ABSTRACT
In this work, we show that Clostridium difficile phage ϕC2 transduces erm(B), which confers erythromycin resistance, from a donor to a recipient strain at a frequency of 10−6 per PFU. The transductants were lysogenic for ϕC2 and contained the erm(B) gene in a novel transposon, Tn6215. This element is 13,008 bp in length and contains 17 putative open reading frames (ORFs). It could also be transferred at a lower frequency by filter mating.
IMPORTANCE
Clostridium difficile is a major human pathogen that causes diarrhea that can be persistent and difficult to resolve using antibiotics. C. difficile is potentially zoonotic and has been detected in animals, food, and environmental samples. C. difficile genomes contain large portions of horizontally acquired genetic elements. The conjugative elements have been reasonably well studied, but transduction has not yet been demonstrated. Here, we show for the first time transduction as a mechanism for the transfer of a novel genetic element in C. difficile. Transduction may also be a useful tool for the genetic manipulation of C. difficile.
INTRODUCTION
Clostridium difficile is an important pathogen that causes diarrhea in humans and animals and has been detected in environmental and food samples (1). C. difficile infection is difficult to resolve and control because antibiotic therapy is a predisposing factor, and spore formation allows the bacterium to persist in the environment. The emergence of several virulent strains in the last 10 years has prompted detailed comparative genomic analyses that have revealed considerable genome plasticity, a large percentage of which is mediated by horizontally acquired elements, such as conjugative transposons (CTn) and phages (2, 3).
Conjugative transposons are modular mobile genetic elements that contain regions required for recombination, conjugative transfer, and regulation. In addition to these core areas, they also contain accessory regions not directly required for transfer and transposition, such as antibiotic resistance genes (4). Elements initially found in C. difficile are able to transfer to other species; for example, Tn5397 can transfer from C. difficile to Bacillus subtilis (5) and Enterococcus faecalis (6).
Some C. difficile phages are capable of lysogeny and integration of phage genome(s) into the host chromosome. Prophage sequences are commonly found in C. difficile isolates, many of which form infectious particles either spontaneously or following mitomycin C induction (7, 8). The five sequenced C. difficile phage genomes are modular; distinct clusters of homologous genes for virion structure, cell lysis, lysogeny control, and DNA replication and recombination were identified. Homologous genes were found in phages of other clostridia, Streptococcus pneumoniae, Staphylococcus aureus, Lactobacillus johnsonii, and Bacillus cereus. The phage genomes are also mosaic, meaning each genome is a unique composite of gene modules interspersed with nonhomologous sequences (9–13). Diversity in phage genomes can arise from recombination events involving host or other phage genomes, plasmids, or transposons. The ubiquity of prophage sequences in C. difficile strains suggests that phage infection is widespread, indicating the potential for transduction. The acquisition of antimicrobial resistance and virulence genes is mediated by phage transduction in several clinically important Gram-positive bacteria, such as S. aureus (14, 15) and E. faecalis (16, 17). Opportunities for phage-mediated gene transfer occur during bacterial colonization or infection, as lysogenization has been demonstrated in vivo for phages of Streptococcus pyogenes and C. difficile (18–20). In this work, we investigated the ability of phage to transduce antimicrobial resistance markers between C. difficile strains and showed that phage ϕC2 mediates the transfer of a novel mobile element conferring erythromycin resistance.
RESULTS
ϕC2 mediates transfer of erythromycin resistance from CD80 to CD062.
Donor and recipient strains were chosen for transduction experiments based on ϕC2 susceptibility, determined previously (21), and on either the presence or absence of antibiotic resistance genes, determined in this study (Table 1). Phage ϕC2 (21) was propagated in the erythromycin- and tetracycline-resistant donor strain CD80 to a concentration of 107 to 108 PFU/ml and used to infect the antibiotic-susceptible strains CD38, CD062, CD839, and CD6938 to test for transduction of resistance genes. Erythromycin-resistant CD062 cells were isolated (using either crude or purified phage) at a frequency of 1.2 × 10−6 ± 1.0 × 10−6 (mean ± standard deviation) per PFU. Crude and purified phage suspensions of ≤6 × 107 PFU/ml and a multiplicity of infection (MOI) of less than 0.02 did not result in transduction. There was no growth on control plates when no recipients were present, showing that the phage preparation did not contain any bacterial contaminants. Tetracycline resistance was not transferred to CD062, and erythromycin or tetracycline resistance transfer did not occur for the other three strains tested. The donor and all Ermr transductants contained erm(B), as determined by PCR with primer pair E5/E6 (see Table S1 in the supplemental material). The amplicon sequences from the donor and transductants were identical. A control mixture of CD80 filtrate that did not contain ϕC2 and CD062 did not result in erythromycin-resistant derivatives of CD062, indicating that ϕC2 is necessary for the transfer of erythromycin resistance from CD80 to CD062. Two transductants, CD062E1 and CD062E2, were randomly selected for further analysis. Both strains were lysogenized with ϕC2, as they produced plaques after mitomycin C induction and contained ϕC2 integrase (results not shown).
