over time, we calculated their relative concentrations in urine samples obtained from 29 healthy volunteers at 2 time points. The interclass correlation coefficients calculated for the biomarkers were as follows: miR-21, 0.02; miR-200c, 0.11; miR-423, 0.33; miR-4640, 0.06; miR-4650, 0; miR-4698, 0.15; and miR-502, 0.15. Although these values suggest that the relative concentrations of miR-21, miR-200c, and miR-423 vary considerably over time, the miRNA signals in patients with AKI were significantly higher than the background noise created by the temporal variation (Fig. 5). This high variation in the healthy population highlights the fact that these miRNAs play an important role, not only in the diseased condition but also in normal human physiology.
Fig. 5. Biological variation in the values for the miRNA biomarker candidates over time for healthy volunteers.
Values for the 7 miRNAs measured in urine samples from healthy individuals taken at 2 different times and showing the biological variation (n = 29). Dashed line represents the mean value of each marker in AKI.
Discussion
Kidney disease is receiving increased global attention owing to its substantially increased prevalence and its high mortality rates (21). Although the traditional biomarkers of AKI, such as blood urea nitrogen and SCr, have been a part of clinical practice for decades, they have limitations with respect to sensitivity, specificity, and timeliness of diagnosis (22, 23). In this study of the first clinical evaluation of 1809 miRNAs in the urine, we have identified 4 miRNAs—miR-21 (P = 0.0005), miR-200c (P < 0.0001), miR-423 (P = 0.001), and miR-4640 (P = 0.0355)—that are capable of differentiating between patients with AKI (n = 98) and patients without AKI (n = 97). We thoroughly evaluated the recovery of miRNAs extracted from urine samples with spiked-in C.el-miR-39 and investigated the efficiency of reverse transcription with the miRTC oligonucleotide. We demonstrated the CV to be <9% across all 224 samples. Of the 9 invariant miRNAs used to normalize the 378 miRNAs, we identified miR-1287 as the most consistent in all the 224 samples tested. Therefore, we used this miRNA, with its mean intraplate CV of 2.5% and interplate CV of 3.5%, as a “reference.”
Since their discovery in human plasma, extracellular miRNAs have been explored as biomarkers of injury (11). Urinary miRNAs have been documented to be unaffected by multiple freeze–thaw cycles and to be stable at room temperature for 24 h (17), properties that make them attractive tools for diagnostics. miRNA profiling has established the urinary miRNA miR-210 as a marker of acute cellular rejection in transplantation patients (17) and as a predictor of survival in critical cases requiring renal-replacement therapy (24). Argyropoulos et al. showed that the urinary miRNA spectrum in diabetic nephropathy patients varies across the various stages of untreated diabetic nephropathy (16); however, no studies to date have described the urinary miRNome in the clinical setting of AKI.
Of the 4 miRNAs identified in this study, miR-21 has already been studied extensively, and its roles in development, immunity, cancers, and different pathologic conditions have been well documented (25). In cancer studies, miR-21 has been established as an oncomiR because of its overproduction in 6 different solid tumors, and it has been proposed as a biomarker of malignancy (25). Experiments with mouse models of fibrosis of the heart (26), lung (27), and kidney (28) have provided strong evidence of a profibrotic role for miR-21. Wild-type mice develop evidently more kidney interstitial fibrosis than mice lacking miR-21, and administration of anti–miR-21 oligonucleotides to wild-type mice can ameliorate this condition. Fibrosis is typically the final stage of various chronic diseases that include chronic kidney disease, and it eventually leads to impairment of organ function and death. Whether urinary concentrations of miR-21 predict the progression of AKI to chronic kidney disease has yet to be investigated, but we previously demonstrated that miR-21 is upregulated in rodents under similar conditions of AKI (18), a result establishing that miR-21 upregulation is mechanistically well conserved across species. An alignment of the sequences of miR-21, miR-200c, and miR-423 from 7 different jawedvertebrate species shows that their mature sequences are well conserved across all of these organisms (see Fig. 1 in the online Data Supplement).
