I. Introduction: The Cell Cycle in Zebrafish
In multicellular organisms, the cell cycle is a fundamental feature of cellular physiology that is critical for normal development, organogenesis and tissue homeostasis. Reflecting this central role, the molecular pathways that regulate cell division in eukaryotes are evolutionarily conserved. Aberrations in the control of the cell cycle are common in degenerative diseases and cancer. Therefore, analysis of the cell cycle in non-mammalian organisms can illuminate the processes underlying human development and disease. Forward genetic screens in yeast and Drosophila have been invaluable for gene discovery and have made important contributions to understanding pathways regulating cell proliferation. Importantly, it has been found that the human orthologs of some genes identified in these organisms are misexpressed in human tumors (Hariharan and Haber 2003). Zebrafish have proven to be an excellent model of early vertebrate development (Driever, Solnica-Krezel et al. 1996; Haffter, Granato et al. 1996) and also of a wide variety of human diseases such as cancer, anemia, cardiovascular defects, neuromuscular conditions, kidney disease and host-pathogen interaction, to name a few examples (Ackermann and Paw 2003; Bassett and Currie 2003; Lambrechts and Carmeliet 2004; Miller and Neely 2004; Drummond 2005; Goessling, North et al. 2007; Hsu, Wen et al. 2007).
The particular advantages that make zebrafish ideal for developmental embryology—including external fertilization of oocytes, transparent embryos, and rapid embryonic development—also provide the opportunity to study early cell divisions, tissue-specific cellular proliferation and, more broadly, the role of cell-cycle genes in development and disease. A number of methods and markers have been successfully applied to investigate the cell cycle in zebrafish embryos, including video microscopy (Kane, Warga et al. 1992; Kane 1999), histone-GFP fusions (Pauls, Geldmacher-Voss et al. 2001), BrdU labeling (Link, Fadool et al. 2000; Baye and Link 2007), Proliferating Cell Nuclear Antigen (PCNA) RNA and protein expression (Wullimann and Knipp 2000; Koudijs, den Broeder et al. 2005), phosphohistone H3 (pH3) immunohistochemistry (Shepard, Amatruda et al. 2005), and minichromosome manintenance protein expression (Ryu and Driever 2006).
Studies of the developing zebrafish embryo have revealed similarities to the early cell divisions of other vertebrates, such as Xenopus. In the zebrafish, the first seven cell divisions are synchronous and cycle rapidly between DNA replication (S phase) and mitosis (M phase) without the intervening gap phases, G1 and G2 (Kimmel, Ballard et al. 1995). The midblastula transition (MBT) ensues during the 10th cell division, which is approximately 3 h post fertilization. MBT is accompanied by loss of division synchrony, increased cell cycle duration, activation of zygotic transcription, and the onset of cellular motility (Kane and Kimmel 1993). Embryonic cells first exhibit a G1 gap phase between the M and S phases during MBT. Recently, Dalle Nogare et al. demonstrated that during cycles 11–13, embryonic cells acquire a G2 phase in a transcription-independent fashion, through inhibition of Cdk1 and its activating phosphatase, Cdc25a (Dalle Nogare, Pauerstein et al. 2008).
Further understanding of cell cycle regulation in zebrafish embryos was obtained by studying their responses to various cell cycle inhibitors, including aphidocolin, hydroxyurea, etoposide, camptothecin, and nocodazole (Ikegami, Rivera-Bennetts et al. 1997; Ikegami, Zhang et al. 1997; Ikegami, Hunter et al. 1999). Exposure to these agents after MBT induces cell cycle arrest, sometimes accompanied by initiation of an apoptotic program. However, prior to MBT, the embryonic cells continue to divide, often with deleterious effects, after exposure to cell cycle inhibitors. These studies indicate that zebrafish embryos do possess cell cycle checkpoints, but they are not functional until after MBT.
Later developmental stages of zebrafish embryogenesis provide the opportunity to study the cell cycle in distinct tissue types. Studies of cell cycle regulation in older embryos (10–36 h post fertilization) have focused on the developing eyes and central nervous system. Lineage analysis of CNS progenitor cells revealed a correlation between morphogenesis and cell cycle number, implying that the nervous system development may be at least partially regulated by the cell cycle (Kimmel, Warga et al. 1994). Whereas most developing vertebrate embryos exhibit a constant lengthening of the cell cycle duration throughout development, meticulous analysis of cell number in the developing zebrafish retina revealed a surprising mechanism of modulated cell cycle control. Li and colleagues (Li, Hu et al. 2000) reported that the retinal cell cycle duration temporarily slows between 16–24 hours post fertilization (hpf), followed by an abrupt change to more rapid cell divisions.
Several studies have elucidated a role for the zebrafish cell-cycle machinery in tissue differentiation during development and in the regenerative response to injury. Bessa et al. found that Meis1, a marker of the eye primordium, promotes G1-S progression and a block of differentiation in the zebrafish eye through regulation of Cyclin D1 and c-myc expression (Bessa, Tavares et al. 2008). Fischer and co-workers showed that loss of caf1b in zebrafish (by mutation or MO injection) leads to an S-phase arrest and eventual apoptosis that can be rescued by p53 deficiency. However, loss of caf1b also leads to a block in differentiation in tissues that express caf1b, implicating caf1b in the switch from proliferation to differentiation (Fischer, Prykhozhij et al. 2007). The effect of loss of early mitotic inhibitor 1 (emi1) on somite formation was evaluated by Zhang et al. These authors found that cell-cycle progression was required for proper somite morphogenesis, but not for formation of the segmentation clock (Zhang, Kendrick et al. 2008). The role of the cell cycle in regeneration has also been assessed. Certain traumas result in loss of hair cell precursors, which results in deafness in vertebrates. Hernández and co-workers used BrdU labeling and transgenic GFP reporter lines to study hair cell regeneration, identifying proliferation- dependent and –independent mechanisms of hair cell renewal (Hernandez, Olivari et al. 2007).
Forward-genetic screens
Several groups have carried out forward-genetic screens to identify mutations that alter cell proliferation in embryos. Shepard et al. used phosphohistone H3 (pH3) as a marker of cell proliferation in a 2-generation haploid genetic screen. They identified seven mutant lines with different alterations in pH3 immunoreactivity. At least two of these lines demonstrate aneuploidy and increased cancer susceptibility as heterozygotes (Shepard, Amatruda et al. 2005; Shepard, Amatruda et al. 2007). Using a similar screening strategy, Pfaff et al. identified a further set of genes required for cell proliferation mutants, among which was SIL ( for Scl-Interrupting Locus), which was identified as a novel, vertebrate-specific regulator of mitotic spindle assembly (Pfaff, Straub et al. 2007). Koudijs and co-workers used Proliferating Cell Nuclear Antigen (PCNA) expression in the CNS as a readout to identify new mutations in repressors of the Hedgehog (Hh) signaling pathway (Koudijs, den Broeder et al. 2005). Finally, another screen for genes that control eye growth uncovered two zebrafish lines mutant for the anaphase-promoting complex/cyclosome (APC/C) (Wehman, Staub et al. 2006). Loss of APC/C results in a loss of mitotic progression and apoptosis; in this study, co-labeling with BrdU and pH3 revealed cells undergoing mitotic catastrophe.
