ABSTRACT
Epigenetic gene regulation has emerged as a major mechanism for gene regulation in all eukaryotes. Histones are small, basic proteins that constitute the major protein component of chromatin, and posttranslational modifications (PTM) of histones are essential for epigenetic gene regulation. The different combinations of histone PTM form the histone code for an organism, marking functional units of chromatin that recruit macromolecular complexes that govern chromatin structure and regulate gene expression. To characterize the repertoire of Toxoplasma gondii histone PTM, we enriched histones using standard acid extraction protocols and analyzed them with several complementary middle-down and bottom-up proteomic approaches with the high-resolution Orbitrap mass spectrometer using collision-induced dissociation (CID), higher-energy collisional dissociation (HCD), and/or electron transfer dissociation (ETD) fragmentation. We identified 249 peptides with unique combinations of PTM that comprise the T. gondii histone code. T. gondii histones share a high degree of sequence conservation with human histones, and many modifications are conserved between these species. In addition, T. gondii histones have unique modifications not previously identified in other species. Finally, T. gondii histones are modified by succinylation, propionylation, and formylation, recently described histone PTM that have not previously been identified in parasitic protozoa. The characterization of the T. gondii histone code will facilitate in-depth analysis of how epigenetic regulation affects gene expression in pathogenic apicomplexan parasites and identify a new model system for elucidating the biological functions of novel histone PTM.
IMPORTANCE
Toxoplasma gondii is among the most common parasitic infections in humans. The transition between the different stages of the T. gondii life cycle are essential for parasite virulence and survival. These differentiation events are accompanied by significant changes in gene expression, and the control mechanisms for these transitions have not been elucidated. Important mechanisms that are involved in the control of gene expression are the epigenetic modifications that have been identified in several eukaryotes. T. gondii has a full complement of histone-modifying enzymes, histones, and variants. In this paper, we identify over a hundred PTM and a full repertoire of PTM combinations for T. gondii histones, providing the first large-scale characterization of the T. gondii histone code and an essential initial step for understanding how epigenetic modifications affect gene expression and other processes in this organism.
INTRODUCTION
Eukaryotic chromosomal DNA is packaged in nucleosomes that consist of DNA wrapped around a histone octamer composed of two monomers of each histone (H2A, H2B, H3, and H4). Histones are small, basic, conserved proteins that represent about 50% of the total weight of chromatin and function to regulate access to information contained in DNA (1). Histones share a common structure, composed of flexible domains (N- and C-terminal tails) and a globular domain that includes the conserved histone fold. Histone N-terminal tails (and the C terminus of H2A) are exposed outside the nucleosome and are targets of numerous posttranslational modifications (PTM), such as acetylation, methylation, phosphorylation, ubiquitination, sumoylation, ADP ribosylation, deimination (or citrullination), and isomerization. New histone posttranslational modifications (PTM) that have recently been discovered include succinylation, crotonylation, and O-GlcNAcylation (2–4). Histone PTM are highly dynamic and operate in an interdependent manner to create the histone code originally proposed by Strahl and Allis in 2000 (5). The histone code of an organism generates a complex network of possible PTM combinations that can change the architecture of chromatin by modifying the interactions between histones, DNA, and associated macromolecular complexes. The combination of PTM can promote or prevent the binding of specific proteins that read the code and activate or repress nuclear processes, such as transcription, DNA repair, and cell cycle control.
