ABSTRACT
Nitrogen regulation in Escherichia coli is a model system for gene regulation in bacteria. Growth on glutamine as a sole nitrogen source is assumed to be nitrogen limiting, inferred from slow growth and strong NtrB/NtrC-dependent gene activation. However, we show that under these conditions, the intracellular glutamine concentration is not limiting but 5.6-fold higher than in ammonium-replete conditions; in addition, α-ketoglutarate concentrations are elevated. We address this glutamine paradox from a systems perspective. We show that the dominant role of NtrC is to regulate glnA transcription and its own expression, indicating that the glutamine paradox is not due to NtrC-independent gene regulation. The absolute intracellular NtrC and GS concentrations reveal molecular control parameters, where NtrC-specific activities were highest in nitrogen-starved cells, while under glutamine growth, NtrC showed intermediate specific activity. We propose an in vivo model in which α-ketoglutarate can derepress nitrogen regulation despite nitrogen sufficiency.
IMPORTANCE
Nitrogen is the most important nutrient for cell growth after carbon, and its metabolism is coordinated at the metabolic, transcriptional, and protein levels. We show that growth on glutamine as a sole nitrogen source, commonly assumed to be nitrogen limiting and used as such as a model system for nitrogen limitation, is in fact nitrogen replete. Our integrative quantitative analysis of key molecules involved in nitrogen assimilation and regulation reveal that glutamine is not necessarily the dominant molecule signaling nitrogen sufficiency and that α-ketoglutarate may play a more important role in signaling nitrogen status. NtrB/NtrC integrates α-ketoglutarate and glutamine signaling—sensed by the UTase (glnD) and PII (glnB), respectively—and regulates the nitrogen response through self-regulated expression and phosphorylation-dependent activation of the nitrogen (ntr) regulon. Our findings support α-ketoglutarate acting as a global regulatory metabolite.
INTRODUCTION
Integrative systems biology approaches provide comprehensive data on the physiological state of the cell that can reveal control parameters and limits of regulatory networks, inform predictive models, and guide metabolic engineering approaches. Ultimately, they can be used to understand adaptive processes. Nitrogen (N) is a major nutrient for cells, and nitrogen regulation and metabolism have been extensively studied (1).
The central nitrogen metabolic circuit is conserved in the vast majority of plants, archaea, and bacteria. Ammonium is the preferred nitrogen source of most bacteria, and nitrogen assimilation involves the enzymes glutamate dehydrogenase (GDH), glutamine synthetase (GS), and glutamate synthase (or glutamate-oxoglutarate amidotransferase [GOGAT]). Glutamate provides 88% of nitrogen for the synthesis of all nitrogen-containing cellular compounds (1) but also serves as a major compound to maintain the K+ pool (2). GDH reversibly aminates α-ketoglutarate (α-KG) to glutamate but has a higher Km for ammonium (>1 mM) than GS (Km < 200 µM) and appears to play a role in nitrogen assimilation only under energy- and carbon (C)-limiting conditions (2). GS aminates glutamate to glutamine, the amine donor for the remaining 12% of nitrogen-containing compounds. During growth in saturating glucose conditions, glutamate is nearly exclusively produced through amidotransfer from glutamine to α-KG by GOGAT (3), so that nitrogen assimilation from ammonium into glutamine and glutamate would rely essentially on GS. GS activity is posttranslationally regulated by adenylylation in response to the glutamine and α-KG levels (4). Nitrogen assimilation into glutamine is regulated primarily through expression and posttranslational modification of GS.
Glutamine is thought to be the main intracellular signal for nitrogen availability in Escherichia coli and most other bacteria. Its levels are sensed by the uridyltransferase/uridylyl-removing enzyme (UT/UR), which sits at the top of a well-studied regulatory cascade involving several proteins (5). At low glutamine levels, UT/UR uridylylates the paralogous transducers/regulators PII and GlnK, which are thought to have some redundant functions. PII-UMP stimulates the adenylate-removing activity of the bifunctional adenylylate transferase/removing enzyme (AT/AR) to increase the catalytic activity of GS. The nonuridylylated PII activates the adenyl transferase activity of GS. PII also controls the activity of the bifunctional enzyme NtrB, reducing its histidine kinase and stimulating its regulated phosphatase activity (6). These PII activities were shown to be modulated in vitro through direct binding of α-KG, ATP, and ADP, presumably to coordinate carbon and energy with nitrogen assimilation (7). The NtrB/NtrC two-component system (also called NRII and NRI) regulates transcription of the ntr regulon, comprising 27 operons involved in nitrogen scavenging, metabolism, and regulation (8). NtrC is a bacterial enhancer binding protein (bEBP) that activates the alternative σ54 RNA polymerase (9, 10). NtrB phosphorylates the NtrC receiver domain in response to nitrogen status, triggering conformational changes that favor formation of higher order NtrC oligomers, which is thought to be a prerequisite for transcription activation (11). However, NtrC can also act as a transcriptional repressor, and the role of its phosphorylation at repressed promoters is unclear. One consequence of this alternative regulatory mechanism is the strict requirement of an enhancer binding protein for the activation of σ54-dependent transcription (12).
The glnALG operon is central to nitrogen metabolism. It codes for GS, NtrB, and NtrC and comprises three promoters (glnAp1, glnAp2, and glnLp) with potentially three NtrC-dependent autofeedback loops (13). The cAMP receptor protein (CRP) also regulates transcription of glnAp1 and glnAp2 in response to C source availability (14).
Many genetic studies on the regulation of N assimilation have used glutamine as the sole N source (e.g., references 2 and 15–19). It was proposed that active import of glutamine by the high-affinity glutamine transporter (glnHPQ) is too slow to support fast growth and the necessary metabolic fluxes from glutamine, and so would account for intracellular N-limiting conditions that result in the strong NtrC-dependent upregulation of the ntr regulon (20). Ammonium is the preferred N source, allowing fast growth and resulting in the repression of the ntr regulon and the transport of glutamine by the glutamine transporter (glnHPQ) (21). More generally, slow growth of enteric bacteria such as Salmonella in a number of nonoptimal nitrogen sources, such as arginine and proline, in conjunction with strong NtrC-dependent transcription, has been defined as nitrogen limiting due to low intracellular glutamine concentrations (22).
Here, we report transcript, protein, and metabolite levels of the key molecules (transcripts, proteins, metabolites) involved in N regulation and metabolism in wild-type Escherichia coli and an isogenic glnG deletion, grown in batch cultures under N-rich (10 mM NH4Cl), glutamine (5 mM glutamine), and N-starved (3 mM NH4Cl) conditions. We reasoned that direct measurements of key intracellular molecule levels would (i) provide a more quantitative description of the N-adaptive physiological states and how these relate to transcription control, (ii) not suffer from potential pleiotropic effects in gene deletion studies, and (iii) provide multilevel control parameters of the N regulatory network. Strikingly, we found that intracellular glutamine levels under glutamine conditions were nearly 6-fold higher than for growth under nitrogen-rich conditions, which is inconsistent with glutamine being a sufficient and dominant signal of nitrogen status regulating NtrC-dependent transcription. We propose a modified regulatory network with a predominant role for α-ketoglutarate.
RESULTS
NtrC is required for optimal growth in both nitrogen-poor and -rich media.
