Abstract
The tyrosine O-prenyltransferase SirD in Leptosphaeria maculans catalyzes normal prenylation of the hydroxyl group in tyrosine as the first committed step in the biosynthesis of the phytotoxin sirodesmin PL. SirD also catalyzes normal N-prenylation of 4-aminophenylalanine and normal C-prenylation at C7 of tryptophan. In this study, we found that 4-mercaptophenylalanine and several derivatives of tryptophan are also substrates for prenylation by dimethylallyl diphosphate. Incubation of SirD with 4- mercaptophenylalanine gave normal S-prenylated mercaptophenylalanine. We found that incubation of the enzyme with tryptophan gave reverse prenylation at N1 in addition to the previously reported normal prenylation at C7. 4-Methyltryptophan also gave normal prenylation at C7 and reverse prenylation at N1; whereas 4-methoxytryptophan gave normal and reverse prenylation at C7, and 7-methyltryptophan gave normal prenylation at C6 and reverse prenylation at N1. The ability of SirD to prenylate at three different sites on the indole nucleus, with normal and reverse prenylation at one of the sites, is similar to behavior seen for dimethylallyltryptophan synthase. The multiple products produced by SirD suggests it and dimethylallyltryptophan synthase use a dissociative electrophilic mechanism for alkylation of amino acid substrates.
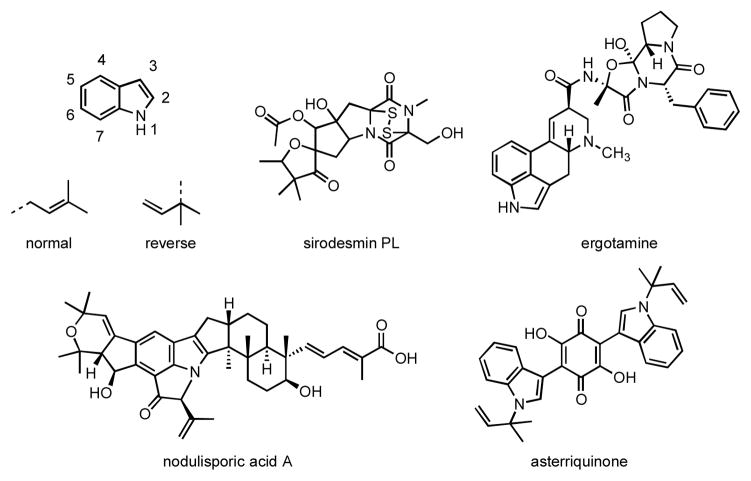
Phytopathogenic fungi produce toxic secondary metabolites that provide selective advantages against other microorganisms. Epipolythiodioxopiperazines (ETPs) and prenylated indole alkaloids constitute two classes of chemically diverse biologically active natural products.1–4 Biosynthetically, the carbon skeletons of many of these compounds are constructed from prenyl diphosphates and tyrosine, tryptophan or derivatives of tryptophan. Well-known examples of ETPs include gliotoxin and sirodesmin PL; examples of prenylated indole alkaloids include the ergot alkaloids, the asterriquinones, and the nodulisporic acids (Figure 1).
Figure 1.
Indole ring numbering, possible prenylation orientations, and representative structures of ETPs and prenylated indole alkaloids.
A family of enzymes has been identified that catalyzes alkylation of the indole ring in tryptophan by an isoprenyl diphosphate, typically dimethylallyl diphosphate (DMAPP).5–8 The isoprenyl moiety can be attached at C1′ (“normal” prenylation) or C3′ (“reverse” prenylation), further increasing structural diversity. These dimethylallyltryptophan synthases (DMATS) catalyze alkylation of the indole ring in tryptophan or tryptophan-containing dipeptides at positions N1, C2, C3, C4, C5, C6, or C7 to give a variety of naturally occurring prenylated indoles.9–16 Both normal and reverse prenylation is seen at N19,17 and at C2.10,18 Reverse prenylation is seen at C3, giving products with an α (AnaPT)11 or a β (CdpC3PT)12 configuration. Recently, FtmPT1 (a normal C2 prenyltransferase) was reported to also catalyze normal prenylation at C3 of tryptophan-containing cyclic dipeptides.19 Only normal prenylation is found at C4,13,18 C5,14 and C6.15 7-DMATS catalyzes normal prenylation at C7 of L-tryptophan;16 whereas, MpnD catalyzes reverse prenylation at C7 of an indolactam.20
4-DMATS catalyzes the normal prenylation of tryptophan at C4 as the first committed step in ergot alkaloid biosynthesis21 and has been extensively studied from Claviceps purpurea13,22,23 and Aspergillus fumigatus.18,24–27 4-DMATS (FgaPT2) from A. fumigatus consists of a 10-stranded antiparallel β-barrel surrounded by α-helices.26 This “ABBA” prenyltransferase (PT) fold has been reported for five other enzymes. FtmPT128 and CdpNPT29 catalyze normal C2 and reverse C3 alkylation of indole, respectively; whereas, NphB (formerly Orf2),30 CloQ,31 and EpzP,32 alkylate naphthalene, benzene, and phenazine rings, respectively.
The mechanism proposed for prenylation by DMATSs is a dissociative electrophilic alkylation of the indole ring by the dimethylallyl cation generated by heterolytic cleavage of the C–O bond in DMAPP, followed by loss of a proton to give the prenylated product.22,33 In the case of 4-DMATS, regiospecific alkylation at C4 of the indole ring by C1′ of the allylic cation generates an arenium intermediate, which rearomatizes by deprotonation at C4 to give dimethylallyltryptophan (DMAT).
Relaxed selectivity for the aromatic substrate is reported for several of the DMATSs. 7-DMATS prenylated both enantiomers of tryptophan, tryptophan-containing linear and cyclic dipeptides, and a variety of tryptophan derivatives, although no activity was observed when 7-DMATS was incubated with tyrosine.16,34 CTrpPT from Aspergillus oryzae DSM1147 was shown to simultaneously prenylate two different sites on the indole ring of tryptophan-containing cyclic dipeptides with normal prenylation at C7 and reverse prenylation at N1 of the indole nucleus.17 We recently reported that 4-DMATS prenylates several 4-substituted tryptophans to give products with a normal prenylation at N1, C3, C5, or C7 or a reverse prenylation at C3.35
An aromatic amino acid prenyltransferase from the pathogenic fungus Leptosphaeria maculans, dimethylallyltyrosine synthase (SirD), was recently reported.36 The enzyme likely has an ABBA PT fold and catalyzes normal O-prenylation of the hydroxyl group in tyrosine as the first pathway specific step in the biosynthesis of sirodesmin PL.37 SirD also catalyzes normal N-prenylation of the aromatic amino group in 4-aminophenylalanine and normal C-prenylation of C7 of the indole ring in L-tryptophan. 37,38 We now report a much broader substrate selectivity for SirD, which includes normal prenylation of the sulfhydryl group in 4-mercaptophenylalanine and normal prenylation of C6 and C7 and reverse prenylation of N1 and C7 of the indole ring in tryptophan derivatives. These results are consistent with a dissociative electrophilic alkylation similar to that reported for 4-DMATS.
