ABSTRACT
The intestinal tract is the largest reservoir of microbes in the human body. The intestinal microbiota is thought to be able to modulate alterations of the gut induced by enteropathogens, thereby maintaining homeostasis. Listeria monocytogenes is the agent of listeriosis, an infection transmitted to humans upon ingestion of contaminated food. Crossing of the intestinal barrier is a critical step of the infection before dissemination into deeper organs. Here, we investigated the role of the intestinal microbiota in the regulation of host protein-coding genes and microRNA (miRNA or miR) expression during Listeria infection. We first established the intestinal miRNA signatures corresponding to the 10 most highly expressed miRNAs in the murine ileum of conventional and germfree mice, noninfected and infected with Listeria. Next, we identified 6 miRNAs whose expression decreased upon Listeria infection in conventional mice. Strikingly, five of these miRNA expression variations (in miR-143, miR-148a, miR-200b, miR-200c, and miR-378) were dependent on the presence of the microbiota. In addition, as is already known, protein-coding genes were highly affected by infection in both conventional and germfree mice. By crossing bioinformatically the predicted targets of the miRNAs to our whole-genome transcriptomic data, we revealed an miRNA-mRNA network that suggested miRNA-mediated global regulation during intestinal infection. Other recent studies have revealed an miRNA response to either bacterial pathogens or commensal bacteria. In contrast, our work provides an unprecedented insight into the impact of the intestinal microbiota on host transcriptional reprogramming during infection by a human pathogen.
IMPORTANCE
While the crucial role of miRNAs in regulating the host response to bacterial infection is increasingly recognized, the involvement of the intestinal microbiota in the regulation of miRNA expression has not been explored in detail. Here, we investigated the impact of the intestinal microbiota on the regulation of protein-coding genes and miRNA expression in a host infected by L. monocytogenes, a food-borne pathogen. We show that the microbiota interferes with the microRNA response upon oral Listeria infection and identify several protein-coding target genes whose expression correlates inversely with that of the miRNA. Further investigations of the regulatory networks involving miR-143, miR-148a, miR-200b, miR-200c, and miR-378 will provide new insights into the impact of the intestinal microbiota on the host upon bacterial infection.
INTRODUCTION
Listeria monocytogenes is a Gram-positive food-borne pathogen that infects both humans and animals and has emerged as a model organism in infection biology and host-bacterium interaction studies (1). Infection starts with the ingestion of contaminated food. Then, L. monocytogenes crosses the intestinal barrier to reach the lymph and the bloodstream, allowing the bacterium to disseminate in the whole organism. To remain hidden from the innate and adaptive immune systems and to dampen the host inflammatory response, L. monocytogenes deploys an arsenal of virulence factors (2). L. monocytogenes peptidoglycan N-deacetylase and O-acetyltransferase play a role in inhibition of signaling cascades that allows Listeria to evade the host innate immune system (3–5). The L. monocytogenes surface protein ActA and the internalin InlK prevent bacterial recognition by the cell autophagic machinery (6–8). InlC, another member of the internalin family, by interacting with IKKα, a subunit of the IκB kinase complex critical for the phosphorylation of IκB and activation of NF-κB, dampens the host innate response induced by Listeria infection (9). Listeriolysin O (LLO) is a secreted hemolysin. Besides its role in allowing escape from the phagosome, LLO causes T lymphocyte apoptosis and a variety of other signaling events (10, 11). Conversely, the host uses a combination of cellular and molecular mechanisms for clearance of the invader and recovery from possible damage (12). Among these, a fast reprogramming of the host transcriptome with expression of immune-related genes is thought to be crucial to mount an efficient antibacterial defense.
Strikingly, the mechanisms by which both the bacterium and the host cell reprogram their transcription during infection involve RNA-mediated regulation through small noncoding RNA molecules. Research highlighting the multiple roles played by regulatory noncoding RNAs in both eukaryotes and prokaryotes has exploded during the last decade. Noncoding RNAs have emerged as major regulators of various biological processes, including virulence (13, 14). In 2009, the first Listeria whole-genome transcriptomic analysis revealed that, when Listeria reaches the host intestinal lumen, an extensive transcriptional reshaping occurs, relying on a sigma B-mediated activation of specific virulence genes. In contrast, in the blood, the function of the master regulator of Listeria virulence genes, PrfA, dominates transcriptional reprogramming. Remarkably, several noncoding RNAs, absent in the nonpathogenic species Listeria innocua, exhibit the same expression patterns as the L. monocytogenes virulence genes (15). Together, these data have unraveled successive and coordinated global transcriptional changes during infection. In eukaryotes, microRNAs (miRNAs or miRs) are single-stranded RNAs that are involved in several functions, such as cellular growth, apoptosis, metabolism, immunity, and cancer (16–18). miRNAs regulate gene expression by binding to partially complementary sites found in target transcripts and then promoting their translational repression and decay via deadenylation, decapping, and exonucleolysis. Various roles of miRNAs in plant and mammal infections have been reported (19–21). A specific host miRNA response involving, among others, miR-155 and miR-146, takes place during infection with Helicobacter pylori, as well as with Citrobacter rodentium and intracellular bacteria like Mycobacteria, L. monocytogenes, Francisella tularensis, and Salmonella enterica (19, 20).
The intestinal translocation is the first step of L. monocytogenes infection. The intestinal tract is the largest reservoir of microbes in the human body. This microbiota provides the host with a wide range of enzymes and metabolites that are key to its physiology and represents a metabolic activity that makes it a virtual organ whose importance is increasingly acknowledged (22–24). Together with the intestinal mucosa and the lymphoid organs, the intestinal microbiota contributes to protection against gut colonization by enteropathogens (25). Strikingly, it is still largely unknown how the microbiota regulates host miRNA expression. Recent studies provided evidence that the microbiota exploits the microRNA pathway to maintain the intestinal homeostasis (26–30). We previously identified 3 intestinal miRNAs (miR-192, miR-200b, and miR-215) which are modulated in the intestinal tissue during L. monocytogenes infection and whose levels are affected by an oral pretreatment by lactobacilli (31).
Here, we undertook a comprehensive analysis of the host gene expression in the ileum of mice infected by L. monocytogenes. Using RNA-sequencing technology, we compared the miRNA profiles of conventional (CV) and germfree (GF) mice during infection and showed that the presence of the microbiota in the lumen influences miRNA expression in the intestinal tissue. Our work thus reveals the existence of an miRNA modulation through which the intestinal microbiota may affect host transcriptional reprogramming during infection by a human pathogen.
RESULTS
The intestinal microbiota provides protection against oral Listeria infection in wild-type mice.