We used wild-type (WT) E. coli strain NCM3722 and nitrogen regimes that allow comparisons with many previous studies of the Ntr system (e.g., see references 8, 23, 24, and 25). NCM3722 is prototrophic and a close reconstruction of the originally sequenced but genetically corrupted MG1655 E. coli K-12 strain, which suffers from various growth defects (23). We produced the NCM3722ΔglnG (ΔglnG) strain, which lacks the coding sequence for NtrC, through phage transduction from donor strain JW3841-1 (BW25113ΔglnG), provided by the Keio collection.
We directly quantified key metabolites, transcripts, and proteins under three different nitrogen regimes: two steady states and one transient physiological state. We used glucose as the carbon and energy sources throughout, in a defined minimal medium. Steady-state samples were taken during exponential growth (optical density at 600 nm [OD600] between 0.4 and 0.6). The initial nitrogen contents were 10 mM NH4Cl, 5 mM glutamine, and 3 mM NH4Cl. Ammonium is the preferred nitrogen source for most bacteria, and 10 mM NH4Cl is nitrogen replete (NH4 rich). Growth on glutamine as the sole nitrogen source (glutamine) is slow, and NtrC-regulated genes are strongly upregulated during glutamine-dependent growth. Hence, growth on glutamine has been extensively used in earlier studies as a model of nitrogen limitation. To capture changes in the physiological state of cells as they pass from nitrogen-replete to nitrogen-poor growth conditions, we carried out ammonium run-out experiments (nitrogen starved) from an initial 3 mM NH4Cl.
As expected, growth of WT and ΔglnG strains in NH4Cl was considerably faster than growth in glutamine (Fig. 1). We observed that following a period of balanced exponential growth in 3 mM NH4Cl, doublings slowed abruptly in WT and ΔglnG strains. For nitrogen-starved conditions, samples were taken 10 min after growth stopped (Fig. 1C).
FIG 1 .
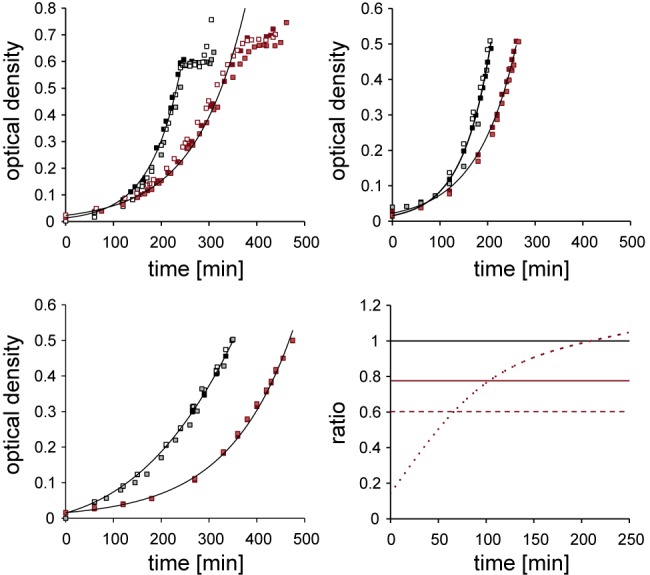
Growth curves of E. coli NCM3722 (black) and NCM3722ΔglnG (red) strains under different nitrogen regimes. Samples for targeted quantitative determination of metabolites, transcripts, and proteins in vivo were from these cultures during exponential growth in ammonium-rich conditions (top left), in glutamine (top right), or 10 min after growth arrest, indicating nitrogen-starved conditions (bottom left). Growth rates for ammonium-starved conditions (Table 1) were derived from logarithmic growth before nitrogen run out. Bottom right, growth rate differences (ΔOD600/Δt) between NCM3722 and NCM3722ΔglnG strains under different regimes: ammonium rich (long dash), glutamine (short dash), ammonium starved (solid).
The doubling time of NCM3722 in defined nitrogen-rich minimal medium was 42 min, which is identical with reported doubling times of wild-type E. coli (26) and very similar to those reported in early studies with wild-type E. coli (e.g., see reference 27) (Table 1). Significant transcriptomic, metabolomic, and proteomic differences have been reported between closely related E. coli strains grown under identical conditions, underlining the importance of using isogenic strains in comparative systems biology studies (28, 29). Our results support the use of NCM3722 as a reference strain for wild-type E. coli (23).
TABLE 1 .
Doubling times (g) and growth rates (μ) of wild-type NCM3722 and NCM3722ΔglnG strains under different growth regimesa
| Genotype | N status | g (min) ± SE | μ (h−1) ± SE |
|---|---|---|---|
| WT | NH4 rich | 42.1 ± 0.9 | 0.99 ± 0.02 |
| WT | Glutamine | 75.24 ± 0 | 0.55 ± 0 |
| WT | NH4 starved | 45.5 ± 3.2 | 1.04 ± 0.08 |
| ΔglnG | NH4 rich | 53.8 ± 0.96 | 0.77 ± 0.01 |
| ΔglnG | Glutamine | 95.7 ± 1.7 | 0.14 ± 0.01 |
| ΔglnG | NH4 starved | 74 ± 2.1 | 0.56 ± 0.02 |
Growth rates for ammonium-starved cells are derived from logarithmic growth before ammonium run-out (see Fig. 1). SE = 1 standard error of the mean.
For all conditions tested, the ΔglnG strain grew significantly slower than the WT. We conclude that NtrC confers fitness under nitrogen-rich conditions, in addition to its well-established role in adaptation to nitrogen starvation (8). The statistically indistinguishable growth rates of the WT in high- and low-ammonium media prior to run-out imply that ammonium uptake under those conditions is not limiting, consistent with ammonia crossing the bacterial membrane at neutral pH and supporting optimal growth. The NtrC-regulated high-affinity ammonium transporter AmtB is activated only at very low (below 30 µM) ammonium concentrations, since passive transport is sufficient at higher concentrations (30). Therefore, the growth phenotypes in Fig. 1 should not be a simple consequence of insufficient ammonium uptake. To test this, we measured ammonium depletion in the media during exponential growth under high and low initial ammonium concentrations (see Fig. S1 in the supplemental material). Ammonium consumption strictly correlated with growth, and ammonium uptake was indistinguishable between high- and low-ammonium conditions during the initial exponential growth phase, consistent with no dependence on AmtB. The early onset of slowed growth of the ΔglnG strain under both ammonium conditions is therefore not easily explained by a lack of intracellular ammonium.
The glutamine paradox.
Growth in glutamine was markedly slower than in media containing ammonium, for both the WT and ΔglnG strains. Unlike ammonia, glutamine cannot freely diffuse across the membrane and is thought to require import by the glutamine transporter encoded by the NtrC-regulated glnHPQ operon. To see if intracellular glutamine was limiting due to slow transport, we measured the intracellular concentrations of glutamine by liquid chromatography-mass spectrometry (LC/MS), after confirming that our sampling approach allowed accurate, rapid, and separate quantification of metabolites present in both the exo- and endometabolomes (see Fig. S2 in the supplemental material).
Intracellular glutamine concentrations were high under glutamine growth (Fig. 2). This calls into question the reported role of intracellular glutamine as a sufficient and dominant signal for nitrogen status, and so we quantified two other key metabolites in nitrogen assimilation, glutamate and α-KG. All samples had similar intracellular glutamate concentrations (Fig. 2), supporting the importance and homeostatic protection of the glutamate pool in enteric bacteria (2), but intracellular α-KG accumulated in nitrogen-starved and glutamine conditions. Interestingly, we also found α-KG in the supernatants of glutamine and nitrogen-starved cultures (data not shown), indicating active export of α-KG to prevent its hyperaccumulation in the cells. We conclude that the key metabolite concentrations of the nitrogen assimilation pathway (glutamine in particular) are not limited under glutamine growth and therefore cannot account for the slow-growth phenotype or the transcriptional activation of NtrC-dependent genes.