RESULTS AND DISCUSSION
Synthesis of Tyrosine and Tryptophan Analogues
The symmetric disulfide of 4-mercapto-L-phenylalanine (1) was prepared by the palladium-catalyzed cross-coupling of N-t-Boc-4-iodo-L-phenylalanine and t-butylthiol,39 followed by simultaneous removal of the Boc and S-t-butyl protecting groups with concentrated HCl.40 Deprotection under oxidative conditions gave the disulfide, which was purified and stored. Immediately before an incubation, the disulfide was treated with β-mercaptoethanol to give 1. 4-Vinyl-L-phenylalanine (2) was synthesized according to the procedure of Yao and coworkers,41 except for changes in the protecting groups. Fmoc was used instead of Cbz and a methyl ester instead of a benzyl ester in order to avoid deprotection by catalytic hydrogenation.
Enantiomerically pure 4-methyl-L-tryptophan (3), 4-methoxy-L-tryptophan (4), and 7-methyl-L-tryptophan (5) were synthesized using the PLP-dependent tryptophan synthase β subunit,42 as previously described.35
Kinetic Studies
Michaelis-Menten kinetic parameters for tyrosine, tryptophan, and their substituted analogues are listed in Table 1. Rates (kcat) and Michaelis constants (Km) were determined from a nonlinear fit of initial velocities versus concentration using GraFit 5.0.11. The values for tyrosine, Km = 0.30 mM and kcat = 1.3 s−1, compared well with the previously reported values.37,38 Km = 1.1 mM and kcat = 2.7 × 10−2 s−1 for 4-mercaptophenylalanine. The catalytic efficiency (kcat/Km) for alkylation of 1 was <1% of the normal reaction. The ~4-fold increase in Km and ~50-fold decrease in kcat of 4-mercaptophenylalanine indicates that the replacement of the side chain oxygen with sulfur decreases substrate binding and reduces the rate of reaction. Since one would have expected that a sulfur nucleophile would facilitate capture of the putative dimethylallyl cation relative to oxygen, the decrease in kcat might result from a conformation for 1 and DMAPP in the active site that reduces the rate of C–O bond cleavage in DMAPP.
Table 1.
Kinetic parameters for SirD with tyrosine, tryptophan, and their analogues.
| Km (mM) | Kcat (s−1) | Kcat/Km (s−1 M−1) | (Kcat/Km)Rel (%) | |
|---|---|---|---|---|
| Tyr | 0.30 ± 0.04 | 1.3 ± 0.1 | 4.3 × 103 | 100 |
| 4-SH-Phe | 1.1 ± 0.2 | (2.7 ± 0.2) × 10−2 | 25 | 0.58 |
| Trp | 0.37 ± 0.06 | (9.5 ± 0.2) × 10−2 | 2.6 × 102 | 5.8 |
| 4-OCH3-Trp | 5.1 ± 2.0 | (1.3 ± 0.3) × 10−2 | 2.5 | 0.06 |
| 7-CH3-Trp | 2.0 ± 0.4 | (1.3 ± 0.1) × 10−2 | 6.5 | 0.15 |
Km and kcat for tryptophan are comparable with the previously reported values.37 Kms are somewhat higher and kcats substantially lower for the 7-methyl and 4-methoxy analogues, respectively, with catalytic efficiencies (kcat/Km) for the 7-methyl- and 4-methoxytryptophan analogues that were <1% of the normal reaction. Maximal velocity was not reached at concentrations before 4-methyltryptophan precipitated (10 mM) under our assay conditions, and values for the kinetic constants were not determined.
Product Studies
4-Mercaptophenylalanine
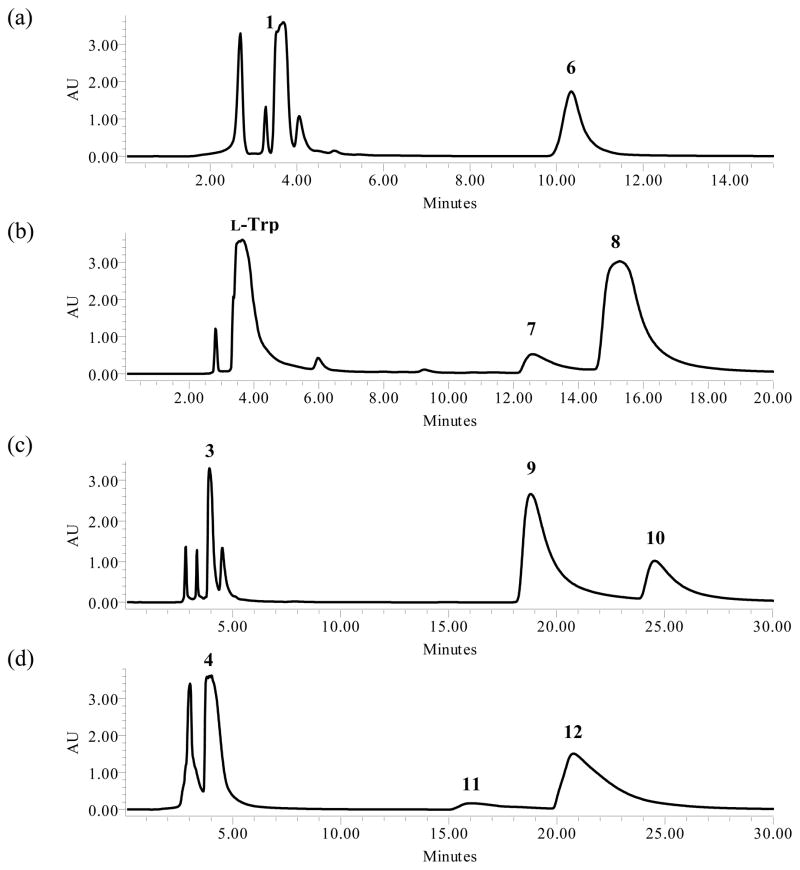
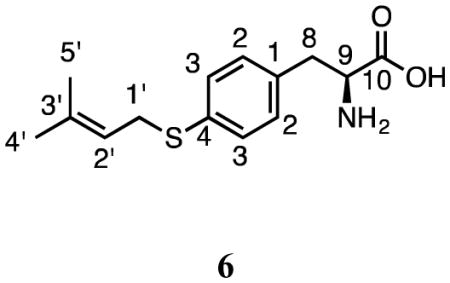
Incubation of 1 and DMAPP with SirD gave a single product (6) (Figure 2a). The UV spectrum of 6 (10.3 min) had a peak at 257.0 nm. Monoprenylation was confirmed by HRMS where the sodium adduct of dimethylallyl mercaptophenylalanine gave a peak at m/z of 288.1041 Da. The structure of 6 was established using a combination of 1H and 2D NMR spectroscopy (Table 2). Normal prenylation was confirmed by the presence of signals for the methylene protons at C1′ (3.55 ppm, d, J = 7.8 Hz), the proton at C2′ (5.24 ppm, t, J = 7.2 Hz), and the methyl groups at C3′ (1.66 and 1.58 ppm). An AB quartet for the aromatic protons at 7.14 and 7.19 ppm, each integrating to two protons indicated that the benzene ring was para-substituted. Thus, prenylation occurred at the sulfur atom. The HMBC spectrum revealed correlations between the C1′ dimethylallyl methylene protons and C2′ (119.2 ppm), C3′ (135.5 ppm), and the para carbon C4 (133.1 ppm). These data demonstrate that SirD catalyzes a normal S-prenylation of 4-mercaptophenylalanine.
Figure 2.
HPLC chromatograms of preparative enzymatic assays for isolation and purification. Each aromatic amino acid was incubated with DMAPP and SirD. (a) 4-Mercaptophenylalanine, (b) tryptophan, (c) 4-methyltryptophan, (d) 4-methoxytryptophan, and (e) 7-methyltryptophan.