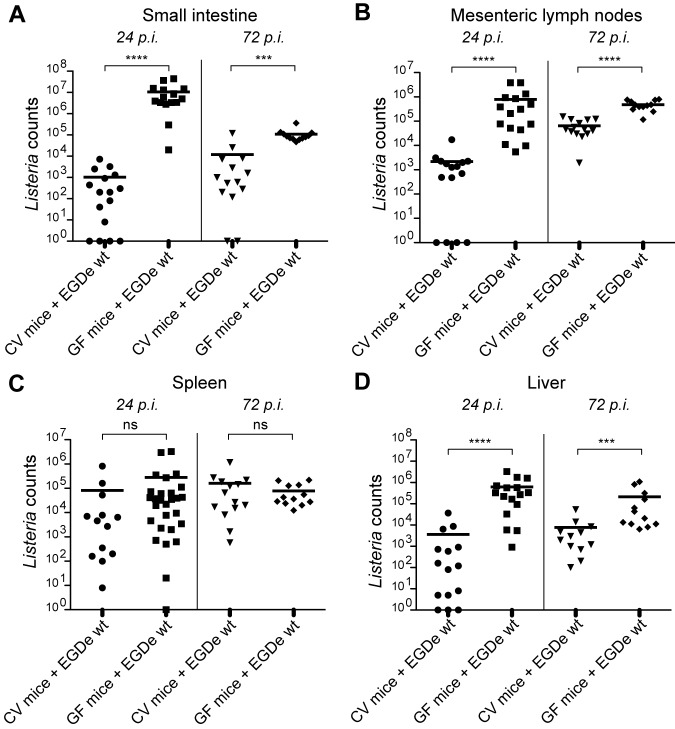
We first compared the susceptibilities of GF and CV wild-type C57BL/6J mice to oral infection with L. monocytogenes strain EGD-e. We determined the L. monocytogenes counts in the intestinal tissue and mesenteric lymph nodes, as well as in the spleen and liver of the infected CV and GF mice, at 24 h and 72 h postinfection (p.i.). We found that at 24 h p.i., the numbers of Listeria were 10,000-fold higher in the small intestine and about 1,000-fold higher in the mesenteric lymph nodes of GF mice than in those of CV mice (Fig. 1A and B, left). In both cases, the higher number of Listeria counts in GF mice was still observed at 72 h p.i., although to a lesser extent (Fig. 1A and B, right). In addition, we found that the number of Listeria was higher in the liver of GF mice than in that of CV mice, while it was the same in the spleen (Fig. 1C and B). Altogether, our results show that GF mice are more susceptible to Listeria infection than CV mice.
FIG 1 .
Germfree mice are hypersensitive to Listeria infection. Listeria counts were assessed in the small intestine (A), mesenteric lymph nodes (B), spleen (C), and liver (D) of conventional (CV) and germfree (GF) wild-type mice uninfected or orally infected with L. monocytogenes EGD-e wild-type (EGDe wt) for 24 h and 72 h (24 h p.i. and 72 h p.i.). Each dot represents one organ. Horizontal bars represent the mean for each condition. Statistical tests were performed using a Mann-Whitney test. Asterisks indicate a P value considered statistically significant (***, P < 0.001; ****, P < 0.0001); ns, nonsignificant difference.
Expression of the dominant miRNAs in both conventional and germfree mice is nearly insensitive to intestinal colonization by Listeria.
The higher susceptibility of the GF mice to bacterial infection has been proposed to be due to a lack of host immune defense (32). We hypothesized that this lower efficiency to mount a proficient defense might correlate with a different miRNA expression, leading to the host failure to adapt its gene transcriptomic response to infection. To explore this hypothesis, we undertook a thorough transcriptional analysis of the host intestine in CV and GF mice, by monitoring both the miRNA and mRNA expression levels. As crossing of the intestinal epithelial barrier is the first step of L. monocytogenes oral infection, total eukaryotic RNAs were extracted from the ileum of uninfected conventional (CV0) and germfree (GF0) mice and from L. monocytogenes-infected conventional and germfree mice at 24 h p.i. (CV24 and GF24, respectively) and 72 h p.i. (CV72 and GF72, respectively). From these samples, transcriptome profiles were generated using Illumina sequencing for miRNA expression or Affymetrix microarrays for mRNA expression.
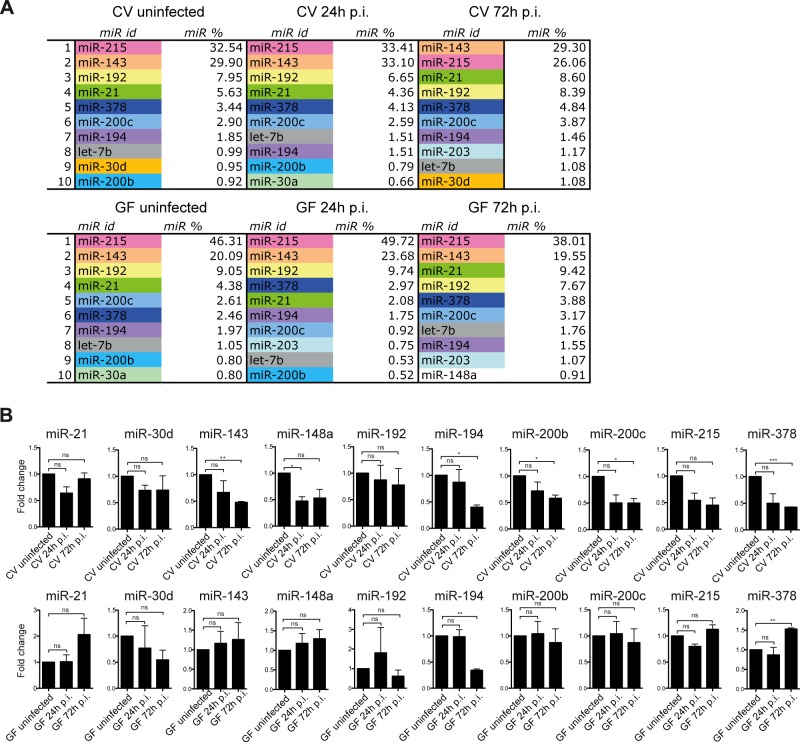
The RNA-sequencing analyses were performed using the ncPRO-seq pipeline (see Fig. S1 in the supplemental material) (33). Under all conditions (CV0, CV24, CV72, GF0, GF24, and GF72), we determined within each of the 6 libraries the percentage of each individual miRNA by dividing the number of corresponding reads by the total number of reads sequenced (total counts). We were thus able to establish a list of the 10 most highly expressed miRNAs in the ileum of mice, infected or not with L. monocytogenes, in the presence or absence of the microbiota (Fig. 2A). miR-143 and miR-215 were the most highly expressed miRNAs under any condition. Alone, they represented between 55% and 72% of the total expressed miRNAs. miR-192, miR-21, and miR-378 represented between 2.5% and 10% of the total intestinal miRNAs and showed a relatively conserved ranking position among the 6 conditions. miR-194 and miR-200c represented between 1 and 4% of total miRNAs under all conditions. Five other miRNAs also occurred repeatedly in the lists of the 10 most highly expressed miRNAs: let-7b, miR-30a, miR-30d, miR-200b, and miR-203. Individually, each of them represented between 0.5% and 1.8%. Last, miR-148a was expressed under one only condition, in germfree mice at 72 h p.i. (less than 1% of the total miRNAs). Altogether, our data suggest that the nature and fraction of the most highly expressed miRNAs in the small intestine are robust features which may define a characteristic miRNA signature of the murine ileum.
FIG 2 .