In this chapter, we provide protocols to characterize the various phases of cell division in zebrafish embryos, and protocols to detect DNA damage, senescence and cell death. Assays discussed in this chapter include: DNA content analysis by flow cytometry, whole-mount embryonic antibody staining, mitotic spindle analysis, BrdU incorporation, cell death analysis, and in situ hybridization with cell cycle regulatory genes. Each assay targets different phases of the cell cycle and in total create a detailed picture of zebrafish embryo cell proliferation. Although our studies have focused on embryonic assays for cell cycle characterization, it is likely that these protocols can be modified to study adult tissues. These protocols can be applied to a variety of experiments, such as characterization of the cell cycle phenotypes of mutants or the analysis of RNA overexpression and morpholino knockdown of cell cycle regulatory genes. Furthermore, the genetic tractability of the zebrafish system (Patton and Zon 2001) makes it an excellent organism in which to pursue forward genetic screens for mutations or chemical screens for novel compounds that alter cell division using one or more of these cell cycle assays.
II. Zebrafish Embryo Cell Cycle Protocols1
A. Analysis of Cell Proliferation and Mitosis
1. DNA Content Analysis
A profile of the cell cycle in disaggregated zebrafish embryos or adult tissue can be obtained through DNA content analysis. In this technique, cells are stained with a dye that fluoresces upon DNA binding, such as Hoechst 33342 or propidium iodide. The intensity of fluorescence is proportional to the amount of DNA in each cell (Krishan 1975). Analysis by fluorescence activated cell sorting (FACS) generates a histogram showing the proportion of cells that have an unreplicated complement of DNA (G1 phase), those that have a fully replicated complement of DNA (G2 or M phase) and those that have an intermediate amount of DNA (S phase).
Protocol:
All steps are performed on ice except for the dechorionation (step 1) and RNAse incubation (step 9).
Dechorionate embryos and wash with E3. Analysis of single embryos is possible, though in practice we typically pool approximately 40 embryos/tube.
Disaggregate embryos (using small pellet pestle) in 500 µl of DMEM (or other tissue culture medium) + 10% fetal calf serum in a matching homogenizing tube.
Bring volume to 1 mL with DMEM/serum and remove aggregates by passing cell suspension sequentially through 105 µm mesh and 40 µm mesh.
Count a sample using a hemocytometer.
Place volume containing at least 2×106 cells in a 15 mL conical tube, and bring volume to 5 mL with 1X PBS.
Spin at 1200 rpm for 10 min. at 4°C.
Carefully aspirate off liquid and gently resuspend cell pellet in 2 mL Propidium Iodide solution.
Add 2 µg of DNAse-free RNAse (Roche). This step is necessary to remove double-stranded RNA, which binds propidium iodide.
Incubate in the dark at room temperature for 30 min.
Place samples on ice and analyze on FACS machine.
Note: Samples can also be fixed in Ethanol, allowing multiple samples or time points to be collected for subsequent analysis.
Harvest cells and prepare single cell suspension in DMEM/serum as above, steps 1–4.
Wash cells in PBS and resuspend at 1–2 × 106 cells/mL.
To 1 mL cells in a 15 mL polypropylene, V-bottom tube add 3 mL ice-cold absolute EtOH. To avoid clumping, add the ethanol dropwise while vortexing the sample.
Fix cells for at least 1 hour at 4 °C. Cells may be stored for several weeks at −20 °C before undergoing PI staining.
Wash cells twice in 1 xPBS. We typically increase the speed of centrifugation to 2500 rpm because the cells do not pellet as readily after EtOH fixation.
Resuspend the pellet in 1 mL Propidium Iodide solution. Add 2 µg of DNAse-free RNAse (Roche) and incubate 3 hr at 4 °C.
Place samples on ice and analyze on FACS machine.
2. Whole Mount Immunohistochemistry with Mitotic Marker Phosphohistone H3
Histone H3 phosphorylation is considered to be a crucial event for the onset of mitosis and this antibody has been widely used in Drosophila and mammalian cell lines as a mitotic marker (Hendzel, Wei et al. 1997). Two members of the Aurora/AIK kinase family, Aurora A and Aurora B, phosphorylate histone H3 at the serine 10 residue (Chadee, Hendzel et al. 1999; Crosio, Fimia et al. 2002). Increased serine 10 phosphorylation of histone H3 has been seen in transformed fibroblasts (Chadee, Hendzel et al. 1999), suggesting that this antibody could make an excellent marker for cell proliferation in the zebrafish as well as detecting cell cycle mutations that may result in transformed phenotypes. In zebrafish, the phospho-histone H3 antibody (pH3) stains mitotic cells throughout the embryo (Figure 1A). pH3 staining in developing organs like the nervous system increases as they undergo proliferation during distinct developmental stages.