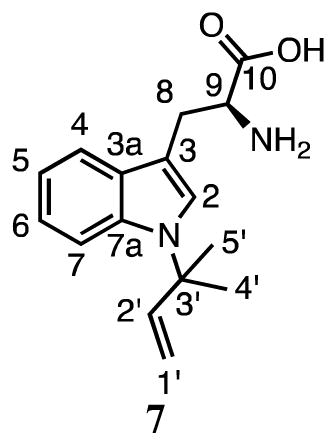
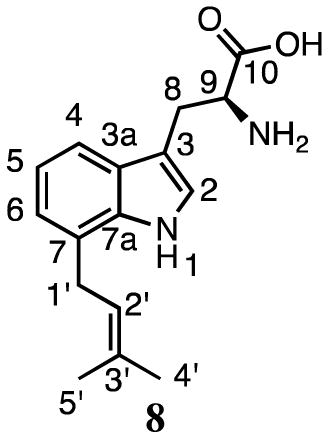
Table 2.
1H and 13C NMR data for products 6 – 14.
| Structure/Compound |
 6 |
 7 |
 8 |
|||
|---|---|---|---|---|---|---|
| Position | δH, multi., J | δC | δH, multi., J | δC | δH, multi., J | δC |
| 1 | - | 137.4 | - | - | exc. | - |
| 2 | 7.14, d, 7.2 | 129.5 | 7.33, s | 128.5 | 7.16, s | 127.1 |
| 3 | 7.19, d, 7.8 | 128.2 | - | n.f. | - | 111.3 |
| 3a | - | - | - | n.f. | - | 129.5 |
| 4 | - | 133.1 | 7.60, d, 7.8 | n.f. | 7.45, d, 7.2 | 118.9 |
| 5 | - | - | 7.05, t, 7.2 | n.f. | 7.01, t, 7.2 | 122.3 |
| 6 | - | - | 7.09, t, 7.8 | n.f. | 6.98, d, 6.6 | 123.6 |
| 7 | - | - | 7.54, d, 8.4 | n.f. | - | 128.1 |
| 7a | - | - | - | n.f. | - | 138.3 |
| 8 | 2.92, d, 11.4 | 32.1 | 3.24, dd, 4.8, 15.0 | 30.3 | 3.29, dd, 4.8, 15.6 | 29.7 |
| 2.58–2.68, m | - | 3.06, dd, 7.8, 15.0 | - | 3.10, dd, 7.8, 15.0 | - | |
| 9 | 3.24, m | 56.7 | 3.75, t, 6.6 | 57.9 | 3.83, dd, 4.8, 7.8 | 57.9 |
| 10 | - | n.f. | - | n.f. | - | n.f. |
| 11 | - | - | - | - | - | - |
| 1′ | 3.55, d, 7.8 | 30.9 | 5.11, d, 10.8 | n.f. | 3.47, d, 7.2 | 31.8 |
| - | - | 5.02, d, 17.4 | - | - | - | |
| 2′ | 5.24, t, 7.2 | 119.2 | 6.03, dd, 10.8, 17.4 | n.f. | 5.36, t, 7.8 | 123.8 |
| 3′ | - | 135.5 | - | n.f. | - | 137.8 |
| 4′ | 1.66, s | 25.1 | 1.62, s | 29.6 | 1.61, s | 27.4 |
| 5′ | 1.58, s | 17.3 | 1.62, s | 29.6 | 1.64, s | 19.7 |
| Structure/Compound |
 9 |
 10 |
 11 |
|||
|---|---|---|---|---|---|---|
| Position | δH, multi., J | δC | δH, multi., J | δC | δH, multi., J | δC |
| 1 | - | - | exc. | - | exc. | - |
| 2 | 7.30, s | 128.8 | 7.14, s | 127.5 | 6.97, s | 126.1 |
| 3 | - | 110.7 | - | 94.6 | - | n.f. |
| 3a | - | 129.3 | - | 127.4 | - | 119.6 |
| 4 | - | 133.7 | - | 131.0 | - | n.f. |
| 5 | 6.79, d, 7.2 | 123.5 | 6.75, d, 7.2 | 123.8 | 6.55, d, 8.4 | 102.2 |
| 6 | 6.96, t, 7.2 | 123.8 | 6.86, d, 7.2 | 123.8 | 7.06, d, 7.8 | n.f. |
| 7 | 7.38, d, 8.4 | 115.2 | - | 125.6 | - | 127.8 |
| 7a | - | 138.3 | - | 138.3 | - | n.f. |
| 8 | 3.48, dd, 5.4, 15.6 | 32.3 | 3.53–3.59, m | n.f. | 3.48, dd, 3.6, 15.0 | 33.5 |
| 3.03, dd, 9.0, 15.6 | - | 3.06, dd, 10.8, 16.2 | - | 3.02, dd, 8.4, 15.0 | - | |
| 9 | 3.68, dd, 5.4, 9.0 | 59.6 | 3.74–3.79, m | 59.1 | 3.88, dd, 3.6, 7.8 | n.f. |
| 10 | - | 179.5 | - | n.f. | - | n.f. |
| 11 | 2.58, s | 22.2 | 2.56, s | 21.7 | 3.84, s | 57.8 |
| 1′ | 5.09, d, 10.8 | 116.1 | 3.47–3.51, m | 31.4 | 5.03, d, 10.8 | 114.4 |
| 5.00, d, 17.4 | - | - | - | 4.98, d, 17.4 | - | |
| 2′ | 6.01, dd, 10.8, 18.0 | 146.6 | 5.33, t, 6.0 | 123.9 | 6.00, dd, 10.8, 17.4 | 149.8 |
| 3′ | - | 61.8 | - | 137.7 | - | 42.0 |
| 4′ | 1.61, s | 29.8 | 1.61, s | 27.3 | 1.35, s | 29.4 |
| 5′ | 1.60, s | 29.8 | 1.63, s | 19.5 | 1.35, s | 29.4 |
| Structure/Compound |
 12 |
 13 |
 14 |
|||
|---|---|---|---|---|---|---|
| Position | δH, multi., J | δC | δH, multi., J | δC | δH, multi., J | δC |
| 1 | exc. | - | - | - | exc. | - |
| 2 | 7.00, s | 126.5 | 7.43, s | 130.3 | 7.23, s | n.f. |
| 3 | - | n.f. | - | 109.5 | - | n.f. |
| 3a | - | 119.3 | - | 133.3 | - | n.f. |
| 4 | - | 154.7 | 7.46, d, 7.8 | 119.1 | 7.48, d, 7.8 | 118.4 |
| 5 | 6.47, d, 7.8 | 102.6 | 7.00, t, 7.8 | 122.5 | 7.03, d, 7.8 | n.f. |
| 6 | 6.84, d, 7.8 | n.f. | 6.96, d, 6.6 | 128.5 | - | n.f. |
| 7 | - | 121.5 | - | 125.5 | - | 121.6 |
| 7a | - | 139.1 | - | 137.2 | - | n.f. |
| 8 | 3.42, dd, 4.2, 15.0 | 31.5 | 3.17–3.23, m | 35.1 | 3.36, dd, 3.6, 15.0 | n.f. |
| 3.02, dd, 8.4, 15.0 | - | 2.97–3.04, m | - | 3.16, dd, 7.8, 15.0 | - | |
| 9 | 3.85, dd, 4.8, 8.4 | 59.2 | 3.66–3.72, m | 58.2 | 3.84, dd, 3.6, 12.0 | 57.9 |
| 10 | - | n.f. | - | n.f. | - | n.f. |
| 11 | 3.80, s | 57.8 | 2.54, s | 26.6 | 2.42, s | 14.7 |
| 1′ | 3.35–3.37, m | 31.2 | 4.04, d, 10.8 | 114.7 | 3.46, d, 7.2 | 33.8 |
| 2′ | 5.30, t, 6.6 | 124.0 | 6.30, dd, 10.8, 17.4 | 150.0 | 5.32, t, 7.2 | 125.8 |
| 3′ | - | 137.5 | - | 62.5 | - | 135.8 |
| 4′ | 1.60, s | 19.7 | 1.64, s | 34.3 | 1.77, s | 19.7 |
| 5′ | 1.59, s | 27.5 | 1.64, s | 34.3 | 1.69, s | 27.5 |
δH and δC in ppm
J in Hz
n.f. = not found
exc. = exchanged
4-Vinylphenylalanine
Incubation of up to 10 mM 2 and DMAPP with SirD yielded no identifiable products (data not shown). We surmise that either the vinyl moiety is too large to be accommodated in the active site or 2 is not properly positioned for prenylation.