Listeria and the intestinal microbiota slightly affect the intestinal miRNA expression pattern. (A) The 10 most highly expressed miRNAs (miR) in the small intestine of conventional (CV) and germfree (GF) mice uninfected or orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.) were determined. In each list, the identity of each individual miR (miR id) is indicated, as well as its percentage (miR %) of the total miRs. (B) Relative expression levels of miR-21, miR-30d, miR-143, miR-148a, miR-192, miR-194, miR-200b, miR-200c, miR-215, and miR-378 in conventional (CV) and germfree (GF) mice orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.). Shown are fold changes after standardization to the small nuclear RNA U6 and using uninfected CV and GF control mice as references. Data are represented as means with standard errors of the means (SEM) of values for individual mice (n ≥ 3 per group) from three independent experiments. Statistical tests were performed using a two-tailed Student’s t test. Asterisks indicate a value considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001); ns, nonsignificant difference.
Presence of the intestinal microbiota contributes to the downregulation of miRNA expression triggered by L. monocytogenes infection.
To further analyze the impact of L. monocytogenes infection on the expression of the 10 most highly expressed intestinal miRNAs, we determined, by quantitative reverse transcription-PCR (RT-qPCR) on biological triplicates, the relative expression levels of miR-215, miR-143, miR-192, miR-21, miR-378, miR-200c, miR-194, miR-30d, miR-200b, and miR-148a in Listeria-infected CV mice and uninfected CV mice. Of the 10 miRNAs tested, the expression of 6 miRNAs, miR-143, miR-148a, miR-194, miR-200b, miR-200c, and miR-378, decreased slightly upon Listeria infection (Fig. 2B, top).
Next, we investigated whether the intestinal microbiota had a role in regulating the expression of 6 miRNAs triggered by Listeria infection (Fig. 2B, bottom). We observed three scenarios. In one case, miR-194, the miRNA expression decreased in germfree mice and in conventional mice 72 h after Listeria infection. In the second situation, we found a downregulation of miRNA expression in conventional mice but not in germfree mice upon infection. Indeed, the expression of miR-143, miR-148a, miR-200b, and miR-200c was similar in Listeria-infected germfree mice and uninfected germfree mice. Strikingly, in the last scenario, the expression of miR-378 was opposite in Listeria-infected conventional and germfree mice. It was higher after infection of germfree mice, while it was lower in infected conventional mice than uninfected mice. Altogether, our data show that even though intestinal miRNA expression appears globally stable, L. monocytogenes infection significantly, albeit slightly, affects miRNA expression within the intestinal tissue. More importantly, we highlight that the presence of the intestinal microbiota at the onset of the infection influences the downregulation of miRNA expression during Listeria infection, suggesting a dialog between the microbiota and the infected host in the reprogramming of the host transcriptome.
Presence of the intestinal microbiota contributes to the whole-genome host transcriptional response upon infection.
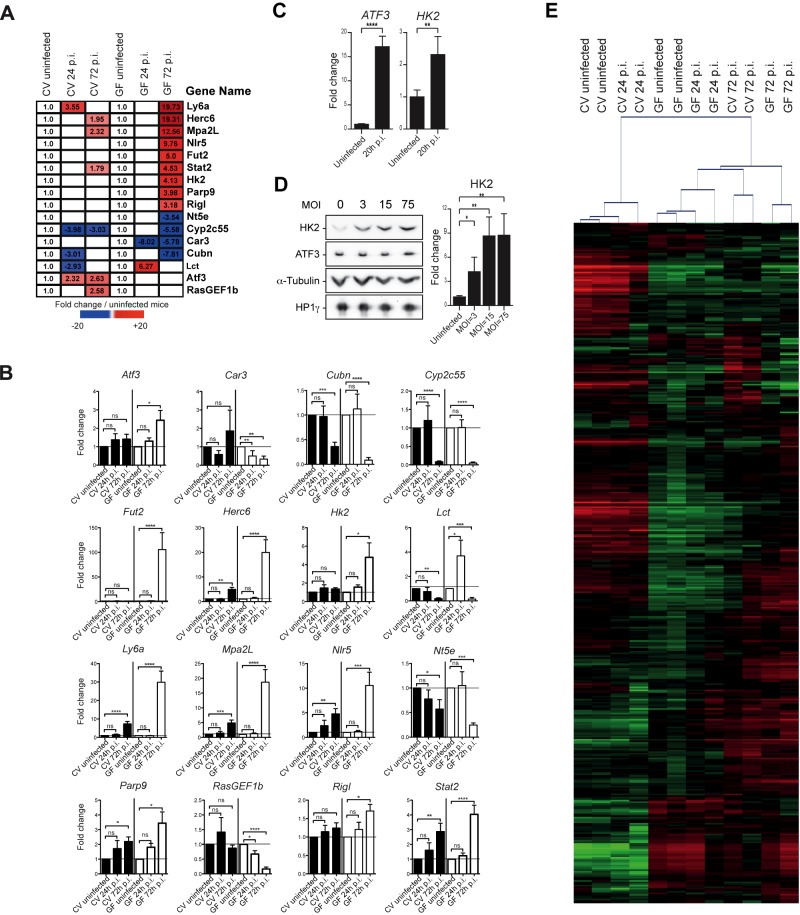
In parallel with the miRNA expression profiles established as described above, we analyzed the expression of the host protein-coding genes under our 6 conditions, CV0, CV24, CV72, GF0, GF24, and GF72, by microarrays (see Fig. S2 in the supplemental material). In the early stages of the infection (24 h p.i.), few genes were significantly (P < 0.05) affected by Listeria infection in conventional and germfree mice, suggesting that this stage may correspond to the priming of the host transcriptomic response. In agreement with previous results (31, 34), in late stages of the infection (72 h p.i.), immune-related genes, e.g., the lymphocyte antigen 6 complex ly6a, macrophage activation 2-like mpa2L, and NOD-like receptor nlr5 genes, were among the most highly induced upon Listeria infection, while the expression of several “xenobiotic” genes (glutathione S-transferase genes, genes belonging to the cytochrome P450 cyp, and solute carrier gene families), as well as other genes, like the cubulin gene (a receptor for intrinsic factor-vitamin B12 complexes), decreased in both conventional and germfree mice (Fig. S2). The expression patterns of a subset of 16 representative genes that were differentially expressed under one of the different conditions and are involved in immune response or metabolic pathways are shown in Fig. 3A. To validate our transcriptomic analysis, we performed RT-qPCR in biological independent triplicates on these 16 genes from the intestine of conventional and germfree mice under the conditions described above, and we observed the same pattern of expression for 13 of them (Fig. 3B). We next undertook to assess whether the observed changes in mRNA levels correlated with changes in protein levels by Western blot analysis. Quantification of several candidate proteins in the total protein extracts from the small intestine of CV and GF mice treated under our experimental conditions proved impossible, all signals being below the detection level (data not shown). We therefore investigated the regulation of two protein targets, ATF3 and HK2, in a simplified in vitro model of cultured intestinal epithelial cells. We selected a human colon adenocarcinoma cell line (LoVo) that can survive long-term infection with high bacterial loads of intracellular EGD-e. At 20 h p.i., the transcript levels of ATF3 and HK2 measured by RT-qPCR were significantly upregulated in LoVo cells (Fig. 3C), similar to the observations in vivo. While the HK2 protein level was found to increase significantly after 22 h of infection (Fig. 3D), ATF3 protein expression did not change upon infection, in spite of a strong upregulation of the cognate transcript. This previously observed discrepancy between transcript and protein abundances (35) may reflect the delay needed to observe protein accumulation after an increase in translation. Alternatively, this might arise from additional levels of gene expression control at the translational stage or from an increase in protein turnover.