The function of the miR-200 family is dependent not only on the tissue type but also on its differentiation state, properties that render its role in cancer very complex (29). Specifically, miR-200c has been suggested as a potential circulating biomarker for pancreatic (30) and ovarian (31) cancers and has been found to be downregulated in renal cell carcinoma in the tissue.
miR-423 has been most widely studied as a biomarker of heart failure (32). Other conditions in which miR-423 deregulation occurs include pulmonary fibrosis (33), lupus nephritis (34), obesity, and Alzheimer disease (35).
miR-4640 was identified in 2011 by next-generation sequencing of small RNAs extracted from breast cancer tissue (36). As a relatively new miRNA under study, miR-4640 has yet to be investigated in the context of physiology and disease. The present study is the first to document a decrease in miR-4640 in human urine after AKI and its potential as a diagnostic biomarker of kidney injury.
All studies of miRNAs in the urine have mainly focused on their roles as noninvasive biomarkers, but there has been little experimental demonstration of the origins of the miRNAs found in urine. Neal et al. reported a lack of correspondence between the plasma and urine miRNA spectra in patients with chronic kidney disease, which prompted the speculation that the kidneys are not involved in the clearance of circulating miRNAs (37). This concept thus implies the possibility that the nonplasma portion of urinary miRNAs has its origin in kidney cells and thus may truly reflect the actual condition of the kidneys. The renal origin of miR-21 can by postulated not only because of the lack of correlation between plasma and urine concentrations (37) but also because miR-21 has been shown to be produced in the renal tubulointerstitial area in mice (38). To our knowledge, no study to date has explored the origin of miR-200c, miR-423, and miR-4640 found in urine.
Normalization has been one of the biggest challenges that previous studies have faced in analyzing extracellular miRNAs, and the use of small nucleolar RNAs (RNU6B, RNU48, and RNU44) or the spike-in of synthetic miRNAs are strategies that have been used to overcome this challenge. These small nucleolar RNAs have been observed to be as variable as miRNAs in cancer samples, however, and their use for normalization is likely to skew any data on relative miRNA concentrations (39). For the present study, we selected 3 invariant miRNAs from our profiling data, and we chose miR-1287 as the internal control because it had the smallest CV among the 244 samples we tested. Additional studies performed at different centers are needed to corroborate this observation, and such studies should include samples from patients with different AKI etiologies.
In summary, we have described a screen of the entire miRNome in human urine and have identified miR-21, miR-200c, miR-423, and miR-4640 as sensitive (and noninvasive) indicators of kidney damage. These miRNAs are potentially attractive biomarker candidates, not only as diagnostic and prognostic indicators of disease but also in the drug-development process—all the way from preclinical safety screening, clinical trial management, and patient stratification to postmarket surveillance. We expect that future work that examines the predictive potential, reproducibility, and robustness of these miRNAs in large multicenter clinical studies will be helpful in determining the value of extracellular miRNAs as biomarkers.
Supplementary Material
Acknowledgments
The authors thank Dr.P.De Jager and the Pheno Genetics Study for access to urine samples from healthy volunteers.
Research Funding: V.S. Vaidya, National Institute of Ennvironmental Health Sciences Outstanding New Environmental Scientist Award (ES017543); J.L. Koyner, National Institute of Diabetes and Kidney Diseases (K23 DK081616); R.A. Betensky, Harvard CTSA Biostatistics Program.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: AKI, acute kidney injury; ICU, intensive care unit; SCr, serum creatinine; miRNA, microRNA; qPCR, quantitative real-time PCR; KIM-1, kidney injury molecule 1; C.el-miR-39, Caenorhabditis elagans miR-39; Cq, threshold cycle; AUC, area under the ROC curve; E, expressed; BE, borderline expressed; NE, not expressed.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: R.A. Betensky, Harvard School of Public Health and Massachusetts General Hospital; V.S. Vaidya, Harvard Program in Therapeutic Science.
Consultant or Advisory Role: R.A. Betensky, NIH, Institute of Medicine, Cowen Research, GLG Consulting, Guidepoint Global Consulting, Edison Pharmaceuticals, ISI Group, Bank of America Merrill Lynch, Novartis Pharmaceuticals, Summer Street Research, Invus Services, Maine Medical Center, Medpanel Consulting, Simulconsult, DeMatteo Monness Consulting, and Columbia University.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: None declared.