Tryptophan
Kremer and Li reported that SirD catalyzes the normal prenylation of tryptophan at C7.37 In our hands, incubation of L-tryptophan and DMAPP with SirD gave two products (Figure 2b). The UV spectra of compounds 7 (12.7 min, 7%) and 8 (15.3 min, 93%) had peaks at 222.7, 284.3 nm and 221.5, 277.2, 297.9 nm, respectively, suggesting the indole chromophores of compounds 7 and 8 were intact. HMRS analysis revealed that the molecular ions of both compounds were 68 Da higher than tryptophan, or its sodium adduct, indicating the addition of one isoprene unit.
1H NMR analysis (Table 2) of compound 8 revealed a spectrum very similar to the data reported for 7-dimethylallyltryptophan.16 The HMBC spectrum showed a correlation between the methylene protons at C1′ (3.47 ppm, d, J = 7.2 Hz) and C7 of the indole ring (128.1 ppm). Together, these data establish that 8 is normal 7-dimethylallyltryptophan.
Compound 7 has a dimethylallyl moiety attached to the indole ring through C3′ as evidenced by signals for the three vinyl protons at 6.03 ppm (dd, J = 10.8, 17.4 Hz), 5.11 ppm (d, J = 10.8 Hz) and 5.02 ppm (d, J = 17.4 Hz), and the methyl groups at 1.62 ppm. Although the NMR sample was too dilute to collect relevant TOCSY, HMQC, or HMBC data, the ROESY spectrum showed a correlation between the proton at C2 (7.33 ppm, s) and the methyl groups of the prenyl moiety. Aromatic protons at C2, C4 (7.60 ppm), C5 (7.05 ppm), C6 (7.09 ppm) and C7 (7.54 ppm) suggested that the dimethylallyl moiety was attached at the indole nitrogen. Collectively, these data indicate that 7 is the product of reverse prenylation of tryptophan at N1. Thus, SirD catalyzes normal prenylation at C7 and reverse prenylation at N1 of the indole nucleus of tryptophan, similar to the cyclo-L-Trp-L-Trp dimethylallyltransferase from A. oryzae DSM1147.17
4-Methyltryptophan
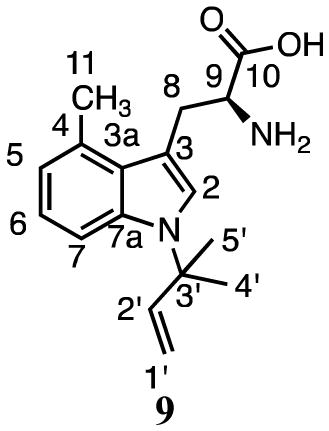
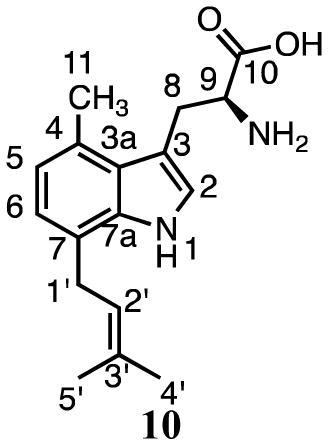
Zou and coworkers38 reported activity for prenylation of 4-methyl-DL-tryptophan at C7 by SirD at a rate ~8% of L-tyrosine. Similar to our results for tryptophan, we found that incubation of 3 and DMAPP with SirD gave two products (Figure 2c). HMRS and UV analysis of compounds 9 (18.8 min, 79%) and 10 (24.6 min, 21%) revealed that they are monoprenylated derivatives of 4-methyltryptophan.
The 1H NMR spectrum of 9 showed three vinyl signals at 6.01 ppm (dd, J = 10.8, 18.0 Hz), 5.09 ppm (d, J = 10.8 Hz) and 5.00 ppm (d, J =17.4 Hz) for the protons at C2′ and C1′, respectively, and methyl groups at 1.61 and 1.60 ppm, all characteristic of a reverse prenylated structure. The ROESY spectrum displayed a correlation between the methyl groups of the prenyl moiety and the proton at C2 (7.30 ppm). The presence of signals at C2, C4 (6.79 ppm), C5 (6.96 ppm), and C6 (7.38 ppm) suggested that prenylation occurred at N1. Thus, compound 9 is reverse N1-(dimethylallyl)-4-methyltryptophan.
Compound 10 was identified as 7-dimethylallyl-4-methyltryptophan by NMR analysis. 1H signals were seen for the methylene protons at C1′ (3.47–3.51 ppm, m), the proton at C2′ (5.33 ppm, t, J = 6.0 Hz), and the methyl groups at 1.61 and 1.63 ppm of the dimethylallyl group (Table 2). Signals for protons attached to C2 (7.14 ppm, s), C5 (6.75 ppm, d, J = 7.2 Hz), and C6 (6.86 ppm, d, J = 7.2 Hz), along with HMBC correlations between the methylene protons at C1′ and C7 in the indole ring (125.6 ppm), confirmed prenylation at C7.
We suspect that Li and coworkers were unable to see the N-prenylated products due to their use of HPLC solvents containing 0.5% trifluoroacetic acid (TFA).37,38 The product from A. fumigatus CdpNPT was originally assigned incorrectly due to disassociation and rearrangement of the dimethylallyl moiety at N1 of the indole ring upon exposure to trichloroacetic acid.43,44 We found that products seen by HPLC analysis of samples from incubation of tryptophan or 3, DMAPP, and SirD, disappeared when the elution solvent contained 0.5% TFA (data not shown).
4-Methoxytryptophan
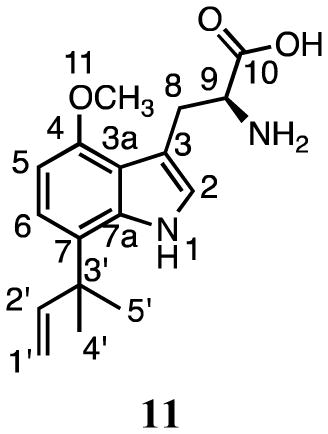
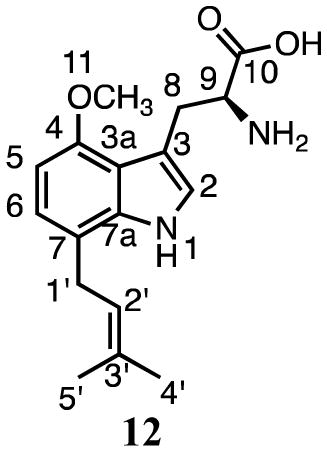
Incubation of 4 and DMAPP with SirD gave compounds 11 (16.1 min, 8%) and 12 (20.8 min, 92%) (Figure 2d). HRMS analysis indicated that the molecular ions were 68 Da higher than the sodium adduct of 4, as expected for monoprenylation. UV spectra for 11 and 12 suggested that the indole nuclei were intact.