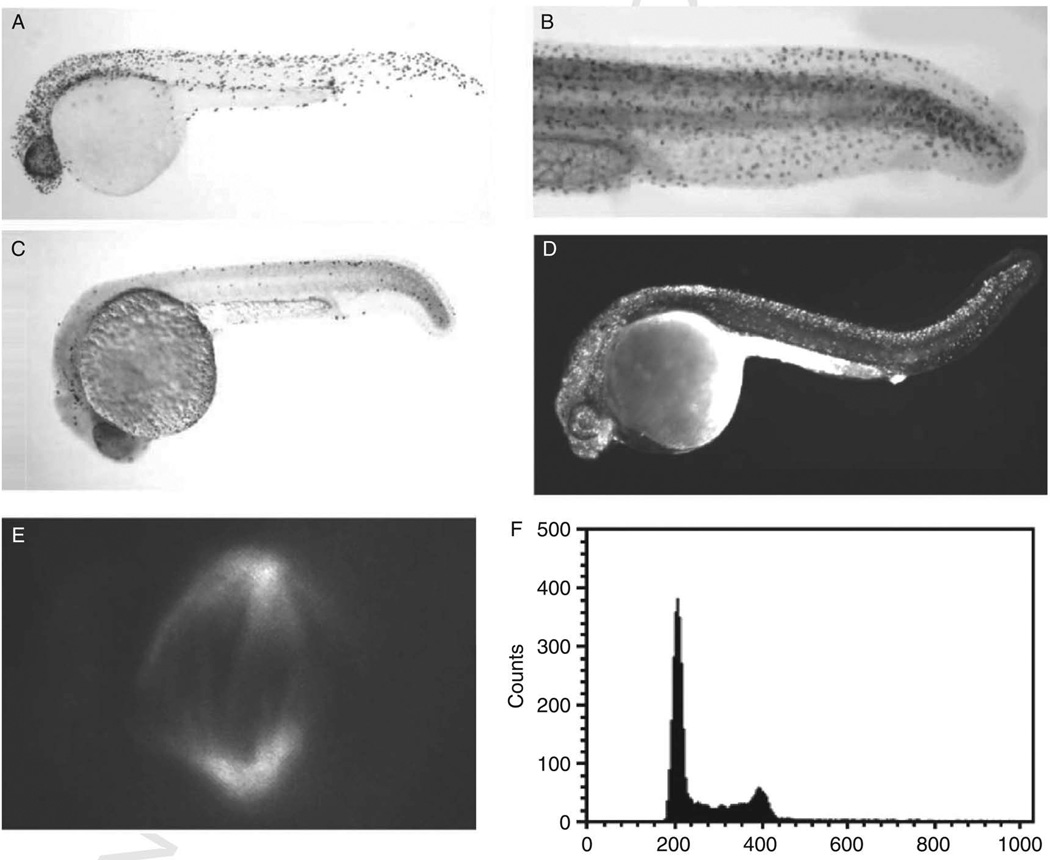
Figure 1. Useful techniques for the study of the cell cycle, proliferation or apoptosis as shown in zebrafish embryos.
A, Antibody staining against phosphorylated histone H3 in wild-type 24hpf embryos. B, BrdU incorporation to mark cells in S phase in the tail of a 28hpf wild-type embryo, C and D, Apoptotic cells can be visualized by TUNEL (C; wild-type 24hpf embryo) or acridine orange (D, 24 hpf crash&burn mutant embryo). E, Anti-alpha tubulin can be used to examine mitotic spindle formation. F, DNA content analysis shows the population of embryonic cells present in all phases of the cell cycle.
Protocol:
Fix embryos overnight at 4 °C in 4% paraformaldehyde (PFA).
Permeabilize embryos for 7 min. in −20 °C acetone.
Wash embryos in H2O followed by two 5-min. washes in PBST.
Incubate for 30 min. at room temperature in block.
Incubate overnight at 4 °C in rabbit anti-phosphohistone H3 at a concentration of 1.33 mg/mL in block. Two different sources of antibody have been used: Santa Cruz Biotechnology and an anti-phosphopeptide polyclonal antibody to the sequence (ARKS[PO4]TGGKAPRKQLC) made and affinity purified by Genemed Synthesis.
Wash 4 × 15 min. in PBST.
Incubate 2 hr. at room temperature in horseradish peroxidase-conjugated secondary goat anti-rabbit IgG (Jackson Immunoresearch) at a concentration of 3 µg/mL in block.
Wash 4 × 15 min. in PBST.
Develop in the dark for 3–5 min. at room temperature in diaminobenzidine (DAB) solution (0.67 mg/mL DAB in 15mL of PBST to which 12 µl of 30% H2O2 has been added).
Wash in PBST and store embryos at 4 °C. in PFA.
3. Mitotic Spindle/Centrosome Detection
Study of the mitotic spindle and centrosomes is an important step in understanding mutants with cell cycle defects, particularly those whose phenotypes appear to be related to problems in mitosis. Genomic instability is one of the main alterations seen in human cancers and such unequal segregation of chromosomes can be caused by problems in mitotic spindle formation or centrosome number (Kramer, Neben et al. 2002). In this protocol, anti-a-tubulin labels the mitotic spindle, anti-g-tubulin the centrosome, and DAPI the DNA.
Protocol:
Fix embryos in PFA for 4 hr. at room temperature.
Dehydrate in methanol at −20°C for at least 30 min.
Rehydrate embryos in graded methanol:PBST series (3:1, 1:1, 1:3) for 5 min. each
Wash 1× 5 min. in PBST.
Place in −20°C acetone for 7 min.
Wash 3 × 5 min. in PBST.
Incubate 1 hr. at room temperature in block.
Incubate in monoclonal mouse α-tubulin antibody (Sigma) at a concentration of 1:500 and in polyclonal rabbit γ-tubulin antibody (Sigma) at a concentration of 1:1000 (both diluted in block) at 4°C overnight.
Wash 4× 15 min. in PBST.
Incubate in rhodamine-conjugated goat anti-mouse secondary (Molecular Probes) at 1:600 dilution and fluorescein-conjugated goat anti-rabbit secondary (Jackson Immunoresearch) at 1:600 dilution for 2 hr. room temperature.
Wash 2 × 15 min. in PBST.
Include a 1:500 dilution of 100 µM DAPI during the third wash to stain DNA.
Wash 2 × 15 min. in PBST
For observation by epifluorescence microscopy, embryos are mounted on glass slides with VectaShield mounting media (Vector Labs) and coverslipped. To permit the specimen to lie flat, is helpful to remove the yolk using forceps or a tungsten needle. Alternatively, for embryos >18 hpf, the tail can be cut off from the embryo and mounted on the slide.
4. BrdU Incorporation
5-bromo-2-deoxyuridine (BrdU) is a nucleoside analog that is specifically incorporated into DNA during S-phase (Meyn, Hewitt et al. 1973) and can subsequently be detected with an anti-BrdU specific antibody. This technique has been used to label replicating cells in zebrafish embryos (Larison and Bremiller 1990) and adults (Rowlerson, Radaelli et al. 1997). The following protocol is designed to label a fraction of proliferating cells in zebrafish embryos, to allow comparison of the replication fraction of different embryos (Figure 1 B). If the embryos are chased for varying amounts of time after the BrdU pulse, then fixed and stained for both BrdU and pH3 (section B), the transit of cells from S phase into G2/M can be assessed.. This is useful in analyzing mutants with mitotic phenotypes.
Protocol
Dechorionate embryos and chill 15 min. on ice in E3.
Prepare cold 10 mM BrdU/15% Dimethylsulfoxide in E3 and chill on ice. Place embryos in BrdU solution and incubate 20 min. on ice to allow uptake of BrdU.
Change into warm E3 and incubate exactly 5 min., 28.5 C. Note: longer incubation times will result in more cells being labeled.
Fix 2 hr., room temperature in PFA. Longer fixation may decrease the staining.