Toxoplasma gondii is an obligate intracellular parasite and a member of the Apicomplexa phylum, which also contains other pathogenic parasites, including Plasmodium, Cryptosporidium, and Babesia. T. gondii is the causative agent of toxoplasmosis, and it is estimated that one-third of the world population is chronically infected with T. gondii. T. gondii has a complex life cycle, with both asexual and sexual life cycle stages. During passage through mammalian hosts, this parasite has two distinct asexual life cycle stages: the tachyzoite, a rapidly replicating form responsible for dissemination of infection, and the bradyzoite, a quiescent form found in tissue cysts that is responsible for transmission of infection as well as creation of a reservoir for the reactivation of infection. Each developmental form of the parasite is morphologically and biochemically distinct, allowing stage-specific adaptation to different environments during transition between hosts (6). Developmental transition to different life stages is accompanied by gene expression changes (7). Control of T. gondii gene expression is, in part, promoted by epigenetic events, and interest in epigenetic regulation in T. gondii has increased with the discovery of compounds, such as apicidin, that act against parasites by inhibiting the activities of histone-modifying enzymes (8, 9). Thus, understanding the particular PTM on histones and identifying the histone-modifying enzymes responsible for histone PTM may lead to development of new antiparasitic drugs.
The T. gondii genome (version 7.0; http://www.ToxodB.org) (10) is predicted to encode a single copy of the core histones, H2A, H3, and H4, whereas H2B is represented by 2 isoforms that differ by 6 amino acids (5 changed amino acids and one deletion) (see Fig. S1 in the supplemental material). T. gondii H2B (TgH2B) isoforms are differentially expressed during the life cycle, with TgH2Ba being upregulated in tachyzoites and TgH2Bb being seen in the sexual life cycle but not in either tachyzoites or bradyzoites (11). Histones H3 and H4 are highly conserved, but H2A and H2B are more divergent, particularly within the N-terminal tail (Fig. S1). The globular or structural domain is highly preserved for all core histones.
In addition to having the canonical histones, T. gondii has five variant histones: centromeric H3 (CenH3), H3.3, H2A.X, H2A.Z, and the parasite-specific H2Bv (Fig. S2). With chromatin immunoprecipitation (ChIP), CenH3 was shown to have a conserved function as the centromeric histone (12). Dalmasso et al. (13) studied the H2A and H2B variants, showing that H2A.Z and H2Bv interact with each other and are located at transcription start sites and that H2A.X appears to be associated with gene silencing and DNA repair. The authors also suggested that given the high expression of H2Bv, in comparison with canonical H2B, H2Bv is the major H2B for T. gondii (13).
Plasmodium falciparum, an apicomplexan parasite responsible for malaria, encodes the four canonical histones and four variant histones, each present as a single-copy gene (14). The apicomplexan histones are very similar, although P. falciparum does not encode H2A.X (Fig. S2). Like T. gondii, P. falciparum encodes the machinery required to reversibly modify histones, and its histone PTM marks have been characterized by mass spectrometry (MS) (14–16). Several marks were identified in both canonical and histone variants in Plasmodium, including acetylation, methylation (mono-, di-, or trimethylation), and ubiquitination. Notably, an abundance of lysine acetylation was present, suggesting a preponderance of chromatin in a transcriptionally active state, with a paucity of marks related to transcription repression (14, 15).
Based upon the high degree of sequence homology between T. gondii, P. falciparum, and human histones, it has been suggested that the same or similar histone PTM are present in T. gondii and that these PTM have similar functions in T. gondii. Some PTM in T. gondii have already been characterized by antibody-based techniques (17–19). While antibody-based techniques have been useful for studying histone PTM, these approaches have some limitations, including potential lack of specificity of antibodies recognizing histone PTM. Advances in mass spectrometric proteomics, including new fragmentation approaches together with high-sensitivity and mass accuracy instrumentation, have made MS the standard technique for the identification of PTM (20–27). Using a combination of complementary mass spectrometry approaches with high-resolution mass spectrometry (LTQ Orbitrap Velos), we have identified the PTM found on histones in the tachyzoites of T. gondii, providing a map of the histone code that governs epigenetic regulation in this organism.
RESULTS
Mass spectrometry identification of T. gondii histone PTM.