The dimethylallyl moiety in compound 11 was attached in a reverse orientation as evidenced by distinctive signals for the vinyl protons at 6.00 ppm (dd, J = 10.8, 17.4 Hz), 5.03 ppm (dd, J = 10.8 Hz), and 4.98 ppm (dd, J = 17.4 Hz), and two methyl groups at 1.35 ppm. The HMBC spectrum showed correlations between the methyl peaks of the prenyl moiety and C7 (127.8 ppm). The ROESY spectrum showed correlations between the methyl peaks of the prenyl moiety and the aromatic proton at C6 (7.06 ppm), and between the methoxy substituent at C4 (57.8 ppm) and the proton at C5 (6.55 ppm). Together, these data confirm that 11 is formed by reverse prenylation at C7 of 4-methoxytryptophan.
Compound 12 contained a dimethylallyl moiety attached in a normal orientation as indicated by signals for the methylene protons at C1′ (3.35–3.37 ppm, m), the proton at C2′ (5.30 ppm, t, J = 6.6 Hz), and the methyl groups at 1.60 and 1.59 ppm. Signals for the proton at C2 (7.00, s) and two other aromatic protons (6.84 ppm, d, J = 7.8 Hz and 6.47 ppm, d, J = 7.8 Hz) showed that prenylation had occurred at either C5 or C7. The ROESY spectrum showed a weak correlation between the methylene protons of the prenyl moiety and the aromatic proton at C6 (6.84 ppm). The HMBC spectrum showed correlations between the methylene protons of the prenyl moiety and C7 (121.5 ppm). Collectively, these data indicate that 12 is formed by normal prenylation at C7 position of 4-methoxytryptophan.
7-Methyltryptophan
Zou and coworkers reported 7-methyl-DL-tryptophan was not a substrate of SirD (relative yield <0.03%).38 Based on our results with 3 and 4, we anticipated that 5 might be a substrate for regioselective prenylation at N1. Interestingly, incubation of 5 and DMAPP with SirD gave two products, 13 (16.3 min, 13%) and 14 (19.6 min, 87%) (Figure 2e). HRMS and UV spectra indicated that the products were monoprenylated with intact indole nuclei.
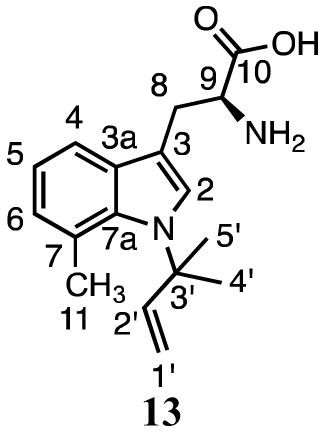
NMR analysis (Table 2) confirmed that compound 13 is reverse N1-(dimethylallyl)-7-methyltryptophan. The orientation of the dimethylallyl unit is clear from the 1H NMR spectrum of 13 where signals were seen for the three vinyl protons at 6.30 ppm (dd, J = 10.8, 17.4 Hz), 4.94 ppm (d, J = 10.8 Hz) and 4.49 ppm (d, J = 17.4 Hz), and the methyl groups at 1.64 ppm (s). The TOCSY spectrum showed correlations between the dimethylallyl methyl groups and the C2 indole proton at 7.43 ppm. Signals for protons at C2, C4 (7.46 ppm), C5 (7.00 ppm), and C6 (6.96 ppm) confirm prenylation of the indole nitrogen.
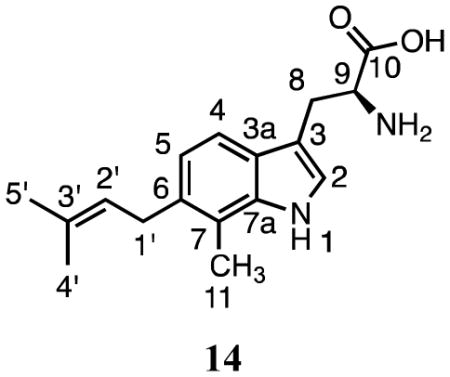
The 1H NMR spectrum of 14 had peaks for the dimethylallyl group at C1′ (3.46 ppm, d, J = 7.2 Hz), C2′ (5.32 ppm, d, J = 7.2 Hz), and the methyl groups at C3′ (1.77 and 1.69 ppm) indicative of normal prenylation. Resonances for the proton at C2 (7.23 ppm, s) and aromatic protons at 7.48 ppm (d, J = 7.8 Hz) and 7.03 ppm (d, J = 7.8 Hz) indicated that prenylation occurred at C4 or C6 in the indole ring. The HMBC spectrum showed a correlation between the methyl group at C7 (2.42 ppm) and C3′ (135.8 ppm). Although we were not able to unambiguously distinguish between prenylation at C4 and C6, a comparison of our 1H NMR spectrum with those published for 4-dimethylallyl-7-methyltryptophan22 and 7-dimethylallyl-6-methyltryptophan15 suggests 14 is prenylated at C6. The reported resonances for C5 (6.99 ppm) and C6 (6.89 ppm) in 4-dimethylallyl-7-methyltryptophan are only 0.1 ppm apart, while those for C4 (7.41 ppm) and C5 (6.86 ppm) in 7-dimethylallyl-6-methyltryptophan are separated by 0.55 ppm. The separation in 14 is 0.45 ppm. Collectively, these data suggest that SirD catalyzes normal prenylation at C6 in 7-methyltryptophan.
Promiscuity and Mechanism
SirD catalyzes normal prenylation of the hydroxyl group in tyrosine as the first committed step in the biosynthesis of the phytotoxin sirodesmin PL. The protein is a presumed member of the ABBA-fold family of aromatic prenyltransferases, whose other members catalyze alkylation of benzene,31 naphthalene,30 and phenazine32 rings and the indole moiety in tryptophan.26,28,29 The most studied member of the family, 4-dimethylallyltryptophan synthase (4-DMATS), gives a single product when incubated with its normal substrates. However, alternate substrates for tryptophan with methyl, methoxy, or amino substituents at C4 give different mixtures of products, which collectively are formed by normal alkylation at N1, C3, C5, and C7 and reverse alkylation at C3.35 This behavior can be explained by a dissociative electrophilic alkylation of tryptophan by the dimethylallyl cation generated by cleavage of the C–O bond in DMAPP, where blocking the preferred alkylation at C4 results in electrophilic attack of other nucleophilic sites in the indole moiety. The regiopromiscuity seen for 4-DMATS provides a logical path for evolution of other dimethylallyltryptophan synthases that are regioselective for alkylation of the indole nucleus at positions other than C4.