Transfer to methanol at −20 °C. overnight. All subsequent steps are performed at room temperature unless otherwise noted.
Rehydrate in graded methanol:PBST series (3:1, 1:1, 1:3) for 5 min. each.
Wash 2× in PBST, 5 min.
Digest embryos in 10 µg/mL Proteinase K, 10 min.
Wash PBST. Refix in PFA for not more than 20 min.
Wash quickly 3 × in H20, then 2× in 2N HCl.
Incubate 1 hr. in 2N HCl. This step denatures the labeled DNA to expose the BrdU epitope.
Remove the 2N HCl solution from the embryos and neutralize in 0.1 M borate buffer, pH 8.5, 20 mins., room temparature.
Rinse several times in PBST. Block for 30 min. in BrdU blocking solution.
Incubate in monoclonal anti-BrdU antibody at a dilution of 1:100 in BrdU block for 2 h at room temperature or overnight at 4 °C. (If carrying out simultaneous BrdU/pH3 staining, add the primary anti-pH3 antibody as described in Section B, except that BrdU block is used).
Wash 5× 10 min in PBST
Incubate 2 h room temperature with horseradish peroxidase or fluorophore-conjugated anti-mouse secondary antibody. (For simultaneous BrdU/pH3 stain, add a fluorescent anti-rabbit antibody as well.)
Wash 5× 10 min in PBST. If using fluorescent secondary, mount embryos as described in Section C, step 14.
If using HRP-conjugated secondary antibody, develop in the dark for 3–5 min. at room temperature in diaminobenzidine (DAB) solution (0.67 mg/mL DAB in 15mL of PBST to which 12 µL of 30% H2O2 has been added). When staining is complete, wash 3 × 5 mins. in PBST, then fix in PFA.
B. Analysis of DNA Damage, Senescence and Apoptosis
1. COMET Assay
The Comet Assay, also known as the Single Cell Microgel Electrophoresis (SCGE) assay, is a highly sensitive technique that is used to detect DNA damage at the single cell level(Singh, McCoy et al. 1988). Cells are embedded into a thin agarose gel, through which a current is run allowing for migration of DNA. Smaller fragments of DNA, resulting from DNA damage, will travel more quickly and appear as a tail to the nucleus “comet head”. The comets can be visualized using a nuclear stain, such as SYBR green, and visualized under a fluorescent microscope. The following protocol is designed to isolate cells from zebrafish embryos and detect any kind of DNA damage. Variations of this technique allows for specific detection of double-stranded break
Protocol
Dechorionate embryos and wash with E3. Typically, about 25–50 embryos are used.
Disaggregate embryos (using small pellet pestle) in 500 µl of DMEM (or other tissue culture medium) + 10% fetal calf serum or lamb serum in a matching homogenizing tube.
Bring volume to 1mL with DMEM/serum.
Count the samples using a hemocytometer.
Spin down cells at 3000 RPM and re-suspend in PBS to a concentration of 1×105 cells/mL.
Combine 10uL of cells with 90uL of molten LMAgarose (Trevigen) pre-warmed to 37°C. Pipette 75uL of the cell agarose mixture onto a CometSlide (Trevigen) pre-warmed to 37°C.
Incubate the slide flat at 4°C for 30 min. in the dark to allow the gel to solidify.
Immerse the slide in Lysis solution (Trevigen) containing 9% DMSO. After this point, it is very important to maintain the slide in low-light conditions.
Dry off the slide, and immerse it in alkaline solution for 30 min.
Prepare a large horizontal electrophoresis apparatus by filling the chamber with fresh alkaline electrophoresis buffer and adjusting the volume of the alkaline electrophoresis buffer such that the current is 300mA when the voltage is set to 25V. Additionally, the chamber should be in prepared and used in a 4°C room.
Place the slide in the electrophoresis apparatus. Run for 30 min. at 4°C in the dark.
Dry off the slide. Rinse by dipping in ddH20.
Incubate the slide in 70% EtOH for 5 min. at RT in the dark.
Air dry the slide for 1 hr.
Pipette 50uL of SYBR Green Staining Solution (Trevigen) onto the microgel on the slide.
View the slide using fluorescence microscopy under a fluorescein filter (figure 3).
Comets can be analyzed using CometScore by Tritek Corp or another similar software program.
2. Detection of Senescence-Associated Beta Galactosidase
The study of cellular senescence was initiated by Hayflick and Moorhead and was first described in 1961(Hayflick and Moorhead 1961). Cellular senescence pertains to the cessation of cell replication and certain morphological and transcriptional changes that occur when cells permanently cease dividing. Although it is unclear whether the events that occur during in vitro cellular senescence also occur during organismal aging (Masoro 2006; Hayflick 2007) studies have revealed strong connections between cellular senescence, cancer, and age-related diseases (Campisi 2005). Cellular senescence most likely arose evolutionarily as a mechanism to defend against tumorigenesis (Shay and Roninson 2004). When a cell is afflicted by stress that may result in transformation (such as oxidative stress, DNA damage or overepxpression of oncogenes) tumor-suppressor genes such as p53 may force the cell to undergo senescence-induced arrest. Arrested cells are functional but are not a risk for tumor initiation. Senescence also occurs as the ends of chromosomes, the telomeres, shorten. During reach replication cycle, if no active telomerase is present (Bodnar, Ouellette et al. 1998), the telomeres shorten, leading eventually to critically short telomeres which may interfere with gene expression and genomic stability (Shay and Wright 2006). Normal cells senesce before telomeres shorten to the point of causing genomic instability, therefore instilling a counting mechanism which confirms Hayflick’s observation in 1961 (Shay and Wright 2006).
Senescent cells lose sensitivity to mitogens or growth factors, repress cell cycle genes, such as cdk2, and become insensitive to apoptotic signals. Morphological changes occur resulting in an enlarged shape and flattened body (Ben-Porath and Weinberg 2005), as well as expression of unique markers, many of unknown function, such as β-galactosidase activity at pH 6.0 (Dimri, Lee et al. 1995). Kishi and co-workers have used senescence-associated as β-galactosidase staining in several studies to characterize senescence in normal and mutant zebrafish embryos and during aging of zebrafish adults (Kishi, Uchiyama et al. 2003; Kishi 2004; Tsai, Tucci et al. 2007; Kishi, Bayliss et al. 2008).
Protocol
We have used the Senescence-Associated Beta-Galacotsidase Detection Kit from Sigma (CS 0030). The following protocol adapts the manufacturer’s instructions specifically for use with zebrafish embryos, and is kindly provided by Jenny Richardson and Dr. Elizabeth Patton, Edinburgh Cancer Research Centre:
Dechorionate embryos and add 1.5mL of 1X fixation buffer (prepared from 10X Sigma Senescence Fixation Buffer). Incubate overnight at 4 °C.