To identify posttranslational modifications (PTM) present in both the canonical and histone variants in T. gondii, we analyzed histones from intracellular tachyzoites of the RH strain grown in human foreskin fibroblasts. To purify histones from nuclei, we used standard hydrochloric acid extraction protocols, since histones are soluble at low pH while most other proteins and DNA precipitate under acidic extraction conditions. An example of the final histone enrichment sample is shown in Fig. 1A, and an overview of the methodology used is shown in Fig. 1B. An alignment of the T. gondii histones to those of humans, Saccharomyces cerevisiae, and Plasmodium falciparum is shown in Fig. S1 in the supplemental material. Figures 2 and 3 summarize the T. gondii histone PTM found on classical and variant histones and compare the T. gondii histone PTM to those reported for Plasmodium and human histones.
FIG 1 .
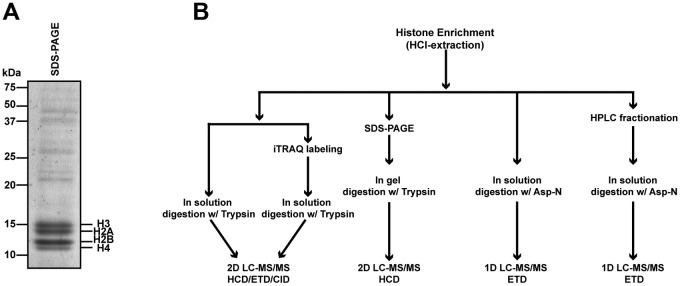
Identification of T. gondii histone posttranslational modifications. (A) Coomassie blue-stained gel showing a representative histone acid extraction sample used for PTM analysis. Relative molecular standards and positions of histones are marked. (B) Scheme of the combined techniques used to identify PTM in T. gondii. Tachyzoites were grown, collected, and purified from human fibroblasts as described in Materials and Methods and Text S1 in the supplemental material. Parasites were lysed, and intact nuclei were purified. The histones were extracted from chromatin under acidic conditions. MS data collection was performed on an LTQ Orbitrap Velos system, and spectra were searched in a custom, combined human Toxoplasma database (60) using Mascot.
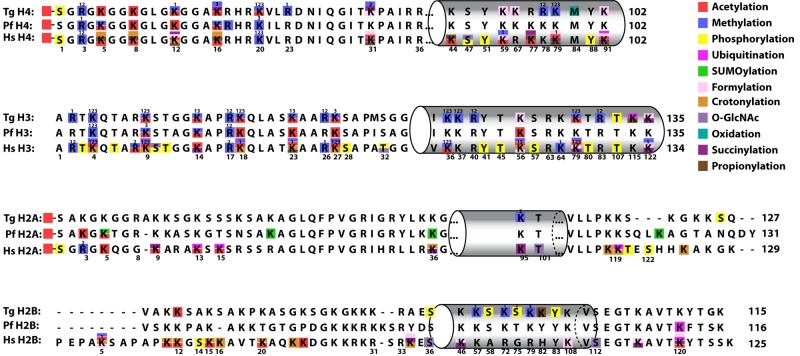
FIG 2 .
Summary of PTM identified on Toxoplasma gondii canonical histones. PTM on canonical histones are shown in comparison with P. falciparum (Pf) and Homo sapiens (Hs) histones. Squares in different colors represent the modifications on three organisms. PTM from Plasmodium were obtained from the literature (14–16). PTM from human cells were obtained from available databases (http://www.uniprot.org, http://www.cellsignal.com, and http://www.actrec.gov.in/histome [61]). Recently identified modifications, such as succinylation, O-GlcNAcylation, propionylation, formylation, and crotonylation, were obtained from the literature (2–4, 47, 48). The gray cylinders indicate the globular domain. Numbers above the squares represent the numbers of methyl groups (“1,”“2,” and “3” indicate mono-, di-, and trimethylation, respectively), while those below the sequence represent the amino acid position in human histones. See Fig. S1 for aligned sequences and the supplemental material for combinations of PTM identified on peptides. In some cases, the exact residue modified by a PTM was not identified. These peptides are not indicated in this schematic but are listed in the supplemental material, which lists each of the peptides identified by mass spectrometry.
FIG 3 .