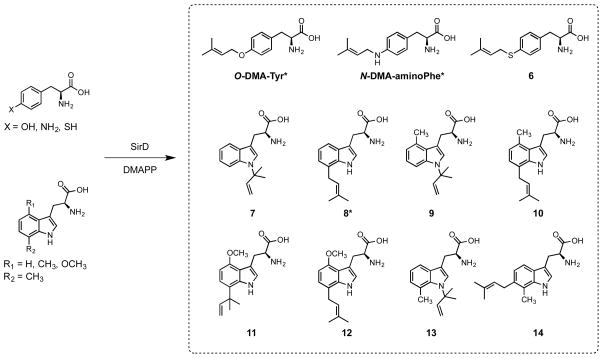
As part of a study to explore structure and function in the ABBA aromatic prenyltransferases, we examined the ability of SirD to catalyze alkylation of a variety of tyrosine and tryptophan analogues. In addition to its normal role in the synthesis of dimethylallyltyrosine, SirD accepts 4-aminophenylalanine,38 4-mercaptophenylalanine, tryptophan,37 4-methyltryptophan,38 4-methoxytryptophan, and 7-methyltryptophan as alternate substrates to generate the products shown in Figure 3. Given the diverse group of aromatic amino acid derivatives that are alkylated by SirD, we were surprised that 4-vinylphenylalanine was not a substrate, even at high concentrations, where alkylation of the vinyl group would be mechanistically similar to alkylation of the vinyl group in protoheme IX by the farnesyltransferase encoded by cyoE in E. coli.45 We surmise that the bound conformations of 4-vinylphenylalanine and DMAPP are incompatible with catalysis.
Figure 3.
Products from incubation of tyrosine or tryptophan analogues and DMAPP with SirD. Structures for compounds with asterisks were determined previously.37,38
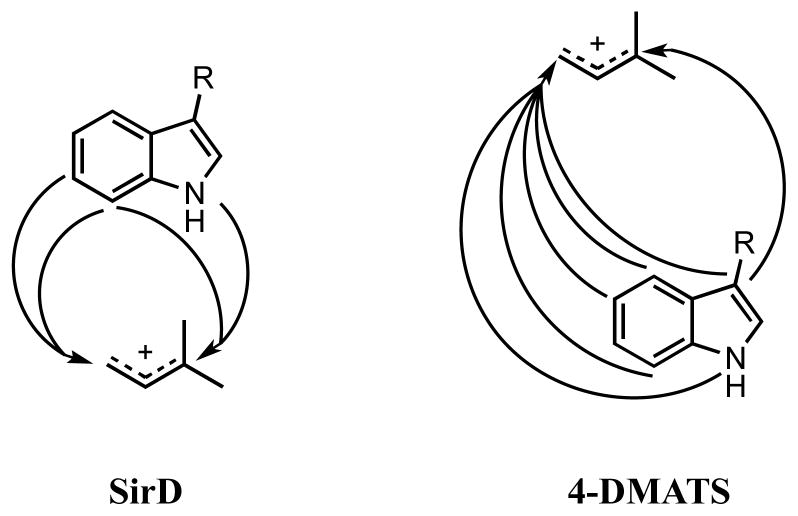
A comparison of the regiochemistry for alkylation of the indole moiety in tryptophan by SirD and 4-DMATS (see Figure 4) emphasizes the regiopromiscuity of both enzymes. All of the SirD products are formed by alkylation of sites on the “southern” (N1, C6, C7) edge of the indole nucleus, as are two of the 4-DMATS products (N1, C7). Four additional 4-DMATS products are from alkylation of the “northern” (C3, C4, C5) edge. Both enzymes catalyze normal and reverse alkylations. The catalytic efficiencies of the alternative substrates for both enzymes are remarkably high, with values of kcat/Km ~10−3 relative to those of the normal substrates for the least efficient analogues. Thus, the ABBA aromatic prenyltransferases appear to have remarkably plastic active sites. Proteins with this fold have evolved to give a family of biosynthetic enzymes that collectively have the ability to alkylate tryptophan at all of the nucleophilic sites in the indole ring.
Figure 4.
Regiochemistry for alkylation of the indole nucleus by SirD and 4-DMATS.
Finally, the electronic properties of the substituents on the indole ring appear to influence the regiochemistry of the alkylation. Incubation of 4-methyltryptophan (σ+methyl = 0.31)46 and DMAPP with SirD gives normal prenylation at C7 and reverse prenylation at N1. In contrast, 4-methoxytryptophan (σ+methoxy = 0.78)46 gives normal and reverse prenylation exclusively at C7. This observation is consistent with an enhancement in the nucleophilicity of C7 relative to N1 by the strongly electron donating methoxy group.
CONCLUSION
SirD catalyzes aromatic S-prenylation of 4-mercaptophenylalanine and N- and C-prenylations of tryptophan and tryptophan analogues at three different sites on the indole ring (N1, C6, C7): normal prenylation at C6, normal and reverse prenylation at C7, and reverse prenylation at N1. Our results provide further evidence for a dissociative electrophilic alkylation by ABBA aromatic prenyltransferases, where substrate orientation within the active site and substituent electronic effects determine the position and type of prenylation.
METHODS
Synthesis of amino acid analogues
Procedures for syntheses of 4-mercapto-L-phenylalanine (1), 4-vinyl-L-phenylalanine (2), 4-methyl-L-tryptophan (3), 4-methoxy-L-tryptophan (4), and 7-methyl-L-tryptophan (5) are described in the Supplemental Information.
Production and Purification of SirD
L. maculans sirD (NCBI GenBank AY553235.1) was optimized for heterologous expression in E. coli and synthesized by Genscript (Figure S2). Modified L. maculans sirD in pUC57 was cloned into pET21a(+) following standard restriction digestion protocols (see Supplemental Information) to give pLmSirD. pLmSirD was transformed into E. coli BL21 (DE3) according to the manufacturer’s instructions. E. coli strain pLmSirD-BL21 (DE3) was incubated in 1 L of LB medium containing 50 μg mL−1 ampicillin at 37 °C with shaking at 225 rpm until an OD600 of 0.6 was reached. The culture was induced with IPTG to a final concentration of 0.6 mM. After 5 h, cells were harvested by centrifugation (4,000 × g, 20 min, 4 °C). Pelleted cells were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, containing 300 mM NaCl, 10 mM imidazole, 1 mg mL−1 of lysozyme and 10 μg mL−1 of RNase A). After incubation at 0 °C for 30 min, the cell suspension was sonicated for 6 × 10 sec at 4 °C. The lysate was clarified by centrifugation (10,000 × g, 20 min, 4 °C). SirD was purified by nickel affinity chromatography using a gradient elution 0–100% of lysis buffer/elution buffer (50 mM NaH2PO4, pH 8.0, containing 300 mM NaCl and 500 mM imidazole). The purified protein was dialyzed against 2 × 4 L of dialysis buffer (20 mM Tris-HCl, pH 8.0, containing 1 mM βME), and then against 4 L of dialysis buffer containing 20% v/v glycerol. The protein was stored at −80 °C until use.
Kinetic Studies
All kinetic assays were performed in a total volume of 100 μL at 30 °C for 10 min unless otherwise noted, and quenched by heating at 100 °C for 30 s. After centrifugation, 10 μL of reaction mixture were spotted and developed by RP-TLC. For tyrosine assays, RP-C18 plates were developed with 25:75 H2O/methanol. For all other substrates, RP-C8 plates were developed with 4:6 25 mM NH4HCO3/methanol. The developed TLC plates were imaged on a storage phosphor screen (Molecular Dynamics) and scanned by a Typhoon 8600 Variable Mode Imager (GE Healthcare). The data were visualized and processed with ImageQuant 5.2. Background radioactivity was determined from incubations without the aromatic substrate. Each kinetic assay was performed in triplicate or quadruplicate. [1-14C]DMAPP and [14C(U)]-L-tyrosine were purchased from American Radiolabeled Chemicals, Inc.