FIG 2 .
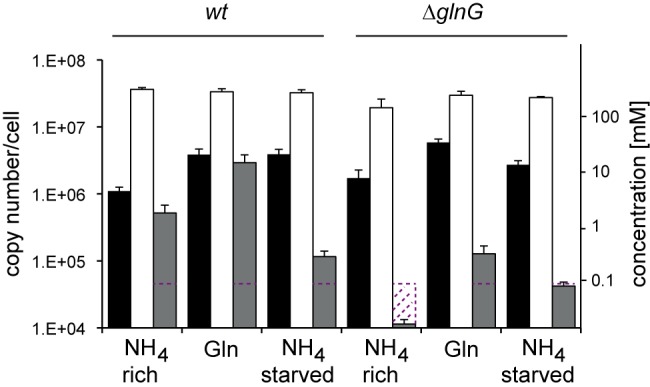
Intracellular concentrations (molecules/cell) of α-ketoglutarate (black), glutamine (white), and glutamine (gray) under different nitrogen regimes as indicated. Note logarithmic scale on the y axes. Red dotted line indicates quantification limit. Error bars indicate one standard error of the mean across minimally three biological replicates.
This apparent paradox calls for an alternative hypothesis to explain the glutamine phenotypes. To better understand the interplay between physiological state and nitrogen regulation by NtrC, we measured the α-KG, glutamate, and glutamine concentrations in the ΔglnG strain. The glutamine concentrations in the ΔglnG strain were at least 10-fold lower than those of the WT under nitrogen-rich and glutamine growth conditions and still measurably lower under starved conditions, although exact measurements of fold change were not possible, as levels were below the limit of quantification for nitrogen-rich and nitrogen-starved samples (Fig. 2). The slower growth in the ΔglnG strain under nitrogen-rich conditions may be a direct consequence of lacking glutamine. However, the relatively high glutamine concentration of the ΔglnG strain under glutamine growth suggests that glutamine limitation does not exclusively account for the severe growth phenotype under these conditions. During the course of the experiments under glutamine but not under ammonium growth, growth rate differentials between the WT and ΔglnG strains (Fig. 1D) indicate that relative growth rates of the ΔglnG strain partly recover compared to those of the WT, suggesting a potential role for NtrC during adaptation to glutamine growth. To better understand the role of NtrC in N adaptation, we turned our attention to the NtrC regulon and its distinct roles in the growth phenotypes, since metabolite concentrations of the central N pathway are not limiting (Fig. 2).
Revisiting the regulation of NtrC-dependent genes.
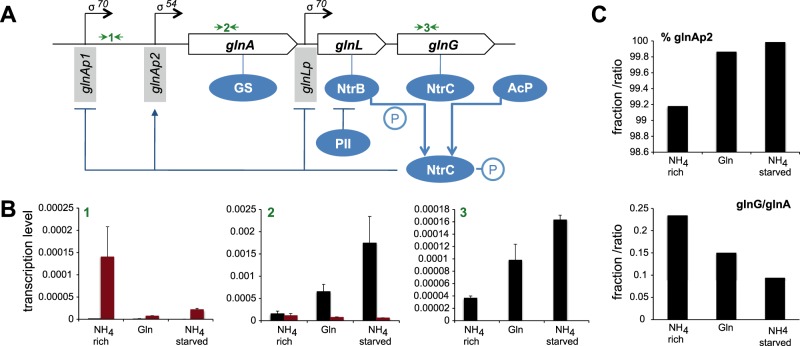
Regulated NtrC expression involves two negative and one positive autoregulatory feedback mechanisms through the glnAp1, glnLp, and glnAp2 promoters, to which NtrC binds within the glnALG operon (Fig. 3A). glnAp2 comprises two high-affinity enhancer NtrC binding sites (1 and 2) and two low-affinity NtrC binding sites further downstream (3 and 4), proposed to act as governor sites to limit glnA transcription at high phosphorylated NtrC (NtrC~P) concentrations (16). Further complexity at the glnA promoter is provided by CRP-cAMP, which can counteract NtrC-dependent regulation by activating glnAp1 and repressing glnAp2, thus integrating C and nitrogen source availability. CRP-cAMP also activates the glnH promoter (glutamine transporter) under high-glutamine and low-carbon conditions (14). The mechanisms of CRP-cAMP regulation at those promoters are complex, and we hypothesized that the high α-KG and glutamine levels under glutamine growth could explain the “glutamine paradox” of strong transcriptional activation of glnA despite clear glutamine sufficiency. We measured transcript levels of glnAp1, glnA, and glnG by real-time PCR to derive promoter activities of glnAp1 and glnAp2 and their relation to glnG transcription (Fig. 3B). The overall contribution of glnAp1 to glnA transcription was highest (0.8%) under nitrogen-rich conditions, indicating that nitrogen-dependent transcription control is dominated by NtrC, at least when cells are grown in glucose. The overall glnA transcript levels showed an inverse activity in response to the nitrogen status. Notably, glnA mRNA levels were lower during glutamine growth than for nitrogen-starved cells, although GS activity measurements suggested the highest output under glutamine (31, 32). The transcription profiles at glnAp1 and glnAp2 indicated that both repression and activation, respectively, were dependent on the nitrogen status and NtrC. The lack of transcription driven from glnAp2 and the elevated transcription levels from glnAp1 promoters in the ΔglnG strain confirmed this. The less well-characterized glnLp promoter showed a similar pattern to glnAp1 when the overall glnA transcript levels were taken into account (glnG/glnA), suggesting that NtrC negatively controls glnLp in response to the physiological status. These results indicate that NtrC, in response to nitrogen status, orchestrates expression of both the key nitrogen assimilation protein GS and itself through direct feedback control of glnAp1, glnAp2, and glnLp. While we cannot completely rule out a CRP-mediated mechanism affecting glnA expression, it appears that transcription from glnAp1 under glutamine growth conditions is regulated by NtrC.
FIG 3 .
Transcription from different NtrC-dependent promoters within the glnALG operon under different nitrogen regimes. (A) Scheme of the glnALG operon architecture and NtrC-mediated feedback loops acting at σ70- and σ54-dependent promoters. Black arrows indicate three primer pairs used for quantitative PCR of glnAp1 (1), glnA (2), and glnG (3). (B) mRNA levels relative to 16 s mRNA of E. coli NCM3722 (black) and NCM3722ΔglnG (red) strains comprising glnAp1 (1), glnA (2), and glnG (3) message. Transcript levels between E. coli NCM3722 and NCM3722ΔglnG strains confirm the regulatory roles of NtrC at those promoters. (C) The percentage of glnAp2 activity of total glnA transcription was derived from glnA transcripts minus glnAp1 transcripts. The ratio of glnG transcripts relative to glnA transcripts reflects on the regulation at the glnLp promoter.
Intracellular concentrations of NtrC and GS.
Phosphorylation of the receiver domain of NtrC shifts the equilibrium from inactive to active NtrC conformers (33). Therefore, transcriptional control by NtrC depends both on NtrC abundance and conformation. NtrB specifically phosphorylates NtrC under nitrogen-limiting conditions but also dephosphorylates NtrC when in complex with nonuridylated PII in a regulated phosphatase reaction. The degree of NtrC phosphorylation depends on the level of glutamine, which determines the uridylylation state of PII, and also on ATP, ADP, and α-KG, which are allosteric effectors modifying PII’s ability to regulate NtrB activities, at least in vitro (7).