TABLE 1 .
C. difficile isolates in this study
| Isolate | Purpose in this study | Relevant genotype/phenotypea; ribotype | Reference |
|---|---|---|---|
| CD80 | Donor | ΔtcdA ΔtcdB Ermr Tetr; 010 | This study |
| CD38 | Recipient (transduction) | tcdA+ tcdB+ | This study |
| CD062 | Recipient (transduction) | ΔtcdA ΔtcdB; 010 | (21) |
| CD839 | Recipient (transduction) | ΔtcdA tcdB+ | (21) |
| CD6938 | Recipient (transduction) | tcdA+ tcdB+ | (21) |
| CD062R11 | Recipient (conjugation) | Rifr (Rifr mutant of CD062) | This study |
| CD062E1 | Transductant | Ermr; 010 | This study |
| CD062R1112 | Transconjugant | Ermr Rifr | This study |
| CD062R1146 | Transconjugant | Ermr Rifr | This study |
| CD062R1170 | Transconjugant | Ermr Rifr | This study |
Ermr, erythromycin resistant; Rifr, rifampin resistant; Tetr, tetracycline resistant.
The erm(B) gene resides in a novel integrative mobile element, Tn6215.
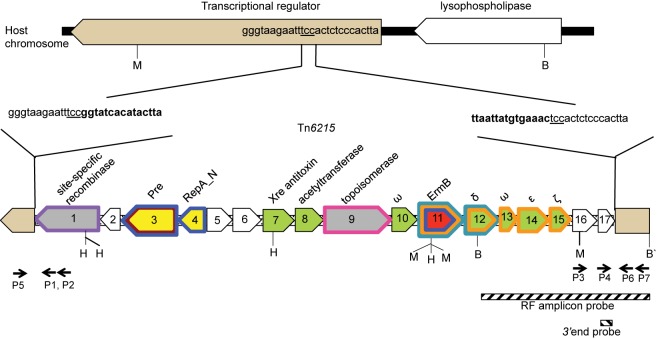
The DNA sequences of the transferable erm(B) gene and the flanking regions were obtained from the donor and transductants, and the DNA region containing the junction between the transferred element and the host genome was located by Southern hybridization (see Fig. S1 in the supplemental material) and obtained by comparing the sequence in the transductants and element-free recipients. The genetic organization of this element is shown in Fig. 1. The element had the genetic organization of a mobilizable transposon (Fig. 1) and was subsequently shown to be capable of transfer by a conjugation-like mechanism; it was assigned transposon number Tn6215 (22). It is 13,008 bp in length, with 17 open reading frames (ORFs) (Table S2). Tn6215 encodes a putative serine site-specific recombinase (SSR) (Fig. 1, ORF1) that is likely to be involved in excision and integration. However, no circular forms of the transposon in the donor or transductant were detected by PCR using outward-facing primers specific to the left and right ends of the integrated transposon. Using primers P5/P6 that are specific for the host target site, two amplicons (543 bp and 13.5 kb) were obtained from the donor and transductants, while only the 543-bp amplicon was detected in the recipient (data not shown). Sequencing of the left and right ends of the 13.5-kb amplicons from the donor and transductants showed Tn6215 integrated within a transcriptional regulator gene (Fig. 1). The sequence of the 543-bp amplicon derived from the donor and transductants was identical to that of the target site in the recipient, indicating that either regenerated integration sites or empty sites were present along with the integrated transposon in the donor and transductants. Sequence comparisons of the transposon-host junctions, the host target site, and the regenerated integration site showed duplication of the target sequence TCC, which is present at either end of the integrated transposon (Fig. 1 and Fig. 2).
FIG 1 .
Genetic structure of Tn6215 and site of host chromosome integration. In recipient strain CD062, a putative transcriptional regulator gene was uninterrupted. In donor strain CD80 and transductant strain CD062E1, Tn6215 was integrated into the putative transcriptional regulator gene and the site of integration, TCC, was duplicated at either end. Predicted functional modules of the 13-kb transposon are color coded as follows: red, resistance; yellow, mobilization and replication; green, stability; gray, recombination; and white, indeterminate. Restriction sites are indicated as follows: B, BsrGI; H, Hyp99I; and M, MfeI. Probes used in this study are shown as hatched bars and are shown under their target regions. Homologues in other mobile elements and plasmids are indicated by colored outlines of putative genes as follows: purple, Tn5397; brown, pMV158; blue, CTnBST; pink, CTnDOT; orange, pSM19035; and turquoise, Tn5398. Primers P1 to P7 flanking host and transposon junctions are indicated by black arrows.
FIG 2 .

Sequences of host target site and resulting host and transposon junctions in transductants. The host target site is underlined, and Tn6215 sequences are in red.