For L-tyrosine, the assay was conducted in 20 mM Tris-HCl buffer, pH 8.0, containing 80 nM SirD, 1 mM DMAPP, and [14C(U)]-L-tyrosine concentrations of 0.04, 0.05, 0.07, 0.08, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35 and 0.4 mM (0.25–25 μCi μmol−1) with 10 min incubations. For 4-mercaptophenylalanine, the assay was conducted in 100 mM Tris-HCl buffer, pH 8.0, containing 10 mM βME, 2% v/v glycerol, 5 μM SirD, 1 mM [1-14C]DMAPP (1 μCi μmol−1), and 4-mercapto-L-phenylalanine concentrations of 0.25, 0.5, 0.75, 1, 2.5, 5, 7.5 and 10 mM with 30 min incubations. For tryptophan, the assay was conducted in 100 mM Tris-HCl buffer, pH 8.0, containing 2% v/v glycerol, 5 μM SirD, 1 mM [1-14C]DMAPP (1 μCi μmol−1), and L-tryptophan concentrations of 0.1, 0.2, 0.3, 0.6 and 1 mM with 20 min incubations. For 4-methoxy and 7-methyltryptophan, concentrations were 0.3, 0.6, 1, 2, 3, 4, 6 and 10 mM with 30 min incubations.
Product Studies
Incubations with 4-mercaptophenylalanine were conducted in 50 mM Tris-HCl buffer, pH 8.0, containing 5 mM MgCl2, 20 mM βME, 5 mM DMAPP, 10 mM compound 1 and 14.5 μM SirD in a total volume of 5 mL. Glycerol present in the SirD dialysis buffer was removed by repeated dialysis/centrifugation using an Amicon Ultra 10 kDa molecular weight cut-off (MWCO) filter. The reaction mixture was incubated at 30 °C for 21 h. SirD was removed by MWCO filtration, samples were concentrated by lyophilization, and the products were purified by HPLC on a Waters 2690 Separation Module using a Microsorb MV™ C18 5 μm column at a flow rate of 1 mL min−1 with isocratic elution using acetonitrile:H2O (25/75). Products were detected with a Waters 996 photodiode array detector with simultaneous monitoring at 254 nm.
Incubations with tryptophan and tryptophan derivatives were identical to those above except as follows: 10 mM MgCl2, 2.92 mM DMAPP, 20 mM tryptophan analogue, 5 μM SirD in a total volume of 5 mL. The reaction mixtures were incubated for 21–42 h and the products were detected at 225 nm. Product ratios were calculated by integrating the area under each product peak visualized at 220 nm (4-methoxy) or 225 nm (tryptophan/4-methyl/7-methyl).
Determination of Structures
Purified products were lyophilized, dissolved in D2O (D, 99.9%), lyophilized again, dissolved in DMSO-d6 (D, 99.96%) or D2O (D, 99.96%) (Cambridge Isotope Laboratories, Inc.) and placed in a Norell 3 mm NMR tube (Sigma). NMR spectra were recorded on an INOVA 600 NMR spectrometer equipped with a HCN cryogenic probe. Unless otherwise noted, the following NMR experiments were performed: 1D 1H, COSY 1H-1H, TOCSY 1H-1H, ROESY 1H-1H, HSQC or HMQC 1H-13C, and HMBC 1H-13C. All spectra were processed with MestReNova 7.1.
Supplementary Material
Acknowledgments
We thank R. Sterner for supplying the tmTrpB vector. We thank J. Muller and J. Skalicky for performing mass spectroscopy and NMR assistance, respectively. This work was supported by NIH grant GM 21328.
Footnotes
ASSOCIATED CONTENT
Additional methods; bacterial strains, plasmids and culture conditions; overproduction and purification of tryptophan synthase; synthesis of DMAPP and amino acid analogues; synthetic schemes for preparation of amino acid analogues (Schemes S1–S3); NMR spectra of aromatic substrate analogues and precursors (Figures S3–S18); NMR spectra of prenylated tyrosine and tryptophan products (Figures S19–S71); structures of products with pertinent 2D NMR correlations (Figure S72); HRMS data for products (Table S1); Michaelis-Menten analysis of kinetic data (Figures S73). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no conflicts of interest.
References
- 1.Gardiner DM, Waring P, Howlett BJ. The epipolythiodioxopiperazine (ETP) class of fungal toxins: Distribution, mode of action, functions and biosynthesis. Microbiology (Reading, U K ) 2005;151:1021–1032. doi: 10.1099/mic.0.27847-0. [DOI] [PubMed] [Google Scholar]
- 2.Li SM. Prenylated indole derivatives from fungi: Structure diversity, biological activities, biosynthesis and chemoenzymic synthesis. Nat Prod Rep. 2010;27:57–78. doi: 10.1039/b909987p. [DOI] [PubMed] [Google Scholar]
- 3.Hulvova H, Galuszka P, Frebortova J, Frebort I. Parasitic fungus Claviceps as a source for biotechnological production of ergot alkaloids. Biotechnol Adv. 2012 doi: 10.1016/j.biotechadv.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Wallwey C, Li SM. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep. 2011;28:496–510. doi: 10.1039/c0np00060d. [DOI] [PubMed] [Google Scholar]
- 5.Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Current Opinion in Chemical Biology. 2009;13:171. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Tello M, Kuzuyama T, Heide L, Noel JP, Richard SB. The ABBA family of aromatic prenyltransferases: broadening natural product diversity. Cell Mol Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saleh O, Haagen Y, Seeger K, Heide L. Prenyl transfer to aromatic substrates in the biosynthesis of aminocoumarins, meroterpenoids and phenazines: The ABBA prenyltransferase family. Phytochemistry (Elsevier) 2009;70:1728–1738. doi: 10.1016/j.phytochem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Li S-M. Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives. Phytochemistry. 2009;70:1746. doi: 10.1016/j.phytochem.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Grundmann A, Kuznetsova T, Afiyatullov SS, Li S-M. FtmPT2, an N-Prenyltransferase from Aspergillus fumigatus, Catalyses the Last Step in the Biosynthesis of Fumitremorgin B. ChemBioChem. 2008;9:2059. doi: 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
- 10.Grundmann A, Li SM. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology (Reading, U K ) 2005;151:2199. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 11.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin Biosynthesis in Neosartorya fischeri: identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 12.Yin WB, Yu X, Xie XL, Li SM. Preparation of pyrrolo[2,3-b]indoles carrying a β-configured reverse C3-dimethylallyl moiety by using a recombinant prenyltransferase CdpC3PT. Org Biomol Chem. 2010;8:2430. doi: 10.1039/c000587h. [DOI] [PubMed] [Google Scholar]
- 13.Gebler JC, Poulter CD. Purification and characterization of dimethylallyl tryptophan synthase from Claviceps purpurea. Arch Biochem Biophys. 1992;296:308–313. doi: 10.1016/0003-9861(92)90577-j. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Liu Y, Xie X, Zheng XD, Li SM. Biochemical Characterization of Indole Prenyltransferases: Filling the last gap of prenylation positions by a 5-dimethylallyltryptophan synthase from Aspergillus clavatus. J Biol Chem. 2012;287:1371–1380. doi: 10.1074/jbc.M111.317982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi S, Takagi H, Toyoda A, Uramoto M, Nogawa T, Ueki M, Sakaki Y, Osada H. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J Bacteriol. 2010;192:2839–2851. doi: 10.1128/JB.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer A, Westrich L, Li SM. A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. Microbiology (Reading, U K ) 2007;153:3409–3416. doi: 10.1099/mic.0.2007/009019-0. [DOI] [PubMed] [Google Scholar]