FIG 3 .
The intestinal microbiota affects the transcriptional host response to Listeria infection. (A) The heat map presents a subset of 16 representative host genes whose expression was significantly affected (false discovery rate, Benjamini and Hochberg approach [FDR-BH], P < 0.05) in the small intestine of conventional (CV) and germfree (GF) mice (n = 2) uninfected or orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.). The first three columns show the fold changes of gene expression levels in CV mice at 24 h p.i. and 72 h p.i. relative to the expression levels in uninfected CV mice. The last three columns show the fold changes of gene expression levels in GF mice at 24 h p.i. and 72 h p.i. relative to the expression levels in uninfected GF mice. White squares indicate genes whose expression was not significantly affected by the Listeria infection. (B) Relative expression levels of the 16 host genes in CV and GF mice orally infected with L. monocytogenes 24 h p.i. and 72 h p.i. Shown are fold changes after standardization to GADPH and using uninfected CV and GF control mice as references. Data are represented as means and SEM of values for individual mice (n ≥ 3 per group) from three independent experiments. (C) Relative abundance of ATF3 and HK2 transcripts in LoVo cells infected with L. monocytogenes for 20 h. Shown are fold changes after standardization to the GAPDH transcript and using uninfected LoVo cells as a reference. Error bars indicate standard deviations. (D) Relative levels of abundance of ATF3 and HK2 proteins in LoVo cells infected with L. monocytogenes for 22 h shown by Western blot analysis of cell extracts of uninfected cells or cells infected at an MOI of 3, 15, or 75 (left). Quantification of the Western blot signals for the HK2 protein (right). Shown are fold changes after standardization to the α-tubulin protein and using uninfected LoVo cells as a reference. Error bars indicate standard deviations. (E) CV and GF mice were orally infected with L. monocytogenes for 24 h p.i. and 72 h p.i. (2 mice per condition). As described in the text, 444 genes were identified as displaying significantly changed expression levels under at least one of the 6 tested conditions. Unsupervised hierarchical clustering based on this list of the 444 genes was performed with the MeV application. The red–green color shows log2 ratios from mean centered gene expression levels and indicates an upregulation and a downregulation compared to the mean, respectively. See Table S1 in the supplemental material for a list of the 444 genes and their corresponding expression values. Statistics were performed using a two-tailed Student’s t test. Asterisks indicate a value considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001); ns, nonsignificant difference.
In the transcriptome analysis, 444 genes were identified as displaying significantly different expression levels across the six sample groups, i.e., CV0, CV24, CV72, GF0, GF24, and GF72 (Table S1). An unsupervised hierarchical clustering of the 444 genes was performed. Three types of expression profiles could be distinguished (Fig. 3E). One linked the CV0 uninfected mice with the CV24 mice, while the profile of GF0 mice was similar to that of GF24 infected mice. The last group corresponded to mice infected for 72 h with L. monocytogenes. Indeed, at 72 h p.i., the profiles of the CV mice were more similar to those of the infected GF mice than to the profiles of the infected CV mice at 24 h. The divergence of CV and GF mouse patterns at 24 h p.i. suggests that the absence of the intestinal microbiota affects the early priming of the host response to Listeria infection, while at 72 h p.i., this transcriptional response may have reached a threshold from which the presence of microbiota is not decisive.
Bioinformatic prediction of miRNA targets together with gene expression data reveals an miRNA-mRNA network in the mouse intestinal response to infection.
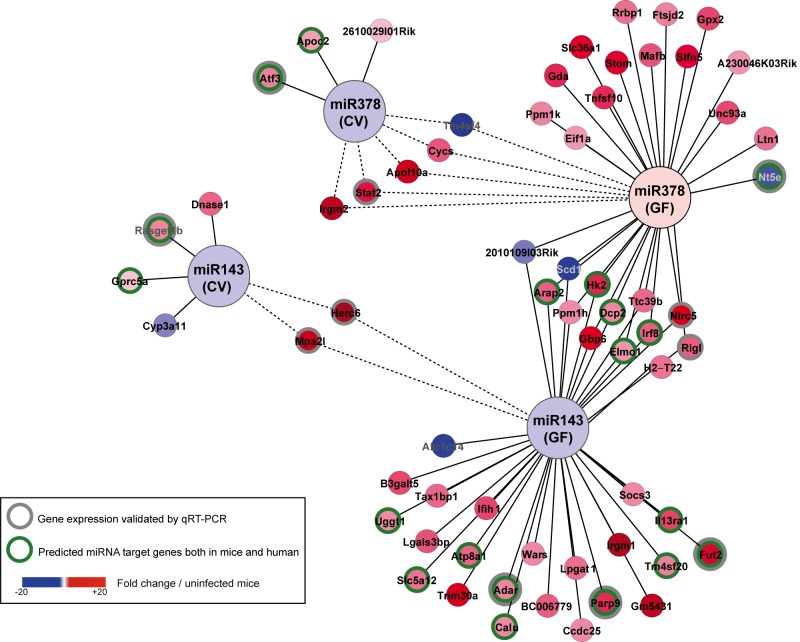
The main mechanism of miRNA function is by base pairing to the 3′-untranslated region (UTR) of the target mRNA, thus inhibiting its translation and stimulating its decay (36). Various parameters have been shown to be critical for the interaction between an miRNA and its mRNA target (37). Accordingly, several target prediction tools have been developed, albeit with unequal performance. Among these, TargetScan is generally acknowledged as having the best accuracy for its target predictions (37). Therefore, using mouse TargetScan, we established a list of predicted mRNA targets for the 5 miRNAs whose expression was affected by L. monocytogenes and the intestinal microbiota. We crossed these 5 distinct lists with the lists of genes that were differentially expressed at 72 h p.i. in conventional and germfree mice. By doing so, we were able to define a network of two miRNAs and candidate mRNA targets; each connection in this network represents a putative regulatory tandem during infection of the mouse intestine (Fig. 4). Strikingly, miR-143 and miR-378 constitute two important nodes with more than 10 putative infection-related targets. Among these, the hexokinase hk2 mRNA has already been described as a target of miR-143 (38). Some of the mRNAs were potential targets for 2 miRNAs, suggesting the existence of fine-tuned regulatory mechanisms. More importantly, we identified within the network more than 15 miRNA-mRNA tandems whose interaction is also conserved in humans (Fig. 4). Last, the expression of several of the predicted target mRNAs correlated inversely with the expression of their tandem miRNAs, supporting a putative miRNA-dependent regulation of these species.