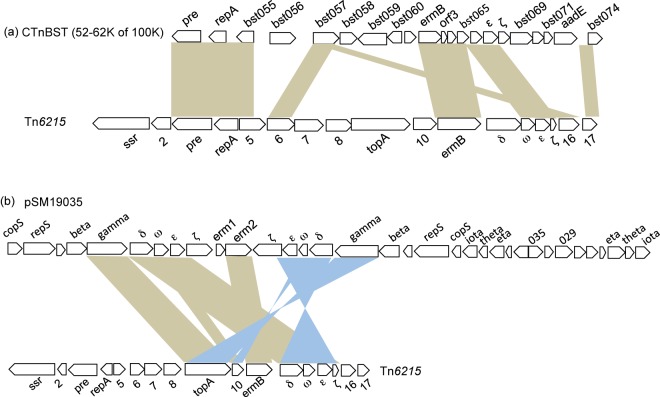
ORF3 is a homologue of the mobilization protein TnpZ from Tn4451 of Clostridium perfringens (see Table S2 in the supplemental material) (23). However, ORF3 is truncated, and the upstream DNA sequence lacks inverted repeats/palindromes for DNA binding (oriT/RSA regions) and, therefore, probably lacks activity compared to TnpZ and the archetypal Pre (24, 25). ORF4 shares the conserved domain RepA_N with replication proteins from plasmids of low G+C Gram-positive bacteria (26). A RepA_N homologue was found in a Bacteroides conjugative transposon, CTnBST, which also contained pre and erm(B) (Fig. 3a) (27). ORF9 is homologous to topoisomerase I, which is required for excision in the conjugative transposons CTnDOT of Bacteroides fragilis and CTnPg1 of Porphyromonas gingivalis (28, 29). Although the amino acid sequence relatedness of the Tn6215 topoisomerase to homologues from CTnDOT and CTnPg1 (Tn6161) was only moderate (29% identity over 644 amino acids [aa] for both) they shared conserved domains for DNA and ATP binding, strand cleavage and joining sites, C4 zinc finger binding, and C-terminal repeats.
FIG 3 .
Nucleotide sequence comparisons of Tn6215 to the Bacteroides conjugative transposon CTnBST (a) and to the S. pyogenes plasmid pSM19035 (b). Sand-colored boxes indicate regions of similarity of >94%, and blue triangles indicate regions of inversions.
Genes associated with toxin-antitoxin systems (TAS) were identified in Tn6215. Two putative type II proteic TAS are present: a GCN5-related N-acetyltransferase (GNAT) family enzyme-Xre system and the ω-ε-ζ system. A GNAT-Xre TAS was found in plasmids by comparative genomic analyses only but has not been experimentally verified (30). The ω-ε-ζ system, together with δ and erm(B), was originally described in pSM19035 (31), a multidrug resistance-encoding plasmid from Streptococcus pyogenes. This TAS is important for plasmid maintenance (31–33), has homologues in Gram-positive and Gram-negative bacteria, and is associated with antibiotic-resistant strains of Enterococcus faecium and methicillin-resistant S. aureus (MRSA) (34, 35). However, the toxin gene (ζ) in Tn6215 is truncated and is predicted to encode only 52/287 amino acids of the complete ζ in pSM19035. Nucleotide sequence comparison of pSM19035 and Tn6215 showed high sequence similarity and similar gene organization of topA, erm(B), δ, ω, ε, and ζ (Fig. 3b). While an erm(B)-δ-ω-ε-ζ module has not been described in transposons, an erm(B)-ω module was found in Tn5398 of C. difficile strain 630, and an erm(B)-ω-ε-ζ (truncated) module was described in a CTn1-like element in C. difficile strains M68 and 2007855 (36). The M68 genome has nucleotide sequence similarity to Tn6215 genes 1 to 5, 7, 10 to 12, 13 to 16, and 17 in distinct regions as determined by BLASTn (nucleotides 1760244 to 1756132, 1753494 to 1752687, 3801235 to 3803491, 3800108 to 3801236, and 1750963 to 1750400, respectively). The previously annotated orf298 of Tn5398 (37) is predicted as δ in this study.
Tn6215 was transferred by filter mating.
To determine whether Tn6215 is conjugative, we carried out filter mating experiments using the ϕC2-free strain CD80 as the donor and the rifampin-resistant derivative of CD062, CD062R11, as the recipient. Transconjugants were selected on brain heart infusion (BHI) medium containing rifampin (25 mg/liter) and erythromycin (10 mg/liter). Erythromycin-resistant transconjugants appeared at a frequency of 1.8 × 10−9 per CFU of the donor. Southern hybridization with the RF amplicon probe (Fig. 1) containing a Tn6215-host junction confirmed the integration of Tn6215 into the transconjugants (see Fig. S1 in the supplemental material) and showed that Tn6215 entered the genome at the same site in the transconjugants as it occupied in the donor (Fig. 1; Fig. S1). Southern hybridization with a probe specific only for the right end of Tn6215 (Fig. 1) showed that there were at least two copies of the element in the donor, CD80, but only one in the CD062 transductants and transconjugants (Fig. S2). Unlike when Tn6215 is transferred by transduction, the transconjugants did not contain a copy of ϕC2.