- 17.Zou HX, Xie XL, Linne U, Zheng XD, Li SM. Simultaneous C7- and N1-prenylation of cyclo-L-Trp-L-Trp catalyzed by a prenyltransferase from Aspergillus oryzae. Org Biomol Chem. 2010;8:3037–3044. doi: 10.1039/c002850a. [DOI] [PubMed] [Google Scholar]
- 18.Unsoeld IA, Li SM. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology (Reading, U K ) 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 19.Wollinsky B, Ludwig L, Xie X, Li SM. Breaking the regioselectivity of indole prenyltransferases: identification of. Org Biomol Chem. 2012;10:9262–9270. doi: 10.1039/c2ob26149a. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Zuo D, Song Y, Wang B, Huang H, Yao Y, Li W, Zhang S, Zhang C, Ju J. Characterization of a single gene cluster responsible for methylpendolmycin and pendolmycin biosynthesis in the deep sea bacterium Marinactinospora thermotolerans. ChemBioChem. 2012;13:547–552. doi: 10.1002/cbic.201100700. [DOI] [PubMed] [Google Scholar]
- 21.Lee SL, Floss HG, Heinstein P. Purification and properties of dimethylallylpyrophosphate:tryptophan dimethylallyl transferase, the first enzyme of ergot alkaloid biosynthesis in Claviceps. sp SD 58. Arch Biochem Biophys. 1976;177:84–94. doi: 10.1016/0003-9861(76)90418-5. [DOI] [PubMed] [Google Scholar]
- 22.Gebler JC, Woodside AB, Poulter CD. Dimethylallyltryptophan synthase. An enzyme-catalyzed electrophilic aromatic substitution. J Am Chem Soc. 1992;114:7354–7360. [Google Scholar]
- 23.Tsai HF, Wang H, Gebler JC, Poulter CD, Schardl CL. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem Biophys Res Commun. 1995;216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 24.Steffan N, Unsoeld IA, Li S-M. Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. ChemBioChem. 2007;8:1298–1307. doi: 10.1002/cbic.200700107. [DOI] [PubMed] [Google Scholar]
- 25.Luk LYP, Tanner ME. Mechanism of Dimethylallyltryptophan Synthase: Evidence for a Dimethylallyl Cation Intermediate in an Aromatic Prenyltransferase Reaction. J Am Chem Soc. 2009;131:13932–13933. doi: 10.1021/ja906485u. [DOI] [PubMed] [Google Scholar]
- 26.Metzger U, Schall C, Zocher G, Unsoeld I, Stec E, Li SM, Heide L, Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A. 2009;106:14309–14314. S14309/14301–S14309/14309. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luk LYP, Qian Q, Tanner ME. A Cope Rearrangement in the Reaction Catalyzed by Dimethylallyltryptophan Synthase? J Am Chem Soc. 2011;133:12342–12345. doi: 10.1021/ja2034969. [DOI] [PubMed] [Google Scholar]
- 28.Jost M, Zocher G, Tarcz S, Matuschek M, Xie X, Li S-M, Stehle T. Structure-Function Analysis of an Enzymatic Prenyl Transfer Reaction Identifies a Reaction Chamber with Modifiable Specificity. Journal of the American Chemical Society. 2010;132:17849. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- 29.Schuller JM, Zocher G, Liebhold M, Xie X, Stahl M, Li SM, Stehle T. Structure and Catalytic Mechanism of a Cyclic Dipeptide Prenyltransferase with Broad Substrate Promiscuity. J Mol Biol. 2012;422:87–99. doi: 10.1016/j.jmb.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 30.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature (London, U K ) 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger U, Keller S, Stevenson CEM, Heide L, Lawson DM. Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway. Journal of Molecular Biology. 2010;404:611. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 32.Zocher G, Saleh O, Heim Joel B, Herbst Dominik A, Heide L, Stehle T. Structure-based engineering increased the catalytic turnover rate of a novel phenazine prenyltransferase. PLoS One. 2012;7:e48427. doi: 10.1371/journal.pone.0048427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian Q, Schultz AW, Moore BS, Tanner ME. Mechanistic Studies on CymD: A Tryptophan Reverse N-Prenyltransferase. Biochemistry. 2012;51:7733–7739. doi: 10.1021/bi3009054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer A, Li SM. Potential of a 7-dimethylallyltryptophan synthase as a tool for production of prenylated indole derivatives. Appl Microbiol Biotechnol. 2008;79:951–961. doi: 10.1007/s00253-008-1505-3. [DOI] [PubMed] [Google Scholar]
- 35.Rudolf JD, Wang H, Poulter CD. Multisite Prenylation of 4-Substituted Tryptophans by Dimethylallyltryptophan Synthase. J Am Chem Soc. 2013;135:1895–1902. doi: 10.1021/ja310734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MSC, Howlett BJ. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol Microbiol. 2004;53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 37.Kremer A, Li SM. A tyrosine O-prenyltransferase catalyses the first pathway-specific step in the biosynthesis of sirodesmin PL. Microbiology (Reading, U K ) 2010;156:278–286. doi: 10.1099/mic.0.033886-0. [DOI] [PubMed] [Google Scholar]
- 38.Zou H-X, Xie X, Zheng X-D, Li S-M. The tyrosine O-prenyltransferase SirD catalyzes O-, N-, and C-prenylations. Applied Microbiology and Biotechnology. 2011;89:1443–1451. doi: 10.1007/s00253-010-2956-x. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopalan S, Radke G, Evans M, Tomich JM. Synthesis of N-t-Boc-4-S-t-butyl-L-thiophenylalanine via palladium catalyzed cross-coupling reaction of N-t-Boc-4-iodo-L-phenylalanine with t-butylthiol or sodium t-butylthiolate. Synth Commun. 1996;26:1431. [Google Scholar]
- 40.Clavier S, Rist O, Hansen S, Gerlach LO, Hoegberg T, Bergman J. Preparation and evaluation of sulfur-containing metal chelators. Org Biomol Chem. 2003;1:4248–4253. doi: 10.1039/b307399h. [DOI] [PubMed] [Google Scholar]
- 41.Yao Z-J, Gao Y, Burke TR., Jr Preparation of L-Nα-Fmoc-4-[di-(tert-butyl)-phosphonomethyl]phenylalanine from L-tyrosine. Tetrahedron: Asymmetry. 1999;10:3727. [Google Scholar]
- 42.Hettwer S, Sterner R. A Novel Tryptophan Synthase β-Subunit from the Hyperthermophile Thermotoga maritima. Journal of Biological Chemistry. 2002;277:8194–8201. doi: 10.1074/jbc.M111541200. [DOI] [PubMed] [Google Scholar]
- 43.Yin W-B, Ruan H-L, Westrich L, Grundmann A, Li S-M. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. ChemBioChem. 2007;8:1154–1161. doi: 10.1002/cbic.200700079. [DOI] [PubMed] [Google Scholar]
- 44.Ruan H-L, Yin W-B, Wu J-Z, Li S-M. Reinvestigation of a cyclic dipeptide N-prenyltransferase reveals rearrangement of N-1 prenylated indole derivatives. ChemBioChem. 2008;9:1044–1047. doi: 10.1002/cbic.200700723. [DOI] [PubMed] [Google Scholar]
- 45.Saiki K, Mogi T, Ogura K, Anraku Y. In vitro heme O synthesis by the cyoE gene product from Escherichia coli. J Biol Chem. 1993;268:26041–26044. [PubMed] [Google Scholar]
- 46.Brown HC, Okamoto Y. Derective effects in aromatic substitution. XXX Electrophilic substituent constants. J Am Chem Soc. 1958;80:4979–4987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.