In order to correlate transcription control with NtrC concentration in vivo, we employed a quantitative targeted proteomics approach using multiple-reaction monitoring mass spectroscopy (MRM-MS). MRM-MS allows precise measurements of protein levels and can be used for absolute quantification if combined with a purified isotopically labeled standard (34). This approach provides arguably the most accurate and reliable quantitation of target proteins (35), particularly when using purified labeled proteins as a standard rather than just labeled synthetic peptides (as this also controls for the efficiency of the tryptic digestion step). We established a robust sampling and MRM-MS workflow to determine the intracellular concentrations of NtrC and GS that could be generally applicable for Gram-negative bacteria (see File S1 in the supplemental material for a detailed protocol and quality control). We purified a number of histidine-tagged, 13C6,15N2-Lys- and 13C6,15N4-Arg-labeled proteins as internal standards (NtrC, GS, IlvE, isocitrate dehydrogenase [IDH], and fumarate and nitrate reductase regulator [FNR]). IlvE, IDH, and FNR served as control proteins to monitor global protein variations that occur in different physiological states but are NtrC independent (8, 36). Intracellular copy numbers have been estimated in E. coli under broadly comparable conditions for IDH, FNR, and GS and compare well with our results (37). Protein standard purities for NtrC and GS were 89% and 92% pure, respectively, and isotopic labeling values were 100% and 97%. Across 18 biological samples, total protein extraction in 7 M urea was 92.91% (standard error [SE] of 0.58%), and trypsin digestion efficiency was 89.25% (SE of 1.95%). The main nonsoluble and trypsin-resistant proteins were determined by Edman degradation to be the outer membrane porins OmpF and PhoE (see File S1 in the supplemental material). We conclude that our MRM-MS samples comprised complete peptide sets of NtrC, GS, IlvE, IDH, and FNR derived from the cytoplasmatic E. coli proteome. We calculated the copy numbers per cell for each protein based on the reported cell volume of E. coli of 1 fl and a cell number of 1.1 × 109/ml/OD600 (Table 2) (38) (with 1,000 molecules per cell corresponding to a concentration of 1.61 µM).
TABLE 2 .
Protein concentrations given as copy number/cell under different nitrogen regimes as determined by MRM-MS with protein standard absolute quantificationa
| Genotype | N status | Copy no./cell ± SE |
||||
|---|---|---|---|---|---|---|
| GS | NtrC | IDH | FNR | IlvE | ||
| WT | NH4 rich | 18,691 ± 536 | 499 ± 7 | 25,446 ± 989 | 8,480 ± 413 | 8,336 ± 406 |
| WT | Glutamine | 66,705 ± 1,084 | 2,091 ± 78 | 35,277 ± 1,686 | 4,183 ± 134 | 4,116 ± 132 |
| WT | NH4 starved | 34,681 ± 1,746 | 1,313 ± 106 | 29,656 ± 3,540 | 2,051 ± 49 | 6,940 ± 544 |
| ΔglnG | NH4 rich | 4,456 ± 221 | 0 ± 0 | 31,838 ± 3,345 | 10,750 ± 947 | 10,568 ± 931 |
| ΔglnG | Glutamine | 3,428 ± 183 | 0 ± 0 | 46,587 ± 1,426 | 5,651 ± 49 | 5,555 ± 230 |
| ΔglnG | NH4 starved | 3,675 ± 171 | 0 ± 0 | 48,615 ± 2,677 | 5,406 ± 694 | 6,302 ± 190 |
GS, glutamine synthetase; NtrC, nitrogen regulation protein C; IDH, isocitrate dehydrogenase; FNR, fumarate and nitrate regulator; IlvE, branched-chain amino acid transferase. SE = 1 standard error of the mean.
GS was most abundant under glutamine growth and least abundant in high ammonium, in line with previous transcriptional reporter gene studies. NtrC was less abundant than GS, supporting a functional role of a putative attenuation site in the glnA-glnL intergenic region (32). NtrC was absent in the ΔglnG strain, and GS levels were drastically reduced in contrast to IDH, FNR, and IlvE, confirming the dominant and direct role NtrC plays in regulating both glnA and itself. For growing cells, glnA and glnG transcript levels correlated well with GS and NtrC protein levels. NtrC and GS levels in starved cells were lower than those seen for cells grown in glutamine, despite higher glnA and glnG transcript levels, indicating a higher specific NtrC transcription activity under nitrogen-starved conditions than in glutamine growth.
We could not directly determine the phosphorylation state of NtrC by MRM-MS, probably because NtrC~P is labile (39). However, the transcript/NtrC ratios provide a relative measure of NtrC-specific transcription activity (NtrCA), while the inverse relationship (NtrC/transcript) relates to NtrC-specific repression activity (NtrCR) at various promoters, as NtrCA depends on adopting an active higher oligomeric state, while NtrCR may not (40, 41). Transcription from the simpler glnK promoter depends exclusively on the activation of NtrC through phosphorylation, and therefore transcription levels from glnKp in relation to NtrC levels report on NtrCA (42). NtrCA was lowest in nitrogen-rich conditions as expected (Fig. 4A).
FIG 4 .
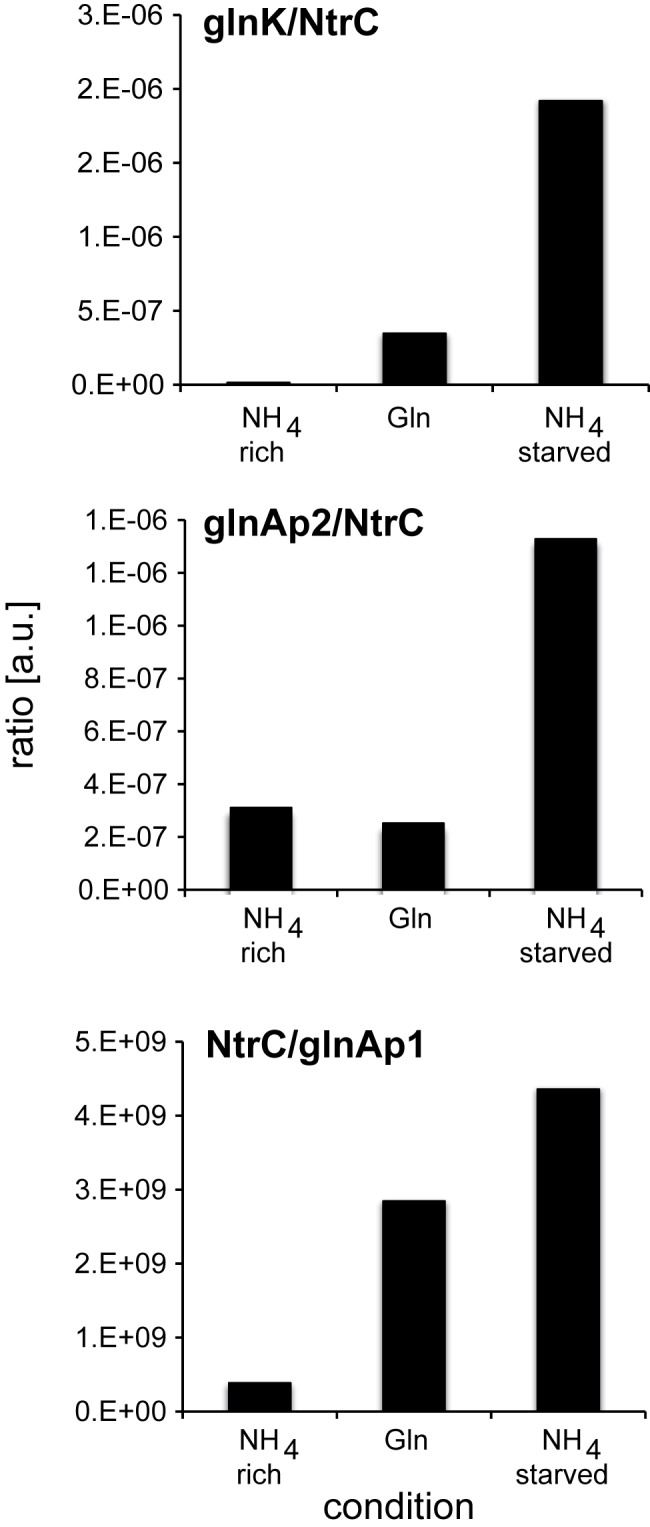
Specific NtrC activation (NtrCA) and repression (NtrCR) activities at the glnK, glnAp2, and glnAp1 promoters. Top and middle, transcript levels from the glnK and plnAp2 promoters per copy number of NtrC molecules reflect on NtrCA; bottom, repression activity at glnAp1 is expressed as NtrC/glnAp2 expression levels (NtrCR). The y axis is in arbitrary units.