DISCUSSION
This is the first report of gene transfer by phage transduction between C. difficile strains. Erythromycin resistance contained within Tn6125 was transferred by ϕC2 from CD80 to CD062. This element could also be transferred by a conjugation-like mechanism by filter mating, resulting in Ermr Tets transconjugants but at a much lower frequency. Transductants were lysogens of ϕC2, whereas transconjugants did not contain the phage. Once transferred, Tn6215 existed as a single copy in both transductants and transconjugants at the same integration site. Although we did not detect circularized Tn6215, empty or regenerated sites in donors and transductants were detected along with integrated Tn6215. The most likely explanation for this is that the transposon excises at a low frequency, resulting in a small number of individual cells that contain a regenerated target site. Presumably the circular form is present in too low a concentration to detect by PCR, as it will be lost from the cell, whereas the regenerated target will remain.
There are two putative type II proteic TAS in Tn6215, a GCN5-related N-acetyltransferase (GNAT) family enzyme-Xre system and the ω-ε-ζ system. However, in the latter system, the Zeta toxin is truncated and contains only 52 amino acids of the N-terminal region, and previous work has shown that more than 79 amino acids are required for function (31). Therefore, the toxin is unlikely to be functional, but the antitoxin Epsilon is intact. A system where just the antitoxin is present has been observed in phage T4, where the Dmd protein (antitoxin) is required for successful infection of hosts containing chromosomal toxin/antitoxin systems (38). A dmd T4 phage mutant was unable to propagate in host cells because phage infection led to the degradation of unstable antitoxins (RlnB or LsoB) and activation of toxins (RlnA or LsoA) that resulted in inhibition of growth in host cells (38). Possibly the Tn6215 epsilon antitoxin has a similar role in stabilizing host cells under conditions that trigger toxin activity.
GNAT can have a range of enzymatic functions, including resistance to antibiotics, regulation of sporulation (39), and cell wall recovery (40), but there is no evidence in the literature of toxicity mediated by GNAT. While some transcriptional regulators and/or antitoxins have a helix-turn-helix (HTH)_XRE domain (41, 42), the putative XRE located next to GNAT suggests that it functions as a transcriptional regulator of the GNAT gene rather than as an antitoxin. Further experiments are needed to determine the functions of the genes encoding the putative TAS GNAT-Xre and Epsilon-Zeta in Tn6215.
Although all four strains tested as recipients were susceptible to ϕC2 infection, only one of the four acquired erm(B) by transduction, indicating that the transduction conditions may not be optimal. Susceptibility to infection by a transducing phage does not necessarily result in transduction, as shown in other studies (17, 43). A possible explanation for the inability to transfer Tn6215 to the other strains is that they lack a transposon integration site. However, PCR and sequencing analysis indicated that the integration site was intact. Further experiments using more donors and recipients in various growth phases and screening for the transfer of more genetic markers are needed to determine how prevalent phage transduction is as a mechanism of horizontal and antibiotic resistance gene transfer in C. difficile.
MATERIALS AND METHODS
Bacterial strains and phage.
All C. difficile isolates were stored either as stocks in brain heart infusion (BHI) broth (Oxoid) with 20% (vol/vol) glycerol (Sigma) at −70°C or as working spore stocks in cooked-meat medium (Oxoid) and were grown in BHI broth or BHI agar supplemented with 5% horse blood in an anaerobic cabinet as previously described (21). The relevant properties of five clinical C. difficile isolates (from Sir Charles Gairdner Hospital, Perth, Western Australia) with known susceptibility to ϕC2 infection (21) are shown in Table 1. PCR detection of erm(B), tet(M), Tn916, Tn5397, tcdA, and tcdB was carried out under the general PCR conditions described below using genomic DNA (gDNA) and the primers listed in Table S1 in the supplemental material.
The temperate phage ϕC2 was propagated in a donor strain, CD80, using log-phase cultures and anaerobe basal agar as previously described (21). Phage suspensions were concentrated by using polyethylene glycol (PEG) and chloroform (crude phage suspension). Purified phage was prepared in a preformed CsCl density gradient (44).
Microdilution for antimicrobial MIC determination.
The determination of the antimicrobial susceptibilities of C. difficile strains was based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (45). Twofold dilutions of antimicrobials (chloramphenicol, erythromycin, and tetracycline; Sigma) were made in BHI broth and in 100-µl aliquots per well of a 96-well plate. The final antimicrobial concentrations ranged from 0 to 256 mg/liter. C. difficile cultures (16 to 18 h) were diluted to a final concentration of ≈105 CFU/ml. The cultures were incubated anaerobically (80% N2, 10% H2, and 10% CO2) at 37°C for 48 h, and the MIC of each antimicrobial for each bacterial strain was determined. The antimicrobials tested were chosen based on reports of resistance genes in C. difficile (5, 46, 47).
Genomic DNA preparation and PCR reactions.