FIG 4 .
miRNA-mRNA network. Using Cytoscape, we highlighted, among the 5 miRNAs whose expression was affected by L. monocytogenes and the intestinal microbiota, a network linking miR-143 and miR-378 and the regulated host genes in the small intestine of conventional (CV) and germfree (GF) mice orally infected with L. monocytogenes 72 h p.i. As described in the text, miR-143 decreased upon infection in both CV and GF mice; in contrast, miR-378 decreased in CV mice but not in GF mice. Nodes represent infection-regulated miRNAs and genes; lines link each miRNA to its putative targets. Some of the predictions in mice were valid in humans (green circles). The color coding indicates the fold change of gene expression in Listeria-infected mice relative to the expression level in uninfected control mice. The differential expression levels of 11 genes were validated by RT-qPCR, as shown in Fig. 3B. Dotted lines show a gene that is similarly regulated in CV and GF mice.
DISCUSSION
Since the first report of tissue-specific microRNA expression in mice in 2002 (39), a constant challenge has been to update the miRNA catalogue, taking advantage of the novel technologies available to detect miRNA sequences (40–43). Except in cancer research, very few studies have assessed the tissue-specific miRNA expression in nonhomeostatic conditions. Moreover, while the key role of miRNAs in the host response to bacterial infection is increasingly recognized, understanding the role of commensal bacteria in this process is still in its infancy (19, 20). Here, we performed the first analysis of the role of the intestinal microbiota in the regulation of both host protein-coding gene and miRNA expression during L. monocytogenes infection.
We identified the 10 most highly expressed miRNAs in the small intestine of conventional wild-type (miR-215, miR-143, miR-192, miR-21, miR-378, miR-200c, miR-194, let-7b, miR-30d, and miR-200b) and germfree (miR-215, miR-143, miR-192, miR-21, miR-200c, miR-378, miR-194, let-7b, miR-200b, and miR-30a) C57BL/6J mice. In agreement with our results, with the exception of miR-378, all have been individually reported at least once as expressed in the gastrointestinal tract (31, 40–44). We confirmed that miR-143, miR-192, miR-194, and miR-215 are dominantly expressed in the gastrointestinal tract (41, 43). We also showed that this expression is not drastically affected by the intestinal microbiota or by infection with L. monocytogenes, therefore establishing a characteristic intestinal miRNA signature. Altogether, our data suggest that potent control mechanisms maintain the levels of dominant miRNA species in the intestinal tissue.
Although the miRNA expression patterns were very similar under our different experimental conditions, we were able to identify microbiota-dependent regulation of miRNA expression upon Listeria infection. The expression of 4 miRNAs, miR-143, miR-148a, miR-200b, and miR-200c, decreased only in conventional mice upon L. monocytogenes infection. The expression pattern of miR-378 was even more striking; indeed, Listeria infection triggered its decrease in conventional mice but its increase in germfree mice. Very recently, the role of the intestinal microbiota in host miRNA regulation has begun to be investigated, especially in the colon (27, 29, 30, 45). Interestingly, downregulation of the expression of a dendritic cell-specific miRNA, miR-10a, by the microbiota has been reported, and its possible involvement in maintaining intestinal homeostasis has been proposed (30). Here, our results suggest that the intestinal microbiota downregulates the expression of some miRNAs upon Listeria infection. This observation has to be considered in parallel with the lower susceptibility of conventional mice to intestinal infections. It is likely that at the beginning of the infection (24 h), the Listeria counts reach a high threshold before inducing the host response and enhancing the transcriptional reprogramming that we observed at 72 h postinfection, when bacterial clearance is effective. Accordingly, the differences in Listeria counts in germfree and conventional mice are larger at 24 h p.i. than at 72 h p.i. Our data are in line with the hypothesis that the ability of the microbiota in priming the host transcriptional response contributes to counteracting a bacterial infection.
The role of several miRNAs, such as miR-146 or miR-155, in the immune response is well established (46–48). Strikingly, neither of these two miRNAs was found in the present study. In contrast, we observed that miR-21, which has been described, together with miR-146 and miR-155, for controlling Toll-like receptor signaling, was highly expressed in the small intestine, an environment where cells are in permanent contact with the surface determinants of the microbiota (49, 50). miR-21 has also been shown to decrease in tuberculosis patients (51). However, to our knowledge, none of the 10 miRNAs described in our study had been previously reported to play a role in maintaining a dialog between the microbiota and the host during bacterial infection. We had previously identified a role in Listeria infection for miR-192, miR-200b, and miR-215 and demonstrated that oral treatment with two Lactobacillus strains modulated the expression of these miRNAs during L. monocytogenes infection (31). Very recently, a role for members of the miR-200 family in Helicobacter infection has been identified. Although they express high basal miR-200 levels, gastric epithelial cells have an increased miR-200b/c expression upon Helicobacter infection, leading to NF-κB activation and initiating a mesenchymal transition by inducing the zinc finger E-box-binding homeobox ZEB (52).
Here, we identified several candidate target genes whose expression correlated inversely with that of miRNAs during infection. Most importantly, we found, by using Human TargetScan 6.0, that some of these predictions in mice were valid in humans, further supporting the direct link between miRNAs and their putative targets. In addition, the function of some of the target mRNAs identified may be associated with the regulation of the infectious process. For instance, Atf3 encodes a transcription factor that is involved in the immune response (53). Gprc5 is a retinoic acid-induced protein that plays a role in epithelial cell differentiation (54). Fut2 is a galactoside 2-alpha-l-fucosyltransferase, an enzyme involved in the fucosylation of epithelial cells (55). Nt5e plays a role in intestinal inflammation, including inflammatory bowel disease (56). Adar encodes a double-stranded RNA-specific adenosine deaminase, an RNA editing enzyme of the miRNA and small interfering RNA (siRNA) pathways (57). More investigations will be required to decipher the direct involvement of a particular miRNA-mRNA tandem in the bacterial-host cross talk during colonization of the intestinal tissue.
To our knowledge, our study is the first comprehensive analysis of the miRNA expression patterns in the small intestine analyzing simultaneously the interplay between a pathogen, the intestinal microbiota, and the infected host. Further investigations of the function of miR-143, miR-148a, miR-200b, miR-200c, and miR-378 will most likely provide new insights into the contribution of the intestinal microbiota to the reprogramming of the host transcriptional landscape during infection by intracellular bacteria.
MATERIALS AND METHODS
Bacterial strain.
L. monocytogenes strain EGD-e was grown in brain heart infusion (BHI) medium (Difco) at 37°C.
Mice.
All experiments involving mice were handled in accordance with the Pasteur Institute guidelines for animal welfare. Only 9- to 12-week-old female C57BL6/J conventional (Charles River) and germfree (CDTA) wild-type mice were used for experiments. Germfree mice were housed in plastic gnotobiotic isolators.
Infection of mice.
L. monocytogenes overnight cultures were diluted in BHI and bacteria were grown to an optical density at 600 nm (OD600) of 1. Bacterial cultures were centrifuged at 3,500 × g for 15 min. After three washes in phosphate-buffered saline (PBS), the L. monocytogenes pellet was resuspended in PBS at a final concentration of 2.5 × 1010 bacteria/ml. Mice were infected orally for 24 h and 72 h with 5 × 109 bacteria diluted in 200 µl of PBS supplemented with 300 µl of CaCO3 (50 mg/ml). Serial dilutions of the inoculum were plated to control the number of bacteria inoculated.
Infection of cells.