Wash embryos 4 times in 1X PBS, 1 hr. each wash.
Make up the Senescence Staining Mixture as per the manufacturer’s protocol. Add 1mL to embryos and incubate for 24 hours at 37 °C.
Wash embryos 3 times in 1X PBS, 10 mins. each wash.
Embryos can be stored at 4 °C in 1xPBS and 0.1% NaN3 or in 70% glycerol at 4 °C.
An alternative protocol was described by Dr. Shuji Kishi and co-workers in a recent paper describing a senescence-based genetic screen (Kishi, Bayliss et al. 2008). The following protocol is adapted from Kishi, S et al., “The identification of zebrafish mutants showing alterations in senescence-associated biomarkers”, PLoS Genet. 2008 Aug 15;4(8):e1000152:
Fix embryos or adult zebrafish in 4% paraformaldehyde in 1xPBS at 4°C (for 3 days in adults and overnight in embryos).
Wash ×3 times for 1 hr. in PBS-pH 7.4 and for a further 1 h in PBS-pH 6.0 at 4°C.
Stain the samples overnight at 37°C in 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, and 1 mg/ml X-gal in PBS adjusted to pH 6.0.
3. Apoptosis Detection by TUNEL Staining
Apoptosis is a form of programmed cell death that eliminates damaged or unneeded cells. It is controlled by multiple signaling pathways that mediate responses to growth, survival or death signals. Cell cycle checkpoint controls are linked to apoptotic cascades and these connections can be compromised in diseases, including cancer. The defining characteristics of apoptosis are membrane blebbing, cell shrinkage, nuclear condensation, segmentation and division into apoptotic bodies that are phagocytosed (Wyllie 1987). The DNA strand breaks that occur during apoptosis can be detected by enzymatically labeling the free ends with modified nucleotides which can then be detected with antibodies (Gavrieli, Sherman et al. 1992).
Protocol:
Embryos are fixed overnight at 4 °C in PFA.
Wash in PBS and transfer to methanol for 30 min. at −20°C.
Rehydrate embryos in a graded methanol:PBST series (3:1, 1:1, 1:3) for 5 min. each.
Wash 1× 5 min. in PBST.
Digest embryos in proteinase K (10 µg/mL) at room temperature (1 min. for embryos younger than 16 hpf, 2 min. for embryos older than 16 hpf).
Wash twice in PBST.
Postfix in PFA for 20 min. room temperature.
Wash 5× 5 min. in PBST.
Postfix for 10 min. at −20°C with prechilled Ethanol:Acetic Acid 2:1.
Wash 3× 5 min. in PBST at room temperature
Incubate for 1 hour at room temperature in 75 ul equilibration buffer (TdT - ApopTag Peroxidase In Situ Apoptosis Detection Kit from Serologics Corporation)
Add small volume of working strength TdT (reaction buffer and TdT at a ratio of 2:1 plus 0.3% Triton) (Serologics Corporation).
Incubate overnight at 37°C.
Stop reaction by washing in working strength stop/wash buffer (1 mL concentrated buffer from Serologics Kit with 34 mL water) for 3–4 hr. at 37°C.
Wash 3 × 5 min. in PBST.
Block with 2 mg/mL BSA, 5% sheep serum in PBST for 1 hr. at room temperature.
Incubate in anti-digoxigenin peroxidase antibody included in kit (full strength).
Wash 4 × 30 min. PBST at room temperature.
Develop in the dark for 5 min. at room temperature in diaminobenzidine (DAB) solution (0.67 mg/mL in 15mL of PBST) and 12 µl 30% H2O2.
Wash in PBST and store embryos at 4 °C in PFA.
4. Apoptosis Detection by Acridine Orange
Another method of apoptotic cell detection that can be performed on living embryos is acridine orange staining. The basis of this method is that the ATP-dependent lysosomal proton pump is preserved in apoptotic but not necrotic cells therefore apoptotic cells will take up the acridine orange dye whereas living or necrotic cells will not (Darzynkiewicz, Bruno et al. 1992). This method is useful for identifying mutants based on an apoptotic phenotype in order to further characterize them in living assays.
Protocol:
Live dechorionated embryos are incubated in a 2 µg/mL solution of acridine orange (Sigma) in 1X E3 for 30 min. at room temperature
Embryos are washed 5× quickly in E3 and then visualized on a stereo dissecting microscope equipped for FITC epifluorescence.
C. In situ hybridization
RNA expression analysis by in situ hybridization of antisense probes in whole-mount zebrafish embryos is a commonly used technique to localize expression of developmental regulatory genes. While the technique is not exceptionally quantitative, it can reveal stark differences in gene expression. More quantitative analysis of gene expression, such as Northern blotting, RT-PCR, or real-time PCR do not permit the examination of alterations in tissue-specific expression or an expression pattern.
Cell division is a highly controlled process that involves regulation at both the transcriptional and post-translational stages. Cyclins are a class of proteins that play critical roles in guiding cells through the G1, S, G2, and M phases of the cell cycle by regulating the activity of the cyclin-dependent kinases. The name cyclin alludes to the fact that their expression levels oscillate between peaks and nadirs that are coordinated with particular phases of the cell cycle (reviewed in (Murray 2004). The tightly regulated expression of these important cell cycle genes incorporates transcriptional, translational, and posttranslational controls. Many genes involved in cell cycle regulation are specifically expressed during the cell cycle phase in which they act.
Zebrafish orthologs of cell cycle regulatory genes such as PCNA and cyclins have been found to possess similar expression patterns throughout the proliferative tissues of developing zebrafish embryos (C. Thisse, B. Thisse, unpublished and www.zfin.org). In situ hybridization for cell cycle regulatory genes can be performed using previously published in situ hybridization protocols (Thisse, Thisse et al. 1993; Thisse, Thisse et al. 1994; Jowett 1999).
III. Screening for Chemical Suppressors of Zebrafish Cell Cycle Mutants
Another way to probe the cell cycle is via chemical agents. Chemical screens could identify novel compounds that are useful tools for studying the cell cycle. Furthermore, mutations in cell cycle genes are commonly found in human cancer. Given the need to improve upon current cancer therapy, one approach is to identify small molecule suppressors that bypass the consequences of specific cell cycle gene mutations. Akin to the use of genetic modifier screens to identify secondary mutations that enhance or suppress a primary defect (St Johnston 2002), chemical suppressor screens would directly identify small molecules that rescue a genetic phenotype. If the phenotype is disease-related, such compounds might represent lead therapeutic agents.