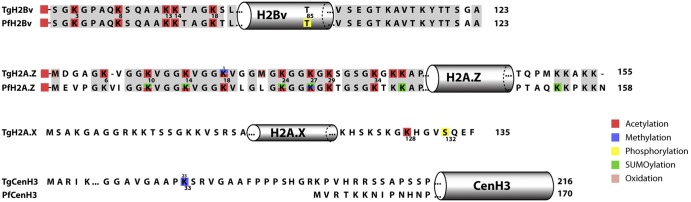
Overview of PTM identified on T. gondii histone variants. PTM on canonical and histone variants are shown in comparison with P. falciparum. Identical amino acids are represented in gray. Squares in different colors represent the modifications on Toxoplasma and Plasmodium. PTM from Plasmodium were obtained from available published literature (14–16). The gray cylinders indicate the approximate locations of the globular domain. Numbers below the Toxoplasma sequence represent the amino acid position in this parasite, while “1,” “2,” and “3” above indicate mono-, di-, and trimethylation, respectively. See Fig. S2 for a complete alignment of variant histones and elsewhere in the supplemental material for combinations of PTM identified on peptides. In some cases, the exact residue modified by a PTM was not identified. These peptides are not indicated in this schematic but are listed in the supplemental material, which lists each of the peptides identified by mass spectrometry.
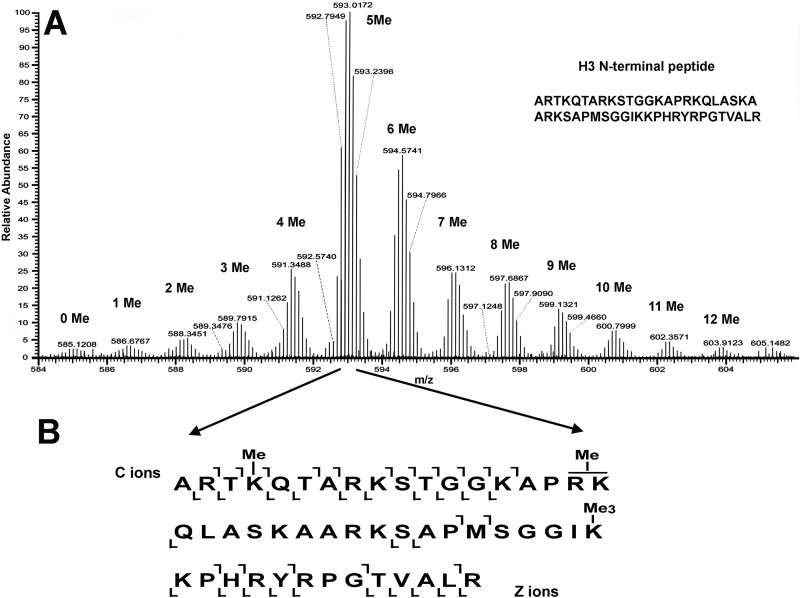
The histone acid extracts were analyzed using (i) off-line high-performance liquid chromatography (HPLC) fractionation followed by Asp-N digestion and middle-down MS analysis, (ii) in-gel trypsin digestion and two-dimensional ultraperformance (to the nanometer scale) LC-tandem MS (2D nanoUPLC-MS/MS analysis), and (iii) in-solution trypsin digestion and 2D nanoUPLC-MS/MS. With electron transfer dissociation (ETD) fragmentation on the LTQ Orbitrap Velos mass spectrometer, we identified several modifications, including acetylation, monomethylation, dimethylation, trimethylation, phosphorylation, and succinylation. Identified peptides had a variety of combinations of these modifications. For example, the T. gondii-specific H3 N-terminal peptide ARTKQTARKSTGGKAPRKQLASKAARKSAPMSGGIKKPHRYRPGTVALR was modified with 0 to 12 methyl groups (Fig. 4A). The abundant distribution of these 12 forms of methylation modification suggests a very complex and sophisticated regulation of methylation. The ETD MS/MS spectrum of the precursor ion at m/z 592.6837 (its monoisotopic mass was determined from the first MS analysis [MS1]) of this H3 N-terminal peptide modified with 5 methyl groups confirms that there is methylation on K4 and trimethylation on K36, as well as methylation on either R17 or K18 (Fig. 4B).