The LoVo human colon adenocarcinoma cell line (CCL-229; ATCC) was grown following ATCC or Invitrogen/Life Technologies recommendations, at 37°C in a humidified atmosphere containing 10% CO2. Cells were seeded to achieve a density at the time of infection of 2.5 × 105 cell/well in 24-well plates for RNA analysis or of 8 × 105 cell/well in 6-well plates for protein analysis. Listeria strain EGD-e was grown in BHI medium to an OD600 of 3, washed in PBS, and diluted in culture medium without serum to achieve a multiplicity of infection (MOI) of 6 for RNA analysis or of 3, 15, or 75 for protein analysis. Bacterial inoculums were added to cell plates and centrifuged for 1 min at 200 × g to synchronize entry. After 1 h, the cells were washed twice and the remaining noninvasive bacteria were killed by adding culture medium containing 20 µg/ml gentamicin. The quantification of intracellular bacteria was performed at 18 h p.i. Infected cells were washed in PBS and then disrupted in sterile water for 10 min at 4°C. Triplicates of serial dilutions were plated on BHI agar; the number of CFU was determined on the next day.
Animal studies.
The small intestine, mesenteric lymph nodes, liver, and spleen were removed. The mesenteric lymph nodes, liver, and spleen were directly disrupted in PBS. The small intestine was treated as previously described (31). Serial dilutions of all organ homogenates were plated on BHI plates and incubated at 37°C. Statistical tests were performed using a Mann-Whitney test on the results of three different experiments.
RNA extraction.
RNAs from the ileal tissue were extracted and purified using the classical TRIzol-chloroform protocol. RNA from LoVo cells was extracted at 20 h p.i. with the miRNeasy minikit (Qiagen), as recommended by the manufacturer, using 1 column per well of a 24-well plate. All samples were treated using a Turbo DNA-free kit (Ambion). The RNA quality was determined using the Experion automated electrophoresis station (Bio-Rad).
Preparation of the sRNA libraries and high-throughput sequencing.
Small RNA (sRNA) libraries from total eukaryotic RNAs extracted from the ileum of uninfected conventional (CV0) and germfree (GF0) mice and from L. monocytogenes-infected conventional and germfree mice at 24 h p.i. (CV24 and GF24, respectively) and 72 h p.i. (CV72 and GF72, respectively) were prepared with the small RNA sample preparation conversion kit, version 1.5, following the manufacturer’s instructions (Illumina). Briefly, the 18- to 30-bp RNA fragments were purified from total RNA on a 15% urea PAGE gel (Bio-Rad). The purified small RNA was ligated with 3′ RNA adaptor (5′-/5rApp/ATCTCGTATGCCGTCTTCTGCTTG/3ddC/), which is specifically modified to target miRNAs and other small RNAs that have a 3′ hydroxyl group. The 5′ RNA adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC) was then ligated to the 5′ phosphate end of the small RNA. Reverse transcription was done to convert the RNA to cDNA, which was then selectively enriched by 12 cycles of PCR. The PCR products were purified on 5% PAGE (Bio-Rad) in a size range of 90 to 105 bp and checked on a Bioanalyzer DNA1000 chip (Agilent). Libraries were sequenced using the Illumina Genome Analyzer II platform to generate 36-base single-end reads. The sRNA-seq analyses were performed using the ncPRO-seq pipeline (33). The reads were aligned on the mm9 genome using Bowtie software. We assessed the quality of the 6 libraries as shown in Fig. S1 in the supplemental material.
Quantitative RT-PCR.
Total eukaryotic RNAs (1 µg) were reverse transcribed using iScript cDNA synthesis (Bio-Rad). The cDNAs were used as the templates for PCR using SYBR green PCR master mix (Applied Biosystems) and detected using the ABI Prism 7900HT real-time PCR system (Applied Biosystems). The expression of eukaryotic genes from individual mice (n ≥ 3 per group) was normalized to the expression of the GADPH (glyceraldehyde-3-phosphate dehydrogenase) gene. For miRNA expression analysis, total eukaryotic sRNAs (1 µg) were reverse transcribed using the miScript reverse transcription kit (Qiagen). The cDNAs were used as the templates for PCR using the miScript SYBR green PCR kit (Qiagen) and detected using the ABI Prism 7900HT real-time PCR system (Applied Biosystems). RT2 qPCR primer assays (Qiagen) and the miScript miRNA PCR array (Qiagen) were used to analyze the expression of candidate murine genes and miRNAs, respectively. qRT-PCR primers pairs for HK2 and ATF3 were selected from qPrimerDepot (http://primerdepot.nci.nih.gov/) and manually designed for GAPDH. For in vivo experiments, statistical tests were performed using a two-tailed Student’s t test on the results of three independent experiments. For in vitro experiments, statistical tests were performed using a two-tailed Student’s t test on the results for 4 samples (two biological duplicates from two independent experiments).
Mouse gene chip analysis.
Labeled cDNA was synthesized from 200 ng of total RNA using the NuGEN Applause WT-amp plus ST systems (NuGEN Technologies) as previously described (31). Briefly, labeled samples were hybridized to Affymetrix MoGene 1.0 ST GeneChips and scanned with an Affymetrix GeneChip Scanner 3000, generating cell intensity files for each array. Gene-level expression values were derived from the CEL file probe-level hybridization intensities using the RMA (model-based robust multichip average) algorithm (58). We calculated the P value of the local-pooled-error test (59) and corrected it using a false discovery rate according to the Benjamini and Hochberg approach (FDR-BH). To detect differentially expressed genes, we applied a cutoff P value of <0.05. Unsupervised hierarchical clustering analysis of the 444 genes displaying significantly changed expression under at least one of the 6 tested conditions was performed using the MultiExperiment Viewer (MeV4) application.
Protein extraction and Western blotting.
In LoVo cells infected as described above and washed once with preheated PBS, proteins were extracted by the direct addition of 100 µl of Laemmli sample buffer supplemented with 2 mM MgCl2 and 200 U/ml Benzonase to the well, scraping, sonication, and denaturation at 95°C. Five microliters of each sample was separated by SDS-PAGE and analyzed by Western blotting using standard procedures. The dilutions for primary antibodies were as follows: mouse monoclonal anti-HXKII antibody (HK2, sc-130358; Santa Cruz), 1/200; anti-α-tubulin antibody (T6074; Sigma), 1/5,000; anti-HP1γ antibody (2MOD-1G6-AS; Millipore), 1/5,000; and rabbit polyclonal anti-ATF3 antibody (sc-188; Santa Cruz), 1/200. Secondary antibodies (goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated antibodies; AbCys) were used at a 1/10,000 dilution. Detection was performed with ECL2 Western blotting substrate (Pierce) on a G:Box system (Syngene). Statistical tests were performed using a two-tailed Student’s t test on three independent biological replicates.
Microarray data accession numbers.
Array data were deposited in the ArrayExpress database with the accession number E-MTAB-1800.