Surprisingly, the NtrCA was markedly higher in nitrogen-starved conditions than in glutamine conditions, suggesting that only a subpopulation of NtrC was active in the latter conditions (Fig. 4). This finding is consistent with the high intracellular glutamine levels that would result in PII-mediated repression of NtrC phosphorylation. Notably, NtrCA levels under glutamine growth were higher than NtrCA levels in nitrogen-rich conditions, despite a 5.6-fold-higher glutamine concentration, indicating that signaling controlling formation of NtrC~P depends also on other factors. The glutamine/α-ketoglutarate ratio is thought to determine primarily the phosphorylation state of NtrC (43). However, the glutamine/α-KG ratios in nitrogen-rich, glutamine-starved, and nitrogen-starved conditions were 0.47, 0.76, and 0.057 (Fig. 2), respectively, indicating no simple correlation between these ratios and NtrCA.
DISCUSSION
NtrC plays a major role in metabolic regulation and adaptation.
The quantitative data on metabolites, proteins, and transcripts allow the assessment of adaptive processes for nitrogen assimilation and regulation in a physiological context. NtrC plays a key role in responding to nitrogen source and nitrogen availability, since growth under all conditions tested here was significantly slower for the ΔglnG strain (Table 1) than for the WT strain. When grown with the preferred nitrogen source, ammonium, glutamine levels in the ΔglnG strain are at least 35-fold lower than in the WT, which could directly account for slow growth. The low constitutive expression level of glnA from the glnAp1promoter (Fig. 3) and resulting low level of GS (Table 2) support this interpretation. The cellular nitrogen demand (based on 0.3 pg [dry weight]/cell for E. coli and 14% nitrogen content) (44), and assuming that glutamate is exclusively anabolised via the GS/GOGAT cycle when grown in glucose-replete conditions (3), is 1.8 × 109 glutamine molecules per cell. The NCM3722 strain had 18,691 GS molecules under these conditions (Table 2), and assuming a kcat value of 50 s−1 as determined in vitro (45), this would be sufficient to support a generation time of 42 min (producing 2.36 × 109 glutamine molecules/42 min), whereas the fewer GS molecules in the NCM3722ΔglnG strain (4,456; producing 5.61 × 108 glutamine molecules/42 min) presumably would not.
One surprising metabolic finding was the high intracellular glutamine level for cells grown on glutamine as a sole nitrogen source (Fig. 2). Therefore, growth under glutamine is not nitrogen limiting in the strict sense of glutamine availability. This observation distinguishes glutamine growth from other nitrogen-limiting nitrogen sources such as aspartate, proline, and arginine, for which intracellular glutamine levels are approximately 10-fold lower than growth on ammonium, at least in the related Salmonella enterica serovar Typhimurium (22). The high-affinity glutamine transporter (glnHPQ) probably imports most of the glutamine under glutamine growth. The glnH promoter architecture (comprising glnHp1 and glnHp2) is very similar to the glnA promoter (glnAp1 and glnAp2) (14) and thought to be activated simultaneously by NtrC from the p1 sites. In agreement with previous findings, we therefore suggest that, similar to GS, the glutamine transporter is highly expressed under glutamine growth (14), thus allowing for rapid glutamine import, since, unlike other amino acid transporters, the high-affinity glutamine transport system (glnHPQ) is not repressed by its substrate but by other N-containing compounds, including ammonium (21).
While slow growth and high levels of NtrC-dependent transcription are common on glutamine, aspartate, proline, and arginine, the molecular basis of transcription under glutamine is probably very distinct. For true nitrogen-limiting substrates (e.g., aspartate, arginine), the import and conversion of amino acids to feed into the central nitrogen metabolic pathway, comprising α-KG, glutamate, and glutamine, is likely to be slow, resulting in nitrogen deficiency, low glutamine levels, and phosphorylation of NtrC via UTase, PII, and NtrB signaling. In contrast, under glutamine growth, the conversion of intracellular glutamine into glutamate via GOGAT should be rapid. It is currently unclear if the high α-KG concentrations when grown under glutamine (Fig. 2) are caused by insufficient efflux of α-KG from the Krebs cycle into nitrogen assimilation and/or if GDH deaminates glutamate to α-KG (46). This would in effect constitute a nitrogen assimilation reverse flux from glutamine to α-KG, in which one glutamine and one α-KG would be converted to two glutamate molecules by GOGAT followed by deamination to α-KG by GDH. In either case, the 5.7-fold-higher α-KG level under glutamine growth compared to growth under ammonium-rich conditions (Fig. 2) suggests a direct in vivo role for α-KG in regulating signaling to NtrC (see below). Glutamine and α-KG levels were both significantly higher for growth in glutamine than for growth in ammonium, while glutamate levels were the same, suggesting that neither C nor N limitation was responsible for slow growth. As far as we know, neither high intracellular glutamine nor α-KG levels are toxic, and so the slow growth could be a consequence of as-yet-unidentified global effects caused by these or downstream metabolites. For instance, α-KG was recently shown to act as a signal mediating global anabolic and catabolic regulation through inhibition of the production of cyclic AMP and therefore to link α-KG levels with cell growth rates (47). We observed that initial growth of the ΔglnG strain in glutamine was particularly slow but reached a growth rate similar to that of the WT after several hours (Fig. 1D), suggesting that slow, presumably global metabolic adaptations to the glutamine regime are involved that are dependent on NtrC.
Signaling to NtrC dominates transcriptional regulation in response to the nitrogen status.