FIG 4 .
Representative MS spectra for T. gondii histone H3 peptide from positions 1 to 49. (A) Spectra showing the abundance of 0 to 12 methyl groups present on this peptide (charge: +9). The majority of the identified peptides show 5 methyl groups on lysines and/or arginines. (B) An example of a methylated TgH3 peptide analyzed by MS/MS shows methylation on K4, trimethylation on K36, and methylation on either R17 or K18. Residues that distinguish TgH3 from HsH3 are S22, M31, S32, and I35 (see also the supplemental material).
Trypsin digestion (in gel or in solution) of histones provided smaller peptides that were analyzed by bottom-up MS/MS. Histone acid extract was separated by SDS-PAGE (Fig. 1A), and the region of the gel containing histones H4, H3, H2A, and H2B were cut and digested with trypsin. The extracted tryptic peptides were analyzed with 2D tandem-reverse-phase (RP/RP) nanoUPLC-MS/MS on the LTQ Orbitrap. Peptides were fragmented by higher-energy collisional dissociation (HCD), electron transfer dissociation (ETD), or collision-induced dissociation (CID) to generate MS/MS spectra. In each run, the sample was on-line fractionated into 15 fractions on the first reverse-phase C18 column (high pH). Peptides in each fraction were then separated on the second reverse-phase C18 column (low pH) and analyzed by tandem mass spectrometry, resulting in 15 LC-MS/MS analyses per sample.
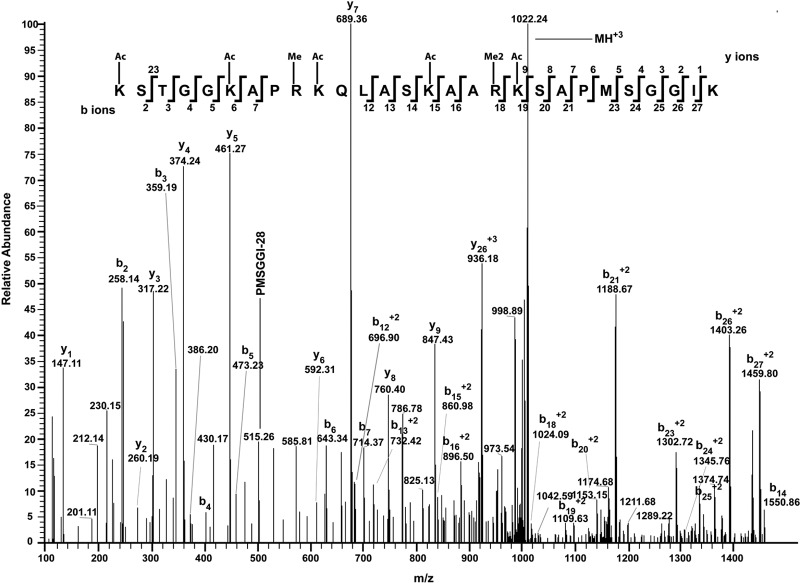
The frequency of lysine residues is very high in histones, yielding very short tryptic peptides that elute early and are too low in mass/charge ratios to be measured using our LC-MS/MS conditions. Therefore, in a separate experiment, iTRAQ labeling on lysine residues of histones was employed prior to in-solution trypsin digestion and 2D nanoUPLC-MS/MS analysis with ETD or HCD (28). Compared to other methods of blocking the free epsilon amino group of lysine, such as propionic anhydride derivatization (12, 29), iTRAQ has several advantages. First, the mild reaction conditions favor retention of the endogenous modifications of histones. Second, unlike chemical propionic anhydride derivatization, iTRAQ labeling introduces a truly artificial (exogenous) modification on histones that can be easily distinguished from its endogenous modifications. Third, the presence of the iTRAQ reporter ion (obtained with HCD) in MS/MS spectra and iTRAQ-labeled peptide sequence pattern after MS/MS enhances our confidence in identifying the labeled peptides. Fourth, iTRAQ derivatization retains the positive charge of lysine, which is favorable to ionization efficiency and ETD fragmentation. Last but not least, iTRAQ introduces a relatively large group on lysine side chains and therefore results in more effective cleavage blocking of trypsin at the labeled residues. For example, an H3 peptide, K(iTRAQ) STGGK(iTRAQ) APR(methyl), was identified in an iTRAQ labeling experiment. Without labeling, this peptide would have been cleaved into smaller pieces by trypsin, and the methylation on Arg would not have been identified.