Zebrafish have recently been utilized in chemical screens to identify compounds that perturb specific aspects of development (Peterson, Link et al. 2000; Khersonsky, Jung et al. 2003; Peterson, Shaw et al. 2004; den Hertog 2005; Bayliss, Bellavance et al. 2006; Anderson, Bartlett et al. 2007). The zebrafish system offers several advantages for chemical screens, providing information on tissue specificity and toxicity, and accounting for compound activation via drug metabolism. Furthermore, cells are not transformed and are in their normal physiological milieu of cell-cell and cell-extracellular matrix interactions. Murphey and co-workers carried out a high-throughput chemical screen to detect small molecules capable of perturbing the cell cycle during zebrafish development, identifying several compounds that were not previously detected in cell-based screens of the same library (Murphey, Stern et al. 2006). As another application of this technique, Stern et al. screened a 16,000-compound library to identify small molecules capable of suppressing the cell proliferation defect in the crash&burn cell cycle mutant (Stern, Murphey et al. 2005). This technology could easily be applied to other cell cycle mutants and could be modified to use cell cycle assays other than pH3 staining. In addition, such chemical suppressor screens could be applied to any zebrafish model of human disease (Dooley and Zon 2000). For these reasons, we provide a detailed protocol below.
The following protocol can be repeated weekly giving a throughput of over 1000 compounds per week for a recessive lethal mutation. In the case of homozygous viable mutants, the throughput could be improved by using fewer embryos (3–5) per well in 96-well plates.
Protocol:
For a chemical screen, large numbers of embryos at approximately the same developmental stage need to be generated. Set up 100 heterozygote pairwise matings with fish separated by a divider. The next morning, remove the divider, allow the fish to mate, and collect the embryos.
- Dilute chemicals into screening medium. The screen is conducted in 48-well plates with a volume of 300 ml per well. Individual chemicals could be added to each well, but to improve throughput, we devised a matrix pooling strategy: The chemical library (courtesy of the Institute of Chemistry and Cell Biology, Harvard Medical School) was arrayed in 384 well plates with the last 4 columns empty, thus containing 320 compounds per plate. Given this plate geometry, 8 by 10 matrix pools were created. A hit detected in both a horizontal and a vertical pool identified the individual compound.
- Transfer 80 ml of screening medium to each well of four 384-well plates using a TECAN liquid handling robot.
- Pin transfer 1ml of each compound (arrayed at 5 mg/mL in DMSO) into each well of screening medium by performing 10 transfers with a 100 nl 384-pin array for each of the four 384-well plates (total of 320 × 4 = 1280 compounds).
- Pooling was performed with a TECAN liquid handling robot by pipeting the diluted chemicals from the 384-well plates to 48-well plates. For vertical pools, 30 mL was transferred from each of 8 wells plus an additional 60 mL of screening medium to bring the total volume to 300 ml. For horizontal pools, 30 mL was transferred from each of 10 wells.
- Aliquot embryos to the 48-well plates at 50% epiboly.
- Prior to aliquoting embryos to wells, examine them under a dissecting microscope and discard all dead, delayed or deformed embryos.
- Pool embryos in a single 100mm tissue culture dish or a 50 mL conical tube.
- Decant the embryo medium, and remove as much liquid from the embryo suspension as possible with a transfer pipet. Pressing the transfer pipet tip to the bottom of the tube or dish allows most liquid to be removed without aspirating the embryos.
- Add approximately 20 embryos to each well by scooping them with a small chemical weighing spatula. With 20 embryos per well and a Mendelian recessive inheritance, there is a 0.3% chance of a well having no mutants. Since a hit requires detection in both a horizontal and a vertical pool, each with 20 embryos, the false-positive rate for identification of complete suppressors is 0.001%.
Place 48-well plates into an incubator at 28.5°C.
One to two hr. later, clean out any dead embryos from each well using a long glass Pasteur pipet bent at a 90 degree angle.
Incubate at 28.5°C overnight.
Dechorionate embryos by adding 150 mL of a 5 mg/mL pronase solution to each well. After 10 min, gently shake plates until embryos come out of the chorions.
Using a transfer pipet fitted with a 10 mL tip, remove as much of the pronase / chemical mixture as possible from each well.
Rinse the embryos once in fresh embryo medium and remove as in step 8.
Add 500 ml of PFA to each well.
Parafilm the edges of the plates to prevent evaporation and fix at 4°C at least overnight but not longer than a week.
Using a transfer pipet, move embryos to 48 well staining grids made of acetone resistant plastic with a wire mesh bottom.
Perform pH3 staining protocol by placing staining grids into 11 by 8.5 cm reservoirs containing 20–30 mL of the appropriate solution. To change solutions, the grid can be lifted out of one reservoir and placed into another reservoir with the next solution. For overnight antibody incubations the reservoir should be sealed with parafilm to prevent evaporation.
After staining is complete, move embryos with a transfer pipet back into 48-well plates that have been precoated with 100 mL of 1% agarose in 1× PBS. The agarose forms a meniscus that keeps embryos in the center of the well where they are easier to score.
Score for absence of mutants or for partial suppression without effect on wild types. In addition to suppressors and enhancers, one can identify compounds that affect both wild types and mutants, thus having a more general effect.
Deconvolute matrix pool to identify individual chemicals.
IV. Conclusions
Given the power of zebrafish in forward vertebrate genetics and organism-based small molecule screens, the system will nicely complement traditional model organisms for studying the cell division cycle. Many of the assays that are commonly used to probe the cell cycle in systems such as yeast, Drosophila, and mammalian cells can be used in the zebrafish. The protocols outlined in this chapter can be utilized to characterize known mutants for alterations in cell proliferation or, alternatively, can be used to screen for more cell cycle mutants. Given that zebrafish embryos are amenable to gene knockdown via antisense morpholino-modified oligonucleotides and overexpression by mRNA injection, these protocols can also be used to study cell cycle genes in the zebrafish without generating a mutant.