NtrCA was 400-fold higher at the glnK promoter under ammonium-starved than ammonium-rich conditions, providing a numerical in vivo approximation of the equilibrium shift induced by NtrB-mediated phosphorylation of NtrC (Fig. 4A). Under ammonium-rich and -starved conditions, we quantified averages of 499 and 1,313 NtrC molecules/cell (0.83 µM and 2.1 µM), respectively. Given the still appreciable number of NtrC molecules present under ammonium-rich conditions, this may suggest that NtrC phosphorylation, rather than increases in copy number, is the determining factor controlling transcription from the glnK promoter, at least under our experimental conditions. It has been proposed that as cells run out of ammonium, the strong glnAp2 promoter would first result in increased NtrC expression, which, above a certain threshold and under inducing conditions, would then activate transcription at the glnK and nac promoters (16). Recalling that NtrC activity requires the formation of higher oligomers from inactive dimers, and the associated higher-order concentration dependency of such self-association, then NtrC concentration should play an important role in NtrCA formation. However, it is unclear if the 2.5-fold-lower NtrC concentration observed under ammonium-rich than under ammonium-starved conditions would be sufficient to exclude prompt expression of glnK following a shift from nitrogen-rich to nitrogen-poor conditions that would not require prior glnG expression.
NtrCA at the glnK and glnAp2 promoters was markedly different under ammonium-rich conditions, while NtrCA levels at the glnAp2 promoter (Fig. 4B) were similar under ammonium-rich and glutamine conditions. Because σ54-dependent transcription—in contrast to potential transcription factor-independent transcription from σ70-type RNA polymerase (48)—strictly depends on active enhancer binding proteins, we propose that even under these conditions, a subpopulation of NtrC exists in an active conformation. This may imply that the molecular kinetics governing transcription at the glnAp2 and glnK promoters are different, because the total transcript levels at glnK and glnAp2 under ammonium-starved conditions are similar (0.00252 and 0.00174 relative to 16S mRNA, respectively).
The apparent low NtrCA level at the glnAp2 promoter under glutamine growth could be a direct consequence of nonphosphorylated NtrC repression through binding to the glnAp2 governor sites p3 and p4, although further experiments would be needed to address this point. NtrCR activities at the glnAp1 promoter (Fig. 4C), between glutamine and when starved for ammonium, reflect NtrCA activities at the glnK and glnAp2 promoters. This correlation may support an in vivo repressor function of NtrC at glnAp1 that would largely be NtrC~P dependent (32).
α-KG may override glutamine signaling to NtrC.
We propose that the elevated α-KG level observed under glutamine conditions is, at least in part, directly responsible for the observed high GS and NtrC levels. In vitro studies show that α-KG can counteract glutamine signaling by directly binding to PII (7). Although the physiological role of this observation was unclear, the dependency of transcription under glutamine growth conditions with elevated NtrCA activities from various NtrC-dependent promoters indicates that a subpopulation of NtrC must be phosphorylated. We propose that, in vivo, elevated α-ketoglutarate concentrations can at least partly override glutamine-dependent signaling to NtrC phosphorylation, most likely through PII binding of α-ketoglutarate resulting in the inhibition of the regulated phosphatase of the PII-NtrB complex on NtrC. As a net result, and contrary to the high intracellular glutamine concentrations when cells are grown with glutamine as the sole nitrogen source, a proportion of NtrC exists in its active, phosphorylated state. Many studies have reported upregulation of NtrC-dependent genes on growth on glutamine as a nitrogen source; the α-ketoglutarate effect would account for this because of the dominant role of NtrC~P in activating the σ54 polymerase at various promoters. Our results indicate that NtrC-dependent gene upregulation on glutamine is not because of the hitherto assumed slow glutamine uptake, which would signal nitrogen deficiency, but because high intracellular glutamine results in metabolic fluxes from glutamine to increase the α-KG level, which acts as a metabolic signal of nitrogen regulation.
Our findings could also explain a previously reported inconsistency between the expected results for glutamine, glutamate, and α-KG and NtrC-dependent transcription. Goss and colleagues (49) pointed out that when E. coli is deleted for the structural genes for GOGAT (gltBD), the glutamine concentrations should be high (since it cannot be depleted rapidly), which could not account for slow growth and the lack of NtrC-dependent activity. When grown on glutamine as a sole nitrogen source, NtrC-dependent activities were highly upregulated (histidase and GS) in WT E. coli as expected but not in gltD mutants. A high glutamine concentration in a gltD mutant could not produce glutamate from glutamine and therefore not reverse nitrogen assimilation flux to α-KG, resulting in derepressed NtrC. Further, growth of the same GOGAT-deficient strains on glutamate as a sole nitrogen source also resulted in highly activated NtrC-dependent activities (49). Neither phenotype (Ntr−) could be explained by glutamine starvation inducing the NtrB/NtrC system, but they are fully consistent with high α-KG concentrations derepressing NtrC in vivo. Because GOGAT and GDH are NtrC independent, the expectation would be that the α-KG concentrations would also be elevated under glutamine growth in the ΔglnG strain, consistent with our measurements (Fig. 2).
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Strain NCM3722 was used as wild-type Escherichia coli (23). The NCM3722ΔglnG strain was constructed by transduction using the P1vir bacteriophage as described (50), with JW3839 from the Keio collection serving as the donor strain and NCM3722 as the recipient strain (51). Cells were grown in Gutnick (33.8 mM KH2PO4, 77.5 mM K2HPO4, 5.74 mM K2SO4, 0.41 mM MgSO4), supplemented with Ho-LE trace elements (52) and 0.4% glucose, and containing either 10 mM NH4Cl (ammonium rich), 3 mM NH4Cl (ammonium starved), or 5 mM glutamine. Glutamine was prepared freshly to avoid spontaneous slow hydrolysis into glutamate and ammonia (53). Steady-state samples were taken at OD600 values between 0.4 and 0.6. For nitrogen-starved conditions, samples were taken 10 min after growth stopped. We monitored growth until the nitrogen-limiting culture ceased to increase in optical density and sampled the amounts indicated below for the multiomic investigation of nitrogen assimilation regulation. Ammonium concentrations were determined using the Aquaquant ammonium quantification kit (Merck Millipore), according to instructions. All experiments were carried out in three independent biological replicates.
Transcription assays.
Cell samples were taken from each time point and RNA stabilized with Qiagen RNA protect reagent. Total RNA was extracted with an Invitrogen RNA purification kit and stored in RNase-free water at −80°C. cDNA was generated from 100 ng RNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Primer and probe mixtures were custom designed from Invitrogen (TaqMan gene expression Assays). Real-time PCR was performed on an ABI 7500 Fast real-time PCR machine. A 16S RNA gene was chosen as the internal control gene and showed exactly the same expression levels across the conditions tested. The relative expression ratios were calculated using the delta-delta method (PerkinElmer).
Proteomics.
A detailed proteomics protocol is provided in File S1 in the supplemental material.
Metabolomics.