Histone H4.
T. gondii H4 differs from human H4 by only 6 amino acids (Fig. S1) and is extensively modified with different PTM combinations (Fig. 2 and S3). As is typical for many histones, H4 loses the initial methionine during protein processing, and the subsequent residue (serine) is acetylated. In addition to acetylation of S1, phosphorylation on S1 of T. gondii H4 was identified. This modification is followed by methylation on R3, a mark previously identified in Plasmodium and Toxoplasma using mass spectrometry and/or biochemical approaches (15, 17). The N-terminal tail of H4 is a highly acetylated region, with acetylation on lysines 5, 8, 12, and 16. Acetylation affects the interaction of histones with DNA, keeping chromatin in a more relaxed state, and this modification is associated with transcription activation.
In T. gondii tachyzoites, the majority of modified H4 peptides are acetylated on two or more residues, suggesting cooperativity or redundancy in the activities of histone acetyltransferase enzymes. Interestingly, some of the H4 residues that are acetylated can also be methylated to different levels (methylated H4K12 [H4K12me] and trimethylated HFK16 [H4K16me3]). H4K12me in yeast was described previously (30), but to our knowledge, this is the first time that H4K16me3 has been identified in any organism.
We also identified methylation on H4K20, a repressive mark in other species (31). H4K20me, dimethylated H4K20 (H4K20me2), and H4K20me3 in Toxoplasma and Plasmodium were characterized using antipeptide antibodies and reported to be located at heterochromatic regions (32). Of the H4 peptides containing H4K20 identified, most were methylated, most commonly with dimethyl. Acetylated H4K20 (H4K20ac) was found in a minority of cases, alone or in combination with other acetylated residues. In addition, we identified H4R23me, a modification not previously described in other species. Acetylation on H4K31 has been reported in Toxoplasma (33) but was not found in our study.
Finally, we identified modified residues inside the globular domain of H4. H4K79 was found methylated in all states (H4K79me, H4K79me2, and H4K79me3) but not acetylated, as previously described (34). Dimethylation and monomethylation were identified on R78, and these modifications have not been described for any other organism. The unique peptide sequences that distinguish TgH4 from Homo sapiens H4 (HsH4) (I54T, I65V, K67R, S69A, R77K, I86V, and S89A) are well represented among the H4 peptides and are shown in yellow in Fig. S3.
Histone H3.
Like H4, histone H3 shares a high degree of sequence similarity with human histone 3, differing by 8 amino acids located in the N-terminal and globular domains and being 1 residue longer at the C terminus (Fig. S1). An overview of the H3 PTM combinations and a comparison of the H3 PTM from Plasmodium and humans are shown in Fig. 2. Numerous TgH3 peptides were identified (Fig. 2, 4 to 6, and S4). T. gondii has, besides the canonical H3, an H3 variant termed H3.3 that differs from it by only 4 amino acids (positions 53, 54, 87, and 89), making it difficult to distinguish between H3 and H3.3 subspecies. The first 30 amino acids of TgH3 are abundantly modified (Fig. 2, 4 to 6, and S4). We identified 108 different H3 peptides, and PTM combinations within the N terminus representing the most abundant peptides within our data set. Many of these peptides encompassed amino acids unique to TgH3, conclusively showing their T. gondii origin (Fig. S4).