C. difficile strains were grown for 18 to 20 h in 5 ml BHI broth supplemented with appropriate antibiotics for gDNA extraction using the Gentra Puregene yeast/bacteria kit (Qiagen). Three DNA polymerases, Crimson Taq, Phusion high-fidelity PCR master mix, and Phire II DNA polymerase (all New England Biolabs), were used for PCR reactions in this study as indicated in Table S1 in the supplemental material. The annealing temperatures and extension times of primers are shown in Table S1. The typical reaction conditions for PCRs using crimson Taq were 1× crimson Taq reaction buffer, 0.2 µM deoxynucleoside triphosphates (dNTPs), 0.125 U crimson Taq DNA polymerase, and 20 ng DNA in a 25-µl volume. The typical cycling conditions were 95°C for 30 s, 35 cycles of 95°C for 30 s, primer-specific temperature for 40 s, 68°C for a primer-specific number of seconds, and a final extension of 68°C for 5 min. The multiplex PCR mixture contained 0.5 µM each primer for the gene of interest, 50 nM each primers PS13/PS14, 1× Crimson Taq reaction buffer, 0.26 µM dNTPs, 0.625 U crimson Taq DNA polymerase, and 20 ng DNA in 25 µl. The cycling conditions were 95°C for 30 s, 35 cycles of 95°C for 30 s, primer-specific temperature for 40 s, 68°C for 1 min, and a final extension of 68°C for 5 min. The typical reaction conditions for PCR using Phusion were 1× Phusion master mix, 0.5 µM of each primer, and 1 to 20 ng DNA in a 20-µl volume. The typical cycling conditions were 98°C for 30 s, 35 cycles of 98°C for 5 s, primer-specific temperature for 10 s, 72°C for a primer-specific number of seconds, and a final extension of 72°C for 5 min. The typical reaction conditions for PCR of DNA ligation reactions using Phire II DNA polymerase were 1× reaction buffer, 200 µM dNTPs, 2 µl ligation reaction buffer, 0.5 µM each primer, and 0.4 µl polymerase. The cycling conditions were 98°C for 30 s, 35 cycles of 98°C for 5 s, primer-specific temperature for 5 s, 72°C for a primer-specific number of seconds, and a final extension of 72°C for 1 min.
Phage transduction and analysis of transductants.
Transduction was carried out anaerobically (80% N2, 10% H2, and 10% CO2), except for the centrifugation steps. Log-phase cultures of the recipients were mixed with either the crude phage suspension or purified phage (see above) to achieve an MOI of 0.02 to 0.3 in a volume of 1 ml. Phage-free filtrates of the donor were used for some experiments; uninfected bacterial lawns of CD80 were resuspended in phage buffer, filtered, and processed with PEG and chloroform as for phage-infected lawns (44). Bacterial and phage concentrations were standardized to 1 × 107 to 5 × 107 CFU/ml and 1 × 107 to 5 × 107 PFU/ml, respectively. A recipient-only control containing only bacteria and phage buffer was included in each experiment. Phage mixed with the recipient was incubated for 1 h at 37°C and then centrifuged at 14,000 × g for 30 s. The supernatants were removed, and the cells were washed in 1 ml BHI broth. The washing was repeated, and the cells were resuspended in 150 µl of BHI broth. The cells were spread plated in 50-µl volumes onto BHI agar plates supplemented with either tetracycline (10 mg/liter) or erythromycin (50 mg/liter) and incubated for 48 to 72 h. Putative transductants were checked for the presence of erm(B) by using PCR primers E5/E6 (see Table S1 in the supplemental material), and two transductants were chosen for further analysis. The transduction frequency was calculated as the number of transductants per PFU. Crude phage suspensions were retrospectively inoculated onto BHIS(TA) (brain heart infusion [Oxoid] supplemented with yeast extract [5 mg/ml; Oxoid], l-cysteine [0.1% wt/vol; Sigma], and sodium taurocholate [0.1% wt/vol; Sigma]) agar and broth to check that they were free of bacterial cells/spores.
Screening for ϕC2 in transductants by mitomycin C induction and plaque assay on strain CD062 was performed as described previously (21). In addition, PCR detection of the ϕC2 integrase gene and integration site in transductants was carried out (see Table S1 in the supplemental material). To control for the integrity of the template, 16S rRNA primers PS13/PS14 were included in the same reaction mixture or used in separate reaction mixtures (48) as described below.
Sequencing of erm(B) mobile element and determination of integration site in transductants.
The mobile element containing erm(B) was amplified from CD80 and CD062E1 in four parts and sequenced by the Sanger method (Eurofins MWG Operon, Germany). Primers SG2F/SG3R and TRR4/TPLHSout1 were used with gDNA, while primers invF2/invR2, invF2.1/invR2.1, and invF3/invR3 were used in ligation-assisted PCR (see Table S1 in the supplemental material). CD80 gDNA (0.5 to 1 µg) was completely digested with either Hpy99I or BsrGI (New England Biolabs) and then self-ligated with T4 DNA ligase (10 U per reaction mixture volume; Fermentas) at 16°C for 18 h, and 16 to 32 ng was used in PCR. Sequences were analyzed and assembled into contigs with CLC Workbench version 6, ORFs were predicted with GeneMark, sequence alignments and homology searches were carried out using BLASTN, BLASTP, and ClustalW, and repeats/palindromes were searched for using einverted (http://emboss.bioinformatics.nl/cgi-bin/emboss/einverted), Inverted Repeats Finder (http://tandem.bu.edu/cgi-bin/irdb/irdb.exe), and REPFIND (http://zlab.bu.edu/repfind/form.html).