References
- 1.Li PK, Burdmann EA, Mehta RL. Acute kidney injury: global health alert. Kidney Int. 2013;83:372–376. doi: 10.1038/ki.2012.427. [DOI] [PubMed] [Google Scholar]
- 2.Star RA. Treatment of acute renal failure. Kidney Int. 1998;54:1817–1831. doi: 10.1046/j.1523-1755.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- 3.Koyner JL, Parikh CR. Clinical utility of biomarkers of AKI in cardiac surgery and critical illness. Clin J Am Soc Nephrol. 2013;8:1034–1042. doi: 10.2215/CJN.05150512. [DOI] [PubMed] [Google Scholar]
- 4.Ho J, Dart A, Rigatto C. Proteomics in acute kidney injury— current status and future promise. [(Accessed October 2013)];Pediatr Nephrol. 2013 Apr 18; doi: 10.1007/s00467-013-2415-x. http://link.springer.com/article/10.1007%2Fs00467-013-2415-x. [DOI] [PubMed]
- 5.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Bonventre JV. Diagnosis of acute kidney injury: from classic parameters to new biomarkers. Contrib Nephrol. 2007;156:213–219. doi: 10.1159/000102086. [DOI] [PubMed] [Google Scholar]
- 7.Dennen P, Parikh CR. Biomarkers of acute kidney injury: Can we replace serum creatinine? Clin Nephrol. 2007;68:269–278. doi: 10.5414/cnp68269. [DOI] [PubMed] [Google Scholar]
- 8.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 9.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 10.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 13.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 14.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Greenhaw J, Shi Q, Su Z, Qian F, Davis K, et al. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol Sci. 2012;125:335–344. doi: 10.1093/toxsci/kfr321. [DOI] [PubMed] [Google Scholar]
- 16.Argyropoulos C, Wang K, McClarty S, Huang D, Bernardo J, Ellis D, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8:e54662. doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11:2221–2227. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 18.Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, et al. Expression, circulation, and excretion profile of microRNA-21, −155, and −18a following acute kidney injury. Toxicol Sci. 2012;129:256–267. doi: 10.1093/toxsci/kfs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya VS, Ozer JS, Dieterle F, Collings FB, Ramirez V, Troth S, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28:478–485. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaidya VS, Waikar SS, Ferguson MA, Collings FB, Sunderland K, Gioules C, et al. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin Transl Sci. 2008;1:200–208. doi: 10.1111/j.1752-8062.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu R, McCulloch C, Dudley R, Lo L, Hsu CY. Temporal changes in incidence of dialysis-requiring AKI. J Am Soc Nephrol. 2013;24:37–42. doi: 10.1681/ASN.2012080800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonventre JV, Vaidya VS, Schmouder R, Feig P, Dieterle F. Next-generation biomarkers for detecting kidney toxicity. Nat Biotechnol. 2010;28:436–440. doi: 10.1038/nbt0510-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaidya VS, Ferguson MA, Bonventre JV. Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48:463–493. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6:1540–1546. doi: 10.2215/CJN.00430111. [DOI] [PubMed] [Google Scholar]
- 25.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang H, Zhang C, Ban T, Liu Y, Mei L, Piao X, et al. A novel reciprocal loop between microRNA-21 and TGFβRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. 2012;44:2152–2160. doi: 10.1016/j.biocel.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003205. 121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res. 2010;3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14:147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 33.Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, et al. A microRNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS One. 2011;6:e21253. doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya M, Bandyopadhyay S. Studying the differential co-expression of microRNAs reveals significant role of white matter in early Alzheimer’s progression. Mol Biosyst. 2013;9:457–466. doi: 10.1039/c2mb25434d. [DOI] [PubMed] [Google Scholar]
- 36.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 37.Neal CS, Michael MZ, Pimlott LK, Yong TY, Li JY, Gleadle JM. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26:3794–3802. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 38.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol. 2011;22:1668–1681. doi: 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–1177. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.