FIG 5 .
Mass spectrometry analysis of PTM for a T. gondii-specific H3 peptide. T. gondii peptide from amino acids 9 to 36 (KSTGGKAPRKQLASKAARKSAPMSGGIK) is unique to T. gondii histone 3. The serine at position 22 in T. gondii histone 3 is a threonine in human H3 (see Fig. S1). Seven PTM were identified in this TgH3 sequence (K9ac, K14ac, R17me, K18ac, K23ac, R26me2, and K27ac). The MS/MS spectra obtained with HCD of the +3 m/z ion at 1021.8967 is shown.
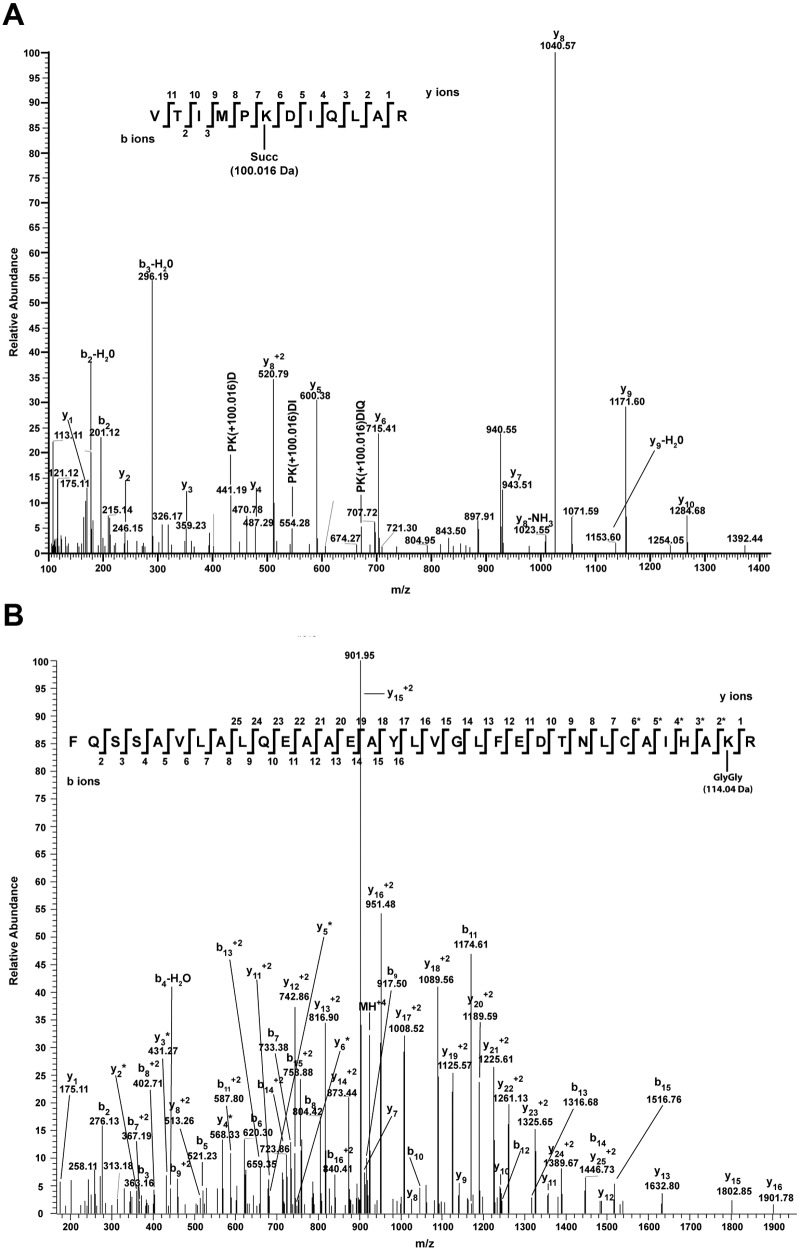
FIG 6 .