The integration site of the erm(B) mobile element in transductants was confirmed by Southern hybridization as described below. Primers P5/P6 (Fig. 1; see Table S1 in the supplemental material) were used to detect the integrated erm(B) mobile element and regenerated or empty integration sites in the donor, recipient, and transductants. Circular molecules of the erm(B) mobile element in transductants were searched for using four primer pairs (P2/P4, P1/P4, P2/P3, and P1/P3) reading out of the ends of the element (Fig. 1; Table S1).
Ribotyping.
Ribotyping of CD80, CD062, and CD062E1 was carried out as previously described (49) except that electrophoresis was done using capillary gel electrophoresis (Qiagen) (50).
Filter mating and analysis of transconjugants compared to transductants.
Filter mating assays were carried out as previously described (51) using a spontaneous rifampin-resistant mutant of CD062 as the recipient. This mutant was selected on BHI agar with 5% horse blood and 25 mg/liter rifampin to generate CD062R11 (Table 1); transconjugants were selected on rifampin (25 mg/liter) and erythromycin (10 mg/liter) after a 5-day incubation. All putative transconjugants were tested by multiplex PCR for 16S rRNA and a gene encoding an ABC-transport system (see Table S1 in the supplemental material) that is present only in the donor, as described below. The integration site of the erm(B) element in transconjugants was determined by PCR with primers specific for the integration site in transductants, P5/P2 for the left end and P4/P7 for the right end (Fig. 1; Table S1).
Genome sequencing to differentiate donor and recipient.
As the donor and recipient strains were highly similar, we needed to find genetic markers for a PCR assay to determine real transconjugants after filter matings. Genes unique to the donor and recipients were found by whole-genome sequencing using the Illumina genome analyzer IIx at low (~100×) coverage. Initially, the sequence reads from the recipients were filtered by excluding those found to align to the donor. This was done using the BWA alignment package (52). The remaining reads were assembled using the A5 pipeline (53), and contigs were annotated against the genome of CD630 using the xBASE annotation pipeline (54–59). A similar process was followed to obtain donor-specific contigs. Candidate marker genes were selected to exclude hypothetical genes without a known function and genes on known transposons or phages. Finally, primers CD80C3U57F/CD80C3U57R (Table S1) were designed for a candidate marker gene with 99% (1,035/1,038) sequence similarity to a gene in CD630 encoding an ABC-type transport system (CD630_08760), present only in the donor, and experimentally verified by PCR as described above. The PCR assay was used to distinguish transconjugants from spontaneous rifampin-resistant mutants of the donor after filter matings.
Southern hybridization.
For detection of the erm(B) mobile element integration site in transductants and transconjugants, the RF amplicon was used as a probe (Fig. 1). The PCR conditions for obtaining the RF amplicon using invF3/invR3 are described in Table S1 in the supplemental material. For determining the copy number of the erm(B) mobile element, a PCR amplicon of P4/C2Tn3′end (Table S1), containing the 3′ end of the mobile element, was used as a probe (Fig. 1). MfeI (Fermentas)-digested gDNA (3 µg) of CD062, CD062R11, CD80, and CD062 transductants and CD062 transconjugants served as the templates for hybridization of digoxigenin (DIG)-labeled probes, at 40°C for the RF amplicon probe and 49°C for the 3′-end probe. Detection was carried out as recommended for the DIG DNA labeling and detection kit (Roche).
GenBank accession number.
The complete sequence of Tn6215 is deposited in GenBank with the accession number KC166248.
SUPPLEMENTAL MATERIAL
Detection of Tn6215 integration site in transductants (a) and transconjugants (b) by Southern hybridization of MfeI-digested genomic DNA probed with the RF amplicon, which contained the right-hand Tn6215-host junction. (a) A 10-kb band in the recipient was replaced by 2.7-kb and 11-kb bands in transductant clones and in the donor. A 5-kb band was also observed in the donor only. Lanes: 1, DIG-labeled marker (Roche); 2, CD062 (recipient); 3 to 7, five single colonies of CD062E1, transductant; 8, CD80 (donor). (b) Transconjugants and transductants had similar profiles. Lanes: 1, DIG-labeled marker (Roche); 2, CD062R (recipient Rifr mutant); 3, CD80; 4, CD062R1112; 5, CD062R1146; 6, CD062R1170. Download
Tn6215 copy numbers in donor, transductants, and transconjugants determined by Southern hybridization of MfeI-digested gDNA to DIG-labeled 3′-end probe. The donor contained two copies, while the transductants (a) and transconjugants (b) contained one copy. (a) Lanes: 1, DIG-labeled marker (Roche); 2, CD062; 3 to 7, CD062E1 to CD062E5; 8, CD80. (b) Lanes: 1, CD062R11; 2, CD80; 3, CD062R1112; 4, CD062R1146; 5, CD062R1170; 6, CD062E1; 7, DIG-labeled marker (Roche). Download
PCR primers and reaction parameters.