Samples for extraction and endometabolome analysis were taken by rapid filtration, using a protocol adopted from Bolten et al. (54) and fully described in Behrends et al. (55). Briefly, 10 ml of cell suspension was harvested by vacuum filtration (filter, PTFE, 0.45-µm pore size, 47 mm diameter, with stand and magnetic filter funnel [Pall, Ann Arbor]) and washed with 5 ml of 1.2× Gutnick medium not containing carbon and nitrogen sources. The filter was then transferred to a precooled (−40°C) 50-ml reaction tube containing 10 ml methanol-acetonitrile-H2O (2:2:1, vol/vol/vol) and frozen in liquid nitrogen. The whole procedure took <40 s. To ensure full lysis, all extracts were subjected to two freeze-thaw cycles and sonication. After the removal of the filter, the extracts were centrifuged to pellet the cellular debris, dried in a vacuum concentrator (Eppendorf; 45°C), and resuspended in 1 ml methanol-acetonitrile-H2O (2:2:1, vol/vol/vol). Eighty percent of the sample was routinely used for nuclear magnetic resonance (NMR), 10% was used for LC/MS, and 10% was kept as backup. For NMR measurements of the endometabolome, the samples were dried in a vacuum concentrator, resuspended in NMR buffer (90% 2H2O, 1 mM sodium 2,2-dimethyl-2-silapentane-2H6-5-sulfonate [DSS], 5 mM NaN3), and transferred to a 5-mm NMR tube. For LC/MS analysis, samples were dried in a vacuum concentrator, resuspended in 50 µl methanol-acetonitrile-H2O (2:2:1, vol/vol/vol), and mixed with 25 µl of 15 µM 13C5,15N2-glutamine in the same solvent for quantification. For analysis of the exometabolome, samples were obtained by centrifuging 500 µl of culture (16,000 × g, 1 min, room temperature). A total of 480 µl of the supernatant was mixed with 120 µl buffer (5 mM DSS and 25 mM NaN3 in 100% 2H2O).
Nuclear magnetic resonance spectroscopy.
Spectra were acquired on a Bruker Avance DRX600 NMR spectrometer (Bruker BioSpin, Rheinstetten, Germany), with a magnetic field strength of 14.1 T and resulting 1H resonance frequency of 600 MHz, equipped with a 5-mm inverse probe following an approach described in Beckonert et al. (56). One-dimensional spectra of extracts were routinely acquired with 768 transients with 8 dummy scans using a standard NOESYPR1d water suppression pulse sequence, while supernatants were acquired with 64 scans. After acquisition, spectra were Fourier transformed and phased in iNMR (Nucleomatica, Molfetta, Italy). The full-resolution data were exported as ASCII and imported into Matlab (MathWorks) using an in-house code for further analysis. Peaks were assigned using spectral information from previous studies (57) and the BioMagResBank online database (58).
Liquid chromatography mass spectrometry.
LC/MS was performed using a method adapted from Spagou et al. (59). Briefly, the samples were chromatographed on an Acquity UPLC (ultra-performance liquid chromatography) system with an Acquity UPLC BEH HILIC (1.7-µm, 2.1- by 100-mm) column (Waters Corp., Milford, MA) at 40°C. Separation of glutamine from glutamate and α-ketoglutarate was achieved with gradient elution of 90/10% ACN-H2O (A) and 50/50% ACN-H2O (B), both containing 0.1% (vol/vol) formic acid and 10 mM ammonium acetate, at a flow rate of 0.4 ml/min. Starting conditions were 99.0% A and 1% B for 1.0 min, changing linearly to 100% B over the next 11 min, after which the solvent composition returned to starting conditions over 0.1 min, followed by reequilibration for 3.9 min prior to the next injection. Spectra were acquired in negative mode on a Waters Xevo TQ-S tandem quadrupole mass spectrometer. Monitored parent-to-fragment ion transitions were 147 to >84 for glutamine, 148 to >84 for glutamate, and 154 to >89 for 13C5,15N2-glutamine. The cone voltage and collision energy were set to 25 V and 15 V, respectively. Data were exported as CDF files and integrated in Matlab using in-house code based on Behrends et al. (60).
SUPPLEMENTAL MATERIAL
Growth and ammonium consumption of NCM3733 in ammonium-rich and ammonium-poor media. Following inoculation into defined media with initial NH4Cl concentrations of 10 mM and 3 mM, growth over time (x axis) was measured by OD600 and NH4Cl concentrations measured from the supernatants to determine consumption (secondary axis), using Aquaquant. Red and black circles show OD600 and NH4Cl values for growth on 3 mM NH4Cl, respectively. Blue and green circles indicate OD600 and NH4Cl for growth in 10 mM NH4Cl. Download
NMR spectra of culture supernatant and cell extract grown in 5 mM Gln and 0.4% glucose. Compared to the supernatant fraction, the lack of detectable glucose in the cell extract fraction indicates no or negligible carryover of medium components into the cell extract fraction. Intensity levels were normalized to the levels of the Gln multiplet at δ2.45 ppm. Ace, acetate; Gln, glutamine; Glu, glutamate; Glc, glucose; Glcα, glucose, α-anomeric proton; Glcβ, glucose, β-anomeric proton; Tre, trehalose. Download
Additional protocols. Download
ACKNOWLEDGMENTS
We thank J. D. Rabinowitz for providing strain NCM3722 and the NBRP-E. coli at NIG for strains from the ASKA(−) and Keio collections.
This work was funded by the BBSRC (BB/G020434/1). Michal Komorowski was also funded by the Polish Academy of Sciences.
Footnotes
Citation Schumacher J, Behrends V, Pan Z, Brown DR, Heydenreich F, Lewis MR, Bennett MH, Razzaghi B, Komorowski M, Barahona M, Stumpf MPH, Wigneshweraraj S, Bundy JG, Buck M. 2013. Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. mBio 4(6):e00881-13. doi:10.1128/mBio.00881-13.
REFERENCES
- 1. Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155–176 [DOI] [PubMed] [Google Scholar]
- 2. Yan D. 2007. Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:9475–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Helling RB. 1994. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 176:4664–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown MS, Segal A, Stadtman ER. 1971. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the P II -regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 68:2949–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okano H, Hwa T, Lenz P, Yan D. 2010. Reversible adenylylation of glutamine synthetase is dynamically counterbalanced during steady-state growth of Escherichia coli. J. Mol. Biol. 404:522–536 [DOI] [PubMed] [Google Scholar]
- 6. Jiang P, Ninfa AJ. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang P, Ninfa AJ. 2009. Alpha-ketoglutarate controls the ability of the Escherichia coli PII signal transduction protein to regulate the activities of NRII (NrB) but does not control the binding of PII to NRII. Biochemistry 48:11514–11521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U. S. A. 97:14674–14679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rombel I, North A, Hwang I, Wyman C, Kustu S. 1998. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harb. Symp. Quant. Biol. 63:157–166 [DOI] [PubMed] [Google Scholar]
- 10. Schumacher J, Zhang X, Jones S, Bordes P, Buck M. 2004. ATP-dependent transcriptional activation by bacterial PspF AAA+ protein. J. Mol. Biol. 338:863–875 [DOI] [PubMed] [Google Scholar]
- 11. Rappas M, Schumacher J, Niwa H, Buck M, Zhang X. 2006. Structural basis of the nucleotide driven conformational changes in the AAA+ domain of transcription activator PspF. J. Mol. Biol. 357:481–492 [DOI] [PubMed] [Google Scholar]
- 12. Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M. 2008. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Microbiol. 68:538–546 [DOI] [PubMed] [Google Scholar]
- 13. Ninfa AJ, Magasanik B. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 83:5909–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mao XJ, Huo YX, Buck M, Kolb A, Wang YP. 2007. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res. 35:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atkinson MR, Ninfa AJ. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431–447 [DOI] [PubMed] [Google Scholar]
- 16. Atkinson MR, Pattaramanon N, Ninfa AJ. 2002. Governor of the glnAp2 promoter of Escherichia coli. Mol. Microbiol. 46:1247–1257 [DOI] [PubMed] [Google Scholar]