SUPPLEMENTAL MATERIAL
Quality assessment of the reads from the six libraries created from purified small RNAs extracted from conventional (CV) and germfree (GF) wild-type mice uninfected or orally infected with L. monocytogenes EGD-e wild-type (EGDe wt) for 24 h and 72 h (24 h p.i. and 72 h p.i.). (A) The table shows the total number of reads in each library. The mean of the total reads was around 6,407,684 ± 1,026,372 reads. (B) Read quality scores. The bar plots show the median Phred quality score at each position of the reads. The Phred quality score of each nucleotide of each read indicates the probability that the specified nucleotide at the specified position may be wrong. As shown on the bar plots, the median Phred scores at each position over all reads of the library were high until the 25th base position, corresponding to low error probabilities. The median quality went down noticeably toward the ends of the reads, which is commonly seen with Illumina reads (quality degrades toward the ends of the reads). All 6 libraries show comparable read quality scores. (C) Lengths of the inserts. The histograms show the distribution of the lengths of the reads after adapter removal, as described in Materials and Methods. Reads of 22 nucleotides in length were dominant in the 6 libraries, with a majority of reads being between 19 and 23 nucleotides in length. This result was in agreement with the expected length of miRNAs, which constitute the main focus of our study. (D) Mapping to the mouse genome assembly mm9. The bar plots show the proportions of reads in the 6 libraries aligned or not to the mouse genome assembly mm9. As shown by the bar plot, between 83% and 92.43% of the reads in each library could be aligned to the mouse genome. Download
Genome-wide host transcriptome analysis. Heat maps display host genes whose expression was significantly affected (false discovery rate, Benjamini and Hochberg approach [FDR-BH], P < 0.05) in the small intestine of conventional (CV) and germfree (GF) mice that were orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.) compared to their expression in uninfected control mice. (A) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control CV mice at 24 h p.i. (B) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control CV mice 72 h p.i. (C) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control GF mice at 24 h p.i. (D) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control GF mice at 72 h p.i. Download
Four hundred forty-four intestinal genes showing differential expression under at least one of the 6 tested conditions. The table indicates the corresponding expression values of the host genes whose expression was significantly affected (false discovery rate, Benjamini and Hochberg approach [FDR-BH], P < 0.05) in the small intestine of conventional (CV) and germfree (GF) mice orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.) compared to the expression values in uninfected control mice.
ACKNOWLEDGMENTS
We thank E. Barillot and E. Heard for discussions and for reading the manuscript. We also thank M.-A. Nahori and the Pasteur animal facilities for technical assistance with in vivo experiments.
This work received financial support from European Research Council (E. R. C.) Advanced grant 233348, ANR Investissement d’Avenir Programme (10-LABX-62), Institut Pasteur, and Institut National de la Santé et de la Recherche Médicale. This work also received financial support from the Fondation Jeantet and Fondation le Roch. C.C. received a fellowship from the Federation of European Biochemical Societies. Pascale Cossart is a Senior International Research Scholar of the Howard Hughes Medical Institute.
Footnotes
Citation Archambaud C, Sismeiro O, Toedling J, Soubigou G, Bécavin C, Lechat P, Lebreton A, Ciaudo C, Cossart P. 2013. The intestinal microbiota interferes with the microRNA response upon oral Listeria infection. mBio 4(6):e00707-13. doi:10.1128/mBio.00707-13.
REFERENCES
- 1. Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 108:19484–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stavru F, Archambaud C, Cossart P. 2011. Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol. Rev. 240:160–184 [DOI] [PubMed] [Google Scholar]
- 3. Aubry C, Goulard C, Nahori MA, Cayet N, Decalf J, Sachse M, Boneca IG, Cossart P, Dussurget O. 2011. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204:731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prévost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rae CS, Geissler A, Adamson PC, Portnoy DA. 2011. Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect. Immun. 79:3596–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dortet L, Mostowy S, Cossart P. 2012. Listeria and autophagy escape: involvement of InlK, an internalin-like protein. Autophagy 8:132–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P. 2011. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 7:e1002168. 10.1371/journal.ppat.1002168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11:1233–1240 [DOI] [PubMed] [Google Scholar]
- 9. Gouin E, Adib-Conquy M, Balestrino D, Nahori MA, Villiers V, Colland F, Dramsi S, Dussurget O, Cossart P. 2010. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the I{kappa}B kinase subunit IKK{alpha}. Proc. Natl. Acad. Sci. U. S. A. 107:17333–17338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carrero JA, Unanue ER. 2012. Mechanisms and immunological effects of apoptosis caused by Listeria monocytogenes. Adv. Immunol. 113:157–174 [DOI] [PubMed] [Google Scholar]
- 11. Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20:360–368 [DOI] [PubMed] [Google Scholar]
- 12. Hawiger J. 2001. Innate immunity and inflammation: a transcriptional paradigm. Immunol. Res. 23:99–109 [DOI] [PubMed] [Google Scholar]
- 13. Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. 2010. RNAs: regulators of bacterial virulence. Nat. Rev. Microbiol. 8:857–866 [DOI] [PubMed] [Google Scholar]
- 14. Toledo-Arana A, Repoila F, Cossart P. 2007. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10:182–188 [DOI] [PubMed] [Google Scholar]
- 15. Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 16. Flynt AS, Lai EC. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schickel R, Boyerinas B, Park SM, Peter ME. 2008. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27:5959–5974 [DOI] [PubMed] [Google Scholar]
- 18. Xiao C, Rajewsky K. 2009. MicroRNA control in the immune system: basic principles. Cell 136:26–36 [DOI] [PubMed] [Google Scholar]
- 19. Eulalio A, Schulte L, Vogel J. 2012. The mammalian microRNA response to bacterial infections. RNA Biol. 9:742–750 [DOI] [PubMed] [Google Scholar]
- 20. Staedel C, Darfeuille F. 2013. MicroRNAs and bacterial infection. Cell. Microbiol. 15:1496–1507. 10.1111/cmi.12159 [DOI] [PubMed] [Google Scholar]
- 21. Staiger D, Korneli C, Lummer M, Navarro L. 2013. Emerging role for RNA-based regulation in plant immunity. New Phytol. 197:394–404 [DOI] [PubMed] [Google Scholar]
- 22. Ivanov II, Honda K. 2012. Intestinal commensal microbes as immune modulators. Cell Host Microbe 12:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249 [DOI] [PubMed] [Google Scholar]
- 25. Stecher B, Hardt WD. 2008. The role of microbiota in infectious disease. Trends Microbiol. 16:107–114 [DOI] [PubMed] [Google Scholar]
- 26. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo-Russo S, Avnit-Sagi T, Cojocaru G, Zreik F, Bentwich Z, Poy MN, Artis D, Walker MD, Hornstein E, Pikarsky E, Ben-Neriah Y. 2011. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 12:239–246 [DOI] [PubMed] [Google Scholar]
- 27. Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. 2011. Microbiota modulate host gene expression via microRNAs. PLoS One 6:e19293. 10.1371/journal.pone.0019293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duerr CU, Hornef MW. 2012. The mammalian intestinal epithelium as integral player in the establishment and maintenance of host-microbial homeostasis. Semin. Immunol. 24:25–35 [DOI] [PubMed] [Google Scholar]