Features of predicted ORFs in Tn6215.
ACKNOWLEDGMENTS
We thank Sara Thean (University of Western Australia) for ribotyping and Keang Pen-G Song (Monash University, Malaysia) for scientific support.
Footnotes
Citation Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. 2013. Phage ϕC2 meditates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 4(6):e00840-13. doi:10.1128/mBio.00840-13.
REFERENCES
- 1. Janezic S, Ocepek M, Zidaric V, Rupnik M. 2012. Clostridium difficile genotypes other than ribotype 078 that are prevalent among human, animal and environmental isolates. BMC Microbiol. 12:48. 10.1186/1471-2180-12-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 3. Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. 2011. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One 6:e23014. 10.1371/journal.pone.0023014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts AP, Mullany P. 2011. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 35:856–871 [DOI] [PubMed] [Google Scholar]
- 5. Mullany P, Wilks M, Lamb I, Clayton C, Wren B, Tabaqchali S. 1990. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J. Gen. Microbiol. 136:1343–1349 [DOI] [PubMed] [Google Scholar]
- 6. Jasni AS, Mullany P, Hussain H, Roberts AP. 2010. Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis. Antimicrob. Agents Chemother. 54:4924–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fortier LC, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73:7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shan J, Patel KV, Hickenbotham PT, Nale JY, Hargreaves KR, Clokie MR. 2012. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl. Environ. Microbiol. 78:6027–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Govind R, Fralick JA, Rolfe RD. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J. Bacteriol. 188:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685 [DOI] [PubMed] [Google Scholar]
- 11. Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190:6734–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horgan M, O’Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, Ross RP. 2010. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene 462:34–43 [DOI] [PubMed] [Google Scholar]
- 13. Sekulovic O, Meessen-Pinard M, Fortier LC. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193:2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tallent SM, Langston TB, Moran RG, Christie GE. 2007. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 189:7520–7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Varga M, Kuntová L, Pantůček R, Mašlaňová I, Růžičková V, Doškař J. 2012. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 332:146–152 [DOI] [PubMed] [Google Scholar]
- 16. Yasmin A, Kenny JG, Shankar J, Darby AC, Hall N, Edwards C, Horsburgh MJ. 2010. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J. Bacteriol. 192:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nezhad Fard R, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52:559–564 [DOI] [PubMed] [Google Scholar]
- 18. Fischetti VA. 2007. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 15:297–300 [DOI] [PubMed] [Google Scholar]
- 19. Revathi G, Fralick JA, Rolfe RD. 2011. In vivo lysogenization of a Clostridium difficile bacteriophage phiCD119. Anaerobe 17:125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meessen-Pinard M, Sekulovic O, Fortier LC. 2012. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl. Environ. Microbiol. 78:7662–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goh S, Chang BJ, Riley TV. 2005. Effect of phage infection on toxin production by Clostridium difficile. J. Med. Microbiol. 54:129–135 [DOI] [PubMed] [Google Scholar]
- 22. Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crellin PK, Rood JI. 1998. Tn4451 from Clostridium perfringens is a mobilizable transposon that encodes the functional mob protein, TnpZ. Mol. Microbiol. 27:631–642 [DOI] [PubMed] [Google Scholar]
- 24. Priebe SD, Lacks SA. 1989. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J. Bacteriol. 171:4778–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lorenzo-Díaz F, Solano-Collado V, Lurz R, Bravo A, Espinosa M. 2012. Autoregulation of the synthesis of the MobM relaxase encoded by the promiscuous plasmid pMV158. J. Bacteriol. 194:1789–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weaver KE, Kwong SM, Firth N, Francia MV. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gupta A, Vlamakis H, Shoemaker N, Salyers AA. 2003. A new Bacteroides conjugative transposon that carries an ermB gene. Appl. Environ. Microbiol. 69:6455–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutanto Y, Shoemaker NB, Gardner JF, Salyers AA. 2002. Characterization of exc, a novel protein required for the excision of Bacteroides conjugative transposon. Mol. Microbiol. 46:1239–1246 [DOI] [PubMed] [Google Scholar]
- 29. Naito M, Sato K, Shoji M, Yukitake H, Ogura Y, Hayashi T, Nakayama K. 2011. Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: determination of the integration site and the genes essential for conjugal transfer. Microbiology 157:2022–2032 [DOI] [PubMed] [Google Scholar]
- 30. Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19. 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zielenkiewicz U, Ceglowski P. 2005. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J. Bacteriol. 187:6094–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dmowski M, Jagura-Burdzy G. 2011. Mapping of the interactions between partition proteins Delta and Omega of plasmid pSM19035 from Streptococcus pyogenes. Microbiology 157:1009–1020 [DOI] [PubMed] [Google Scholar]