VI. Reagents and Supplies
| Alkaline Solution | 0.6g NaOH, 250uL 200mM EDTA pH10, to 50mL ddH20 |
| Alkaline Electrophoresis | |
| Buffer | 12g NaOH, 2mL 500mM EDTA pH 8, to 1L ddH20 |
| Anti-BrdU | Roche cat # 1170 376 |
| Block | 2% blocking reagent (Roche 1096 176), 10% fetal calf serum, 1% dimethylsulfoxide in PBST. |
| BrdU Block | 0.2% blocking reagent (Roche 1096 176), 10% fetal calf serum, 1% dimethylsulfoxide in PBST. The lower concentration of blocking reagent improves detection. |
| DAPI | 4',6-Diamidino-2-phenylindole |
| DMSO | Dimethylsulfoxide |
| E3 | 5mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4. |
| Mesh | Small Parts, inc. 105 µm mesh is cat # U-CMN-105D. 40 µm mesh is cat # U-CMN-40D. |
| PBS | Phosphate-buffered saline, pH 7.5. |
| PBST | 1X PBS with 0.1% (v/v) Tween-20 |
| Pellet pestle & tubes | Fisher, cat # K749520-0090 |
| PFA | 4% paraformaldehyde buffered with 1× PBS |
| Propidium Iodide | 0.1% Sodium Citrate, 0.05mg/mL propidium iodide, 0.0002% Triton X-100 (added fresh) |
| Sigma Senescence Fixation Buffer 10X (Catalog Number F1797) | |
| Contains 20% formaldehyde, 2% glutaraldehyde, 70.4 mM Na2HPO4, 14.7 mM KH2PO4, 1.37 M NaCl, and 26.8 mM KCl | |
| Sigma Senescence Staining Mixture | |
| (Prepare just prior to use) | |
| Mix the following for preparation of 10 ml of the Staining Mixture: | |
| 1 mL of Staining Solution 10X Buffer (Catalog Number S5818) | |
| 125 µL of Reagent B (Catalog Number R5272) | |
| 125 µL of Reagent C (Catalog Number R5147) | |
| 0.25 mL of X-gal Solution (Catalog Number X3753) | |
| 8.50 mL of ultrapure water | |
| Screening medium | E3 supplemented with 1% DMSO, 20 µM metronidazole, 50 units/mL penicillin, 50 mg/mL streptomycin, and 1mM Tris pH 7.4. |
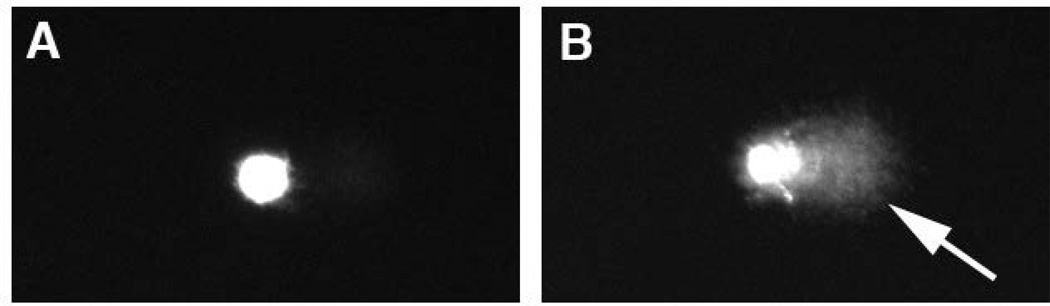
Figure 2. The COMET assay reveals double-strand DNA breaks.
Images of zebrafish embryo cells from a single-cell microgel electrophoresis experiment. A, unirradiated control cell. B, a cell after exposure to 2000R (20 Gy) gamma irradiation. The arrow indicates the comet tail, composed of fragmented DNA (Verduzco and Amatruda, unpublished).
Acknowledgments
We thank Len Zon and members of the Zon laboratory for useful discussions. Original versions of many of these protocols were worked out by Jennifer L. Shepard, Ryan Murphey, Howard M. Stern, Kathryn L. Pfaff and J.F.A. D.V. was supported by NIH Training Grant 5 T32 GM008203 and J.F.A. was supported by grants from the Lance Armstrong Foundation, the Amon G. Carter Foundation, the Welch Foundation and NIH/NCI grant 1R01CA135731.
Footnotes
Items in boldface indicate reagents and supplies listed in Section V.
References
- Ackermann GE, Paw BH. Zebrafish: a genetic model for vertebrate organogenesis and human disorders. Front Biosci. 2003;8:d1227–d1253. doi: 10.2741/1092. [DOI] [PubMed] [Google Scholar]
- Anderson C, Bartlett SJ, Gansner JM, Wilson D, He L, Gitlin JD, Kelsh RN, Dowden J. Chemical genetics suggests a critical role for lysyl oxidase in zebrafish notochord morphogenesis. Mol Biosyst. 2007;3(1):51–59. doi: 10.1039/b613673g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DI, Currie PD. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum Mol Genet. 2003;12(Spec No 2):R265–R270. doi: 10.1093/hmg/ddg279. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. The disarrayed mutation results in cell cycle and neurogenesis defects during retinal development in zebrafish. BMC Dev Biol. 2007;7:28. doi: 10.1186/1471-213X-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss PE, Bellavance KL, Whitehead GG, Abrams JM, Aegerter S, Robbins HS, Cowan DB, Keating MT, O'Reilly T, Wood JM, Roberts TM, Chan J. Chemical modulation of receptor signaling inhibits regenerative angiogenesis in adult zebrafish. Nat Chem Biol. 2006;2(5):265–273. doi: 10.1038/nchembio778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37(5):961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Bessa J, Tavares MJ, Santos J, Kikuta H, Laplante M, Becker TS, Gomez-Skarmeta JL, Casares F. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135(5):799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274(35):24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatiotemporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22(3):874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare DE, Pauerstein PT, Lane ME. G2 acquisition by transcription-independent mechanism at the zebrafish midblastula transition. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13(8):795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- den Hertog J. Chemical genetics: Drug screens in Zebrafish. Biosci Rep. 2005;25(5–6):289–297. doi: 10.1007/s10540-005-2891-8. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10(3):252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Drummond IA. Kidney development and disease in the zebrafish. J Am Soc Nephrol. 2005;16(2):299–304. doi: 10.1681/ASN.2004090754. [DOI] [PubMed] [Google Scholar]
- Fischer S, Prykhozhij S, Rau MJ, Neumann CJ. Mutation of zebrafish caf-1b results in S phase arrest, defective differentiation, and p53-mediated apoptosis during organogenesis. Cell Cycle. 2007;6(23):2962–2969. doi: 10.4161/cc.6.23.4950. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W, North TE, Zon LI. New waves of discovery: modeling cancer in zebrafish. J Clin Oncol. 2007;25(17):2473–2479. doi: 10.1200/JCO.2006.08.9821. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Haber DA. Yeast, flies, worms, and fish in the study of human disease. N Engl J Med. 2003;348(24):2457–2463. doi: 10.1056/NEJMon023158. [DOI] [PubMed] [Google Scholar]
- Hayflick L. Biological aging is no longer an unsolved problem. Ann N Y Acad Sci. 2007;1100:1–13. doi: 10.1196/annals.1395.001. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106(6):348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hernandez PP, Olivari FA, Sarrazin AF, Sandoval PC, Allende ML. Regeneration in zebrafish lateral line neuromasts: expression of the neural progenitor cell marker sox2 and proliferation-dependent and-independent mechanisms of hair cell renewal. Dev Neurobiol. 2007;67(5):637–654. doi: 10.1002/dneu.20386. [DOI] [PubMed] [Google Scholar]
- Hsu CH, Wen ZH, Lin CS, Chakraborty C. The zebrafish model: use in studying cellular mechanisms for a spectrum of clinical disease entities. Curr Neurovasc Res. 2007;4(2):111–120. doi: 10.2174/156720207780637234. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Hunter P, Yager TD. Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev Biol. 1999;209(2):409–433. doi: 10.1006/dbio.1999.9243. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Rivera-Bennetts AK, Brooker DL, Yager TD. Effect of inhibitors of DNA replication on early zebrafish embryos: evidence for coordinate activation of multiple intrinsic cell-cycle checkpoints at the mid-blastula transition. Zygote. 1997;5(2):153–175. doi: 10.1017/s0967199400003828. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Zhang J, Rivera-Bennetts AK, Yager TD. Activation of the metaphase checkpoint and an apoptosis programme in the early zebrafish embryo, by treatment with the spindle-destabilising agent nocodazole. Zygote. 1997;5(4):329–350. doi: 10.1017/s0967199400003919. [DOI] [PubMed] [Google Scholar]
- Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- Kane DA. Cell cycles and development in the embryonic zebrafish. Methods Cell Biol. 1999;59:11–26. doi: 10.1016/s0091-679x(08)61817-8. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119(2):447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kane DA, Warga RM, Kimmel CB. Mitotic domains in the early embryo of the zebrafish. Nature. 1992;360(6406):735–737. doi: 10.1038/360735a0. [DOI] [PubMed] [Google Scholar]
- Khersonsky SM, Jung DW, Kang TW, Walsh DP, Moon HS, Jo H, Jacobson EM, Shetty V, Neubert TA, Chang YT. Facilitated forward chemical genetics using a tagged triazine library and zebrafish embryo screening. J Am Chem Soc. 2003;125(39):11804–11805. doi: 10.1021/ja035334d. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development. 1994;120(2):265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- Kishi S. Functional aging and gradual senescence in zebrafish. Ann N Y Acad Sci. 2004;1019:521–526. doi: 10.1196/annals.1297.097. [DOI] [PubMed] [Google Scholar]
- Kishi S, Bayliss PE, Uchiyama J, Koshimizu E, Qi J, Nanjappa P, Imamura S, Islam A, Neuberg D, Amsterdam A, Roberts TM. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 2008;4(8):e1000152. doi: 10.1371/journal.pgen.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S, Uchiyama J, Baughman AM, Goto T, Lin MC, Tsai SB. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38(7):777–786. doi: 10.1016/s0531-5565(03)00108-6. [DOI] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Keijser A, Wienholds E, Houwing S, van Rooijen EM, Geisler R, van Eeden FJ. The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet. 2005;1(2):e19. doi: 10.1371/journal.pgen.0010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16(5):767–775. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66(1):188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Carmeliet P. Genetics in zebrafish, mice, and humans to dissect congenital heart disease: insights in the role of VEGF. Curr Top Dev Biol. 2004;62:189–224. doi: 10.1016/S0070-2153(04)62007-2. [DOI] [PubMed] [Google Scholar]
- Larison KD, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109(3):567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS., Jr Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio) Dev Dyn. 2000;219(3):391–401. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1063>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Link BA, Fadool JM, Malicki J, Dowling JE. The zebrafish young mutation acts non-cell-autonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127(10):2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Dietary restriction-induced life extension: a broadly based biological phenomenon. Biogerontology. 2006;7(3):153–155. doi: 10.1007/s10522-006-9015-0. [DOI] [PubMed] [Google Scholar]
- Meyn RE, Hewitt RR, Humphrey RM. Evaluation of S phase synchronization by analysis of DNA replication in 5-bromodeoxyuridine. Exp Cell Res. 1973;82(1):137–142. doi: 10.1016/0014-4827(73)90255-3. [DOI] [PubMed] [Google Scholar]
- Miller JD, Neely MN. Zebrafish as a model host for streptococcal pathogenesis. Acta Trop. 2004;91(1):53–68. doi: 10.1016/j.actatropica.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68(4):213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116(2):221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2(12):956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211(12):603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97(24):12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22(5):595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27(16):5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlerson A, Radaelli G, Mascarello F, Veggetti A. Regeneration of skeletal muscle in two teleost fish: Sparus aurata and Brachydanio rerio. Cell Tissue Res. 1997;289(2):311–322. doi: 10.1007/s004410050878. [DOI] [PubMed] [Google Scholar]
- Ryu S, Driever W. Minichromosome maintenance proteins as markers for proliferation zones during embryogenesis. Cell Cycle. 2006;5(11):1140–1142. doi: 10.4161/cc.5.11.2779. [DOI] [PubMed] [Google Scholar]
- Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene. 2004;23(16):2919–2933. doi: 10.1038/sj.onc.1207518. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5(7):577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- Shepard JL, Amatruda JF, Finkelstein D, Ziai J, Finley KR, Stern HM, Chiang K, Hersey C, Barut B, Freeman JL, Lee C, Glickman JN, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21(1):55–59. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J, Finley KR, Pfaff KL, Hersey C, Zhou Y, Barut B, Freedman M, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci U S A. 2005;102(37):13194–13199. doi: 10.1073/pnas.0506583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1(7):366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Halpern ME, Postlethwait JH. Goosecoid expression in neurectoderm and mesendoderm is disrupted in zebrafish cyclops gastrulas. Dev Biol. 1994;164(2):420–429. doi: 10.1006/dbio.1994.1212. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119(4):1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Tsai SB, Tucci V, Uchiyama J, Fabian NJ, Lin MC, Bayliss PE, Neuberg DS, Zhdanova IV, Kishi S. Differential effects of genotoxic stress on both concurrent body growth and gradual senescence in the adult zebrafish. Aging Cell. 2007;6(2):209–224. doi: 10.1111/j.1474-9726.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Wehman AM, Staub W, Baier H. The anaphase-promoting complex is required in both dividing and quiescent cells during zebrafish development. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.10.043. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Knipp S. Proliferation pattern changes in the zebrafish brain from embryonic through early postembryonic stages. Anat Embryol (Berl) 2000;202(5):385–400. doi: 10.1007/s004290000115. [DOI] [PubMed] [Google Scholar]
- Wyllie AH. Apoptosis: cell death in tissue regulation. J Pathol. 1987;153(4):313–316. doi: 10.1002/path.1711530404. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kendrick C, Julich D, Holley SA. Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development. 2008;135(12):2065–2070. doi: 10.1242/dev.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]