- 29. Singh N, Shirdel EA, Waldron L, Zhang RH, Jurisica I, Comelli EM. 2012. The murine caecal microRNA signature depends on the presence of the endogenous microbiota. Int. J. Biol Sci 8:171–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xue X, Feng T, Yao S, Wolf KJ, Liu CG, Liu X, Elson CO, Cong Y. 2011. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J. Immunol. 187:5879–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Archambaud C, Nahori MA, Soubigou G, Bécavin C, Laval L, Lechat P, Smokvina T, Langella P, Lecuit M, Cossart P. 2012. Impact of lactobacilli on orally acquired listeriosis. Proc. Natl. Acad. Sci. U. S. A. 109:16684–16689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith K, McCoy KD, Macpherson AJ. 2007. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19:59–69 [DOI] [PubMed] [Google Scholar]
- 33. Chen CJ, Servant N, Toedling J, Sarazin A, Marchais A, Duvernois-Berthet E, Cognat V, Colot V, Voinnet O, Heard E, Ciaudo C, Barillot E. 2012. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics 28:3147–3149 [DOI] [PubMed] [Google Scholar]
- 34. Lecuit M, Sonnenburg JL, Cossart P, Gordon JI. 2007. Functional genomic studies of the intestinal response to a foodborne enteropathogen in a humanized gnotobiotic mouse model. J. Biol. Chem. 282:15065–15072 [DOI] [PubMed] [Google Scholar]
- 35. Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7:548. 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saito T, Saetrom P. 2010. MicroRNAs—targeting and target prediction. N. Biotechnol. 27:243–249 [DOI] [PubMed] [Google Scholar]
- 38. Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, Li B, Li Y, Li D, Wang ED, Liu MF. 2012. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 31:1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735–739 [DOI] [PubMed] [Google Scholar]
- 40. Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. 2004. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA 10:1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, Dengler U, Hall J, Filipowicz W, Weiler J. 2007. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 35:e52. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takada S, Berezikov E, Yamashita Y, Lagos-Quintana M, Kloosterman WP, Enomoto M, Hatanaka H, Fujiwara S, Watanabe H, Soda M, Choi YL, Plasterk RH, Cuppen E, Mano H. 2006. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 34:e115. 10.1093/nar/gkj093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gaedcke J, Grade M, Camps J, Søkilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Møller S, Beissbarth T, Ried T, Litman T. 2012. The rectal cancer microRNAome—microRNA expression in rectal cancer and matched normal mucosa. Clin. Cancer Res. 18:4919–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu S, Dong TS, Dalal SR, Wu F, Bissonnette M, Kwon JH, Chang EB. 2011. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One 6:e16221. 10.1371/journal.pone.0016221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142:914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Artis D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8:411–420 [DOI] [PubMed] [Google Scholar]
- 50. Quinn SR, O’Neill LA. 2011. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 23:421–425 [DOI] [PubMed] [Google Scholar]
- 51. Kleinsteuber K, Heesch K, Schattling S, Kohns M, Sander-Jülch C, Walzl G, Hesseling A, Mayatepek E, Fleischer B, Marx FM, Jacobsen M. 2013. Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4(+) T cells and peripheral blood from tuberculosis patients. PLoS One 8:e61609. 10.1371/journal.pone.0061609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baud J, Varon C, Chabas S, Chambonnier L, Darfeuille F, Staedel C. 2013. Helicobacter pylori initiates a mesenchymal transition through ZEB1 in gastric epithelial cells. PLoS One 8:e60315. 10.1371/journal.pone.0060315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nguyen TT, Johnsen IB, Knetter CF, Drabløs F, Fitzgerald KA, Lien E, Anthonsen MW. 2010. Differential gene expression downstream of Toll-like receptors (TLRs): role of c-Src and activating transcription factor 3 (ATF3). J. Biol. Chem. 285:17011–17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ito J, Ito M, Nambu H, Fujikawa T, Tanaka K, Iwaasa H, Tokita S. 2009. Anatomical and histological profiling of orphan G-protein-coupled receptor expression in gastrointestinal tract of C57BL/6J mice. Cell Tissue Res. 338:257–269 [DOI] [PubMed] [Google Scholar]
- 55. Terahara K, Nochi T, Yoshida M, Takahashi Y, Goto Y, Hatai H, Kurokawa S, Jang MH, Kweon MN, Domino SE, Hiroi T, Yuki Y, Tsunetsugu-Yokota Y, Kobayashi K, Kiyono H. 2011. Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem. Biophys. Res. Commun. 404:822–828 [DOI] [PubMed] [Google Scholar]
- 56. Doherty GA, Bai A, Hanidziar D, Longhi MS, Lawlor GO, Putheti P, Csizmadia E, Nowak M, Cheifetz AS, Moss AC, Robson SC. 2012. CD73 is a phenotypic marker of effector memory Th17 cells in inflammatory bowel disease. Eur. J. Immunol. 42:3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O’Connell MA. 2009. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 28:3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 59. Jain N, Thatte J, Braciale T, Ley K, O’Connell M, Lee JK. 2003. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics 19:1945–1951 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality assessment of the reads from the six libraries created from purified small RNAs extracted from conventional (CV) and germfree (GF) wild-type mice uninfected or orally infected with L. monocytogenes EGD-e wild-type (EGDe wt) for 24 h and 72 h (24 h p.i. and 72 h p.i.). (A) The table shows the total number of reads in each library. The mean of the total reads was around 6,407,684 ± 1,026,372 reads. (B) Read quality scores. The bar plots show the median Phred quality score at each position of the reads. The Phred quality score of each nucleotide of each read indicates the probability that the specified nucleotide at the specified position may be wrong. As shown on the bar plots, the median Phred scores at each position over all reads of the library were high until the 25th base position, corresponding to low error probabilities. The median quality went down noticeably toward the ends of the reads, which is commonly seen with Illumina reads (quality degrades toward the ends of the reads). All 6 libraries show comparable read quality scores. (C) Lengths of the inserts. The histograms show the distribution of the lengths of the reads after adapter removal, as described in Materials and Methods. Reads of 22 nucleotides in length were dominant in the 6 libraries, with a majority of reads being between 19 and 23 nucleotides in length. This result was in agreement with the expected length of miRNAs, which constitute the main focus of our study. (D) Mapping to the mouse genome assembly mm9. The bar plots show the proportions of reads in the 6 libraries aligned or not to the mouse genome assembly mm9. As shown by the bar plot, between 83% and 92.43% of the reads in each library could be aligned to the mouse genome. Download
Genome-wide host transcriptome analysis. Heat maps display host genes whose expression was significantly affected (false discovery rate, Benjamini and Hochberg approach [FDR-BH], P < 0.05) in the small intestine of conventional (CV) and germfree (GF) mice that were orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.) compared to their expression in uninfected control mice. (A) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control CV mice at 24 h p.i. (B) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control CV mice 72 h p.i. (C) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control GF mice at 24 h p.i. (D) Values correspond to the fold changes of relative gene expression levels in Listeria-infected mice and control GF mice at 72 h p.i. Download
Four hundred forty-four intestinal genes showing differential expression under at least one of the 6 tested conditions. The table indicates the corresponding expression values of the host genes whose expression was significantly affected (false discovery rate, Benjamini and Hochberg approach [FDR-BH], P < 0.05) in the small intestine of conventional (CV) and germfree (GF) mice orally infected with L. monocytogenes for 24 h and 72 h (24 h p.i. and 72 h p.i.) compared to the expression values in uninfected control mice.