- 17. van Heeswijk WC, Molenaar D, Hoving S, Westerhoff HV. 2009. The pivotal regulator GlnB of Escherichia coli is engaged in subtle and context-dependent control. FEBS J. 276:3324–3340 [DOI] [PubMed] [Google Scholar]
- 18. Shiau SP, Schneider BL, Gu W, Reitzer LJ. 1992. Role of nitrogen regulator I (NtrC), the transcriptional activator of glnA in enteric bacteria, in reducing expression of glnA during nitrogen-limited growth. J. Bacteriol. 174:179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sasse-Dwight S, Gralla JD. 1988. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl. Acad. Sci. U. S. A. 85:8934–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bender RA, Magasanik B. 1977. Regulatory mutations in the Klebsiella aerogenes structural gene for glutamine synthetase. J. Bacteriol. 132:100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Willis RC, Iwata KK, Furlong CE. 1975. Regulation of glutamine transport in Escherichia coli. J. Bacteriol. 122:1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ikeda TP, Shauger AE, Kustu S. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259:589–607 [DOI] [PubMed] [Google Scholar]
- 23. Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothstein DM, Pahel G, Tyler B, Magasanik B. 1980. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc. Natl. Acad. Sci. U. S. A. 77:7372–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thakur CS, Brown ME, Sama JN, Jackson ME, Dayie TK. 2010. Growth of wildtype and mutant E. coli strains in minimal media for optimal production of nucleic acids for preparing labeled nucleotides. Appl. Microbiol. Biotechnol. 88:771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kashket ER, Brodie AF. 1962. Effects of near-ultraviolet irradiation on growth and oxidative metabolism of bacteria. J. Bacteriol. 83:1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vijayendran C, Polen T, Wendisch VF, Friehs K, Niehaus K, Flaschel E. 2007. The plasticity of global proteome and genome expression analyzed in closely related W3110 and MG1655 strains of a well-studied model organism, Escherichia coli-K12. J. Biotechnol. 128:747–761 [DOI] [PubMed] [Google Scholar]
- 29. Maharjan RP, Ferenci T. 2005. Metabolomic diversity in the species Escherichia coli and its relationship to genetic population structure. Metabolomics 1:235–242 [Google Scholar]
- 30. Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T. 2012. Need-based activation of ammonium uptake in Escherichia coli. Mol. Syst. Biol. 8:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reitzer LJ, Magasanik B. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785–792 [DOI] [PubMed] [Google Scholar]
- 32. Pahel G, Rothstein DM, Magasanik B. 1982. Complex glnA-glnL-glnG operon of Escherichia coli. J. Bacteriol. 150:202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Volkman BF, Lipson D, Wemmer DE, Kern D. 2001. Two-state allosteric behavior in a single-domain signaling protein. Science 291:2429–2433 [DOI] [PubMed] [Google Scholar]
- 34. Picotti P, Aebersold R. 2012. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9:555–566 [DOI] [PubMed] [Google Scholar]
- 35. Elschenbroich S, Kislinger T. 2011. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Mol. Biosyst. 7:292–303 [DOI] [PubMed] [Google Scholar]
- 36. Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu P, Vogel C, Wang R, Yao X, Marcotte EM. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25:117–124 [DOI] [PubMed] [Google Scholar]
- 38. Kubitschek HE, Friske JA. 1986. Determination of bacterial cell volume with the Coulter counter. J. Bacteriol. 168:1466–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamberov ES, Atkinson MR, Chandran P, Ninfa AJ. 1994. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J. Biol. Chem. 269:28294–28299 [PubMed] [Google Scholar]
- 40. Yan D, Kustu S. 1999. “Switch I” mutant forms of the bacterial enhancer-binding protein NtrC that perturb the response to DNA. Proc. Natl. Acad. Sci. U. S. A. 96:13142–13146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. 2006. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 20:1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Atkinson MR, Blauwkamp TA, Bondarenko V, Studitsky V, Ninfa AJ. 2002. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J. Bacteriol. 184:5358–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Magasanik B. 1989. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie 71:1005–1012 [DOI] [PubMed] [Google Scholar]
- 44. Neidhardt FC, Curtiss R, III, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 45. Witmer MR, Palmieri-Young D, Villafranca JJ. 1994. Probing the catalytic roles of n2-site glutamate residues in Escherichia coli glutamine synthetase by mutagenesis. Protein Sci. 3:1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharkey MA, Oliveira TF, Engel PC, Khan AR. 2013. Structure of NADP-dependent glutamate dehydrogenase from Escherichia coli: reflections on the basis of coenzyme specificity in the family of glutamate dehydrogenases. FEBS J. 280(18):4681–4692. 10.1111/febs.12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T. 2013. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500:301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buck M, Bose D, Burrows P, Cannon W, Joly N, Pape T, Rappas M, Schumacher J, Wigneshweraraj S, Zhang X. 2006. A second paradigm for gene activation in bacteria. Biochem. Soc. Trans. 34:1067–1071 [DOI] [PubMed] [Google Scholar]
- 49. Goss TJ, Perez-Matos A, Bender RA. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183:6607–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atlas RM. 2010. Handbook of microbiological media, vol 1 CRC Press, Boca Raton, FL [Google Scholar]
- 53. Bender RA, Macaluso A, Magasanik B. 1976. Glutamate dehydrogenase: genetic mapping and isolation of regulatory mutants of Klebsiella aerogenes. J. Bacteriol. 127:141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C. 2007. Sampling for metabolome analysis of microorganisms. Anal. Chem. 79:3843–3849 [DOI] [PubMed] [Google Scholar]
- 55. Behrends V, Williams KJ, Jenkins VA, Robertson BD, Bundy JG. 2012. Free glucosylglycerate is a novel marker of nitrogen stress in Mycobacterium smegmatis. J. Proteome Res. 11:3888–3896 [DOI] [PubMed] [Google Scholar]
- 56. Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. 2007. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2:2692–2703 [DOI] [PubMed] [Google Scholar]
- 57. Behrends V, Ebbels TM, Williams HD, Bundy JG. 2009. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl. Environ. Microbiol. 75:2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. 2008. BioMagResBank. Nucleic Acids Res. 36:D402–D408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Spagou K, Wilson ID, Masson P, Theodoridis G, Raikos N, Coen M, Holmes E, Lindon JC, Plumb RS, Nicholson JK, Want EJ. 2011. HILIC-UPLC-MS for exploratory urinary metabolic profiling in toxicological studies. Anal. Chem. 83:382–390 [DOI] [PubMed] [Google Scholar]
- 60. Behrends V, Tredwell GD, Bundy JG. 2011. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 415:206–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth and ammonium consumption of NCM3733 in ammonium-rich and ammonium-poor media. Following inoculation into defined media with initial NH4Cl concentrations of 10 mM and 3 mM, growth over time (x axis) was measured by OD600 and NH4Cl concentrations measured from the supernatants to determine consumption (secondary axis), using Aquaquant. Red and black circles show OD600 and NH4Cl values for growth on 3 mM NH4Cl, respectively. Blue and green circles indicate OD600 and NH4Cl for growth in 10 mM NH4Cl. Download
NMR spectra of culture supernatant and cell extract grown in 5 mM Gln and 0.4% glucose. Compared to the supernatant fraction, the lack of detectable glucose in the cell extract fraction indicates no or negligible carryover of medium components into the cell extract fraction. Intensity levels were normalized to the levels of the Gln multiplet at δ2.45 ppm. Ace, acetate; Gln, glutamine; Glu, glutamate; Glc, glucose; Glcα, glucose, α-anomeric proton; Glcβ, glucose, β-anomeric proton; Tre, trehalose. Download
Additional protocols. Download