- 33. Soberón NE, Lioy VS, Pratto F, Volante A, Alonso JC. 2011. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segrosome complexes. Nucleic Acids Res. 39:2624–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 56:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosvoll TC, Lindstad BL, Lunde TM, Hegstad K, Aasnaes B, Hammerum AM, Lester CH, Simonsen GS, Sundsfjord A, Pedersen T. 2012. Increased high-level gentamicin resistance in invasive Enterococcus faecium is associated with aac(6′)ie-aph(2″)Ia-encoding transferable megaplasmids hosted by major hospital-adapted lineages. FEMS Immunol. Med. Microbiol. 66:166–176 [DOI] [PubMed] [Google Scholar]
- 36. Brouwer MS, Roberts AP, Mullany P, Allan E. 2012. In silico analysis of sequenced strains of Clostridium difficile reveals a related set of conjugative transposons carrying a variety of accessory genes. Mob. Genet. Elem. 2:8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farrow KA, Lyras D, Rood JI. 2001. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 147:2717–2728 [DOI] [PubMed] [Google Scholar]
- 38. Otsuka Y, Yonesaki T. 2012. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 83:669–681 [DOI] [PubMed] [Google Scholar]
- 39. Forouhar F, Lee IS, Vujcic J, Vujcic S, Shen J, Vorobiev SM, Xiao R, Acton TB, Montelione GT, Porter CW, Tong L. 2005. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J. Biol. Chem. 280:40328–40336 [DOI] [PubMed] [Google Scholar]
- 40. Reith J, Mayer C. 2011. Characterization of a glucosamine/glucosaminide N-acetyltransferase of Clostridium acetobutylicum. J. Bacteriol. 193:5393–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fivian-Hughes AS, Davis EO. 2010. Analyzing the regulatory role of the HigA antitoxin within Mycobacterium tuberculosis. J. Bacteriol. 192:4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Furusawa G, Dziewanowska K, Stone H, Settles M, Hartzell P. 2011. Global analysis of phase variation in Myxococcus xanthus. Mol. Microbiol. 81:784–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Battaglioli EJ, Baisa GA, Weeks AE, Schroll RA, Hryckowian AJ, Welch RA. 2011. Isolation of generalized transducing bacteriophages for uropathogenic strains of Escherichia coli. Appl. Environ. Microbiol. 77:6630–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goh S, Riley TV, Chang BJ. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 71:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. CLSI 2009. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 7th ed. CLSI document M11-A7 CLSI, Wayne, PA [Google Scholar]
- 46. Mullany P, Wilks M, Tabaqchali S. 1995. Transfer of macrolide-lincosamide-streptogramin B (MLS) resistance in Clostridium difficile is linked to a gene homologous with toxin A and is mediated by a conjugative transposon, Tn5398. J. Antimicrob. Chemother. 35:305–315 [DOI] [PubMed] [Google Scholar]
- 47. Lyras D, Storie C, Huggins AS, Crellin PK, Bannam TL, Rood JI. 1998. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob. Agents Chemother. 42:1563–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Persson S, Torpdahl M, Olsen KE. 2008. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (CDTA/cdtB) genes applied to a Danish strain collection. Clin. Microbiol. Infect. 14:1057–1064 [DOI] [PubMed] [Google Scholar]
- 49. Stubbs SL, Brazier JS, O’Neill GL, Duerden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J. Med. Microbiol. 57:1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137–141 [DOI] [PubMed] [Google Scholar]
- 52. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tritt A, Eisen JA, Facciotti MT, Darling AE. 2012. An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304. 10.1371/journal.pone.0042304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol.. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. 2008. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36:D543–D546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of Tn6215 integration site in transductants (a) and transconjugants (b) by Southern hybridization of MfeI-digested genomic DNA probed with the RF amplicon, which contained the right-hand Tn6215-host junction. (a) A 10-kb band in the recipient was replaced by 2.7-kb and 11-kb bands in transductant clones and in the donor. A 5-kb band was also observed in the donor only. Lanes: 1, DIG-labeled marker (Roche); 2, CD062 (recipient); 3 to 7, five single colonies of CD062E1, transductant; 8, CD80 (donor). (b) Transconjugants and transductants had similar profiles. Lanes: 1, DIG-labeled marker (Roche); 2, CD062R (recipient Rifr mutant); 3, CD80; 4, CD062R1112; 5, CD062R1146; 6, CD062R1170. Download
Tn6215 copy numbers in donor, transductants, and transconjugants determined by Southern hybridization of MfeI-digested gDNA to DIG-labeled 3′-end probe. The donor contained two copies, while the transductants (a) and transconjugants (b) contained one copy. (a) Lanes: 1, DIG-labeled marker (Roche); 2, CD062; 3 to 7, CD062E1 to CD062E5; 8, CD80. (b) Lanes: 1, CD062R11; 2, CD80; 3, CD062R1112; 4, CD062R1146; 5, CD062R1170; 6, CD062E1; 7, DIG-labeled marker (Roche). Download
PCR primers and reaction parameters.
Features of predicted ORFs in Tn6215.