Abstract
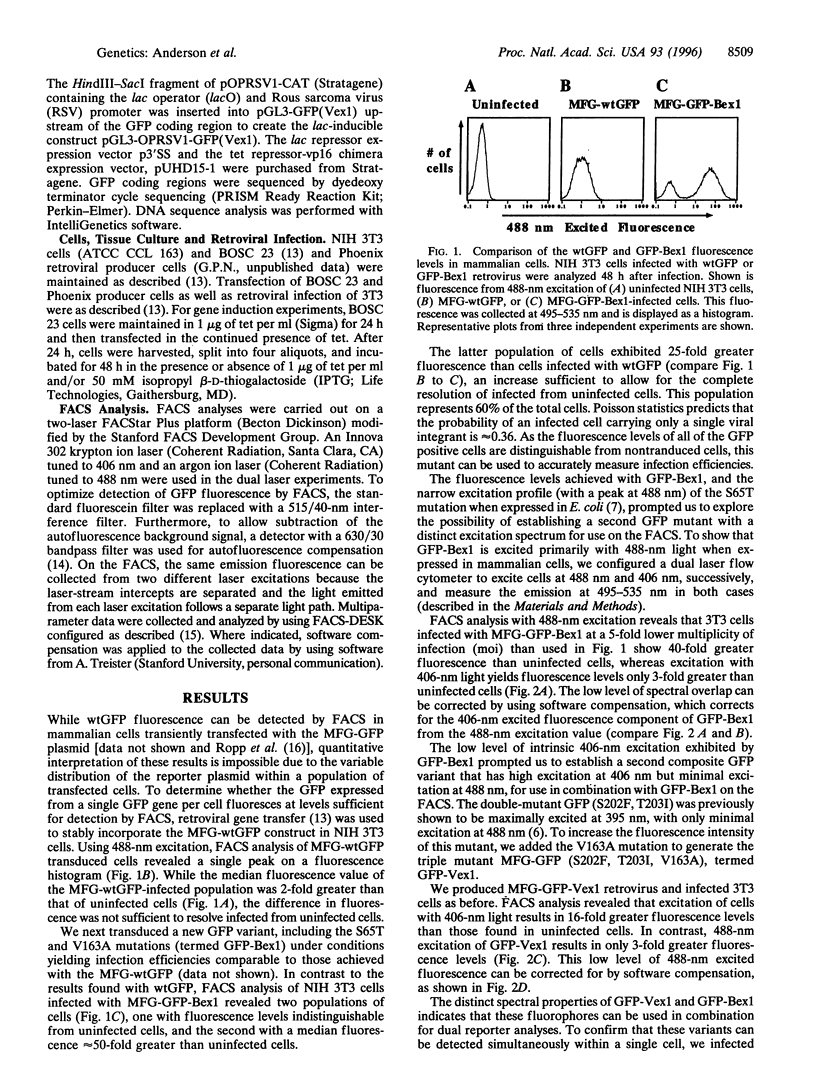
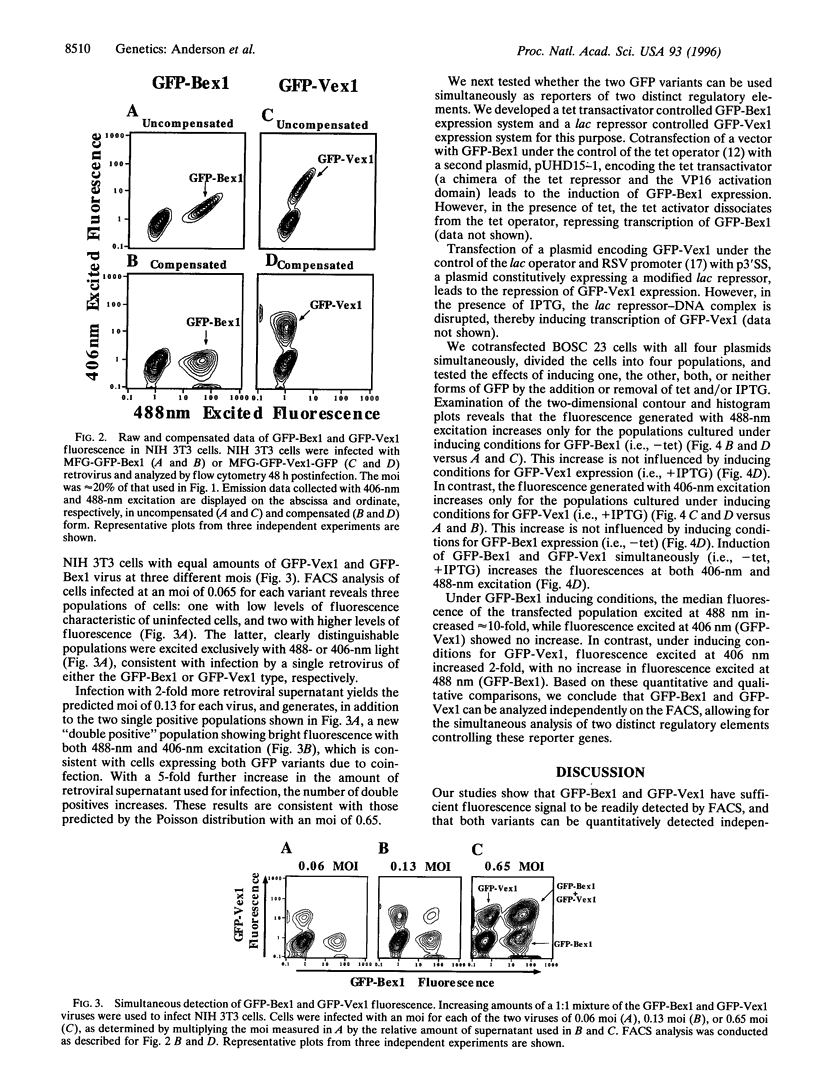
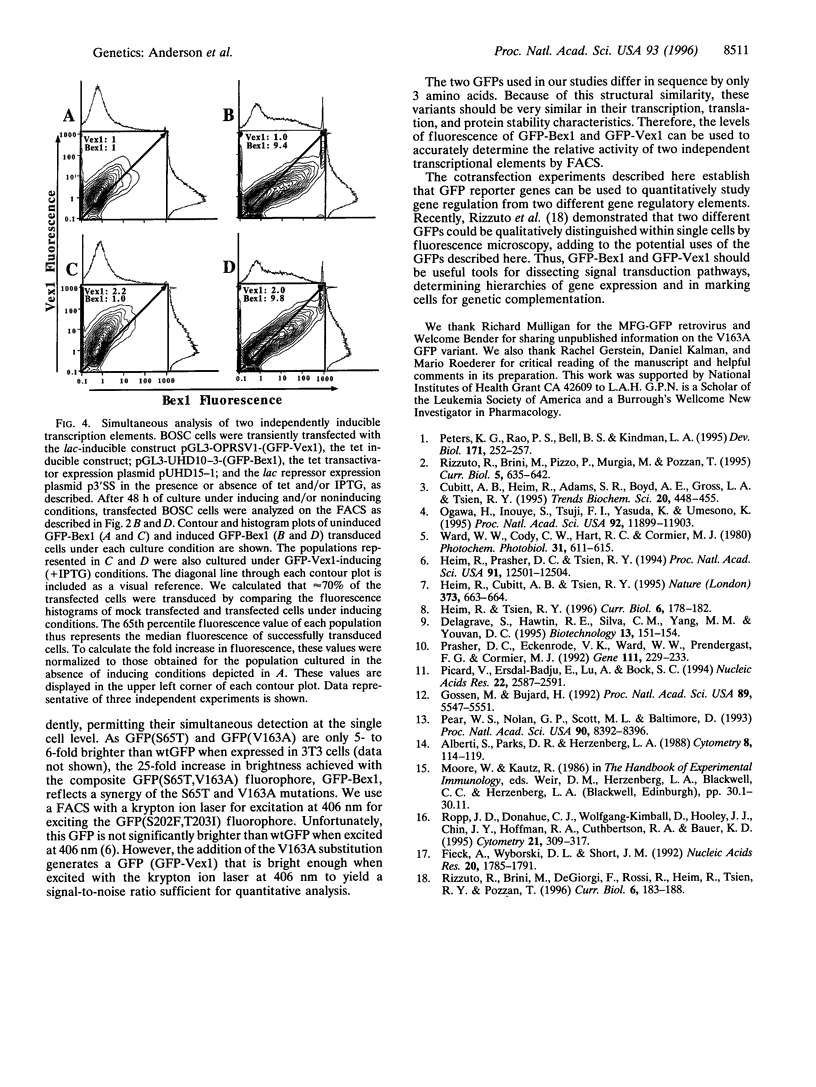
Green fluorescent protein (GFP) is widely used as a reporter gene in both prokaryotes and eukaryotes. However, the fluorescence levels of wild-type GFP (wtGFP) are not bright enough for fluorescence-activated cell sorting or flow cytometry. Several GFP variants were generated that are brighter or have altered excitation spectra when expressed in prokaryotic cells. We engineered two GFP genes with different combinations of these mutations, GFP(S65T,V163A) termed GFP-Bex1, and GFP(S202F,T203I,V163A) termed GFP-Vex1. Both show enhanced brightness and improved signal-to-noise ratios when expressed in mammalian cells and appropriately excited, compared with wtGFP. Each mutant retains only one of the two excitation peaks of the wild-type protein. GFP-Bex1 excites at 488 nm (blue) and GFP-Vex1 excites at 406 nm (violet), both of which are available laser lines. Excitation at these wavelengths allows for the independent analyses of these mutants by fluorescence-activated cell sorting, permitting simultaneous, quantitative detection of expression from two different genes within single mammalian cells.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberti S., Parks D. R., Herzenberg L. A. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987 Mar;8(2):114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- Cubitt A. B., Heim R., Adams S. R., Boyd A. E., Gross L. A., Tsien R. Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995 Nov;20(11):448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Delagrave S., Hawtin R. E., Silva C. M., Yang M. M., Youvan D. C. Red-shifted excitation mutants of the green fluorescent protein. Biotechnology (N Y) 1995 Feb;13(2):151–154. doi: 10.1038/nbt0295-151. [DOI] [PubMed] [Google Scholar]
- Fieck A., Wyborski D. L., Short J. M. Modifications of the E.coli Lac repressor for expression in eukaryotic cells: effects of nuclear signal sequences on protein activity and nuclear accumulation. Nucleic Acids Res. 1992 Apr 11;20(7):1785–1791. doi: 10.1093/nar/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y. Improved green fluorescence. Nature. 1995 Feb 23;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Heim R., Prasher D. C., Tsien R. Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Tsien R. Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996 Feb 1;6(2):178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Inouye S., Tsuji F. I., Yasuda K., Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W. S., Nolan G. P., Scott M. L., Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K. G., Rao P. S., Bell B. S., Kindman L. A. Green fluorescent fusion proteins: powerful tools for monitoring protein expression in live zebrafish embryos. Dev Biol. 1995 Sep;171(1):252–257. doi: 10.1006/dbio.1995.1276. [DOI] [PubMed] [Google Scholar]
- Picard V., Ersdal-Badju E., Lu A., Bock S. C. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994 Jul 11;22(13):2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasher D. C., Eckenrode V. K., Ward W. W., Prendergast F. G., Cormier M. J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992 Feb 15;111(2):229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Brini M., De Giorgi F., Rossi R., Heim R., Tsien R. Y., Pozzan T. Double labelling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr Biol. 1996 Feb 1;6(2):183–188. doi: 10.1016/s0960-9822(02)00451-7. [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Brini M., Pizzo P., Murgia M., Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995 Jun 1;5(6):635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Ropp J. D., Donahue C. J., Wolfgang-Kimball D., Hooley J. J., Chin J. Y., Hoffman R. A., Cuthbertson R. A., Bauer K. D. Aequorea green fluorescent protein analysis by flow cytometry. Cytometry. 1995 Dec 1;21(4):309–317. doi: 10.1002/cyto.990210402. [DOI] [PubMed] [Google Scholar]