T. gondii histone H3 is succinylated and ubiquitinated. (A) MS/MS spectrum of the succinylated H3 from T. gondii peptide from positions 117 to 128 (VTIMPKDIQLAR). A succinylated lysine at position 122 was identified. The succinyl mark has a mass of 100.016 Da, as represented in the figure. The MS/MS spectra were obtained with HCD of the +2 m/z ion at 742.9096. (B) MS/MS spectrum of the ubiquitinated H3 from T. gondii peptide from positions 84 to 116 (FQSSAVLALQEAAEAYLVGLFEDTNLCAIHAKR). A ubiquitinated lysine at position 115 was identified. The Gly-Gly residue has a mass of 114.04 Da. The MS/MS spectra were obtained with HCD of the +4 m/z ion at 923.9716. yi* = yi − 57.02.
The first methionine is removed from the H3 protein, and R2 is methylated. Methylation on R2 is mediated by CARM1 and has been correlated with transcription activation in other organisms (35). H3K4 was found to be mono-, di-, or trimethylated, with mono- or dimethylation being approximately 100 times more represented than trimethylation among the identified peptides. H3K4me3 is a very conserved modification, present at promoter regions of actively transcribed genes in Toxoplasma and other organisms (18). In contrast, H3K4me1 and H3K4me2, although associated with transcription activation, are located in coding regions (see http://www.toxodb.org for ChIP with microarray technology [ChIP-chip] data illustrating this point). Additional H3 PTM associated with transcriptional activation, such as K9ac, K14ac, R17me, K18ac, K23ac, and K27ac, were identified (36), with H3K9ac and H3K14ac being among the most highly represented PTM. These PTM were found together with other PTM known to be associated with gene activation.
H3K9 was also methylated. H3K9me is considered a transcriptional activation mark, while H3K9me2 and H3K9me3 are associated with gene silencing (37). Usually, the methylated forms were found on peptides containing H3K14ac, a PTM associated with gene activation in other eukaryotes, indicating a possible alternative function for one of these PTM in T. gondii. H3K14 was found to be acetylated (H4K14ac) in most cases; however, it can also be monomethylated (H4K14me) or trimethylated (H3K14me3). The methylated form of H3K14 has been identified only in H3.2 from Pisum sativum (http://www.uniprot.org), but the function of this PTM is not known.
Other H3 PTM identified included methylation of K18 (me, me2, me3), K23 (me, me3), R26 (me, me2), K27 (me3), K36 (me, me2, me3), K37 (me, me2, and me3), and R40 (me, me2). The function of most of these PTM is not established, although R26me is purported to act in transcriptional activation, preventing histone deacetylase (HDAC) binding to chromatin (38), while K27me3 is reported to be involved in transcription repression (39). K36me2 is associated with DNA repair (40), and K37me1 is associated with the DNA replication origin (41).
In human H3, at least 8 phosphorylation sites are reported, but only one phosphorylated residue was identified for T. gondii H3, T107 in the globular domain. Phosphorylated T107 (T107p) is a new modification, with unknown function. Other well-conserved modifications, T11p, S10p, and S28p, associated with mitosis (42), were not identified in this study.
TgH3 also demonstrated other PTM inside its globular domain. Methylation of K79 (H3K79me or H3K79me3) has been described for many organisms as a PTM associated with gene activation or repression (37). The presence of methylated H3K79 in T. gondii is notable, since DOT1, the methyltransferase responsible for this PTM in other organisms, appears to be absent in the genome sequence of T. gondii. As occurs in other organisms (43), H3K79 can be acetylated. Methylation on R83, a PTM with unknown function, was also identified inside the globular domain. Finally, dimethyl and trimethyl marks were identified on the variant CenH3 lysine 33 (Fig. 3 and S6). Centromeric H3 from Toxoplasma is larger than in humans and yeast and divergent in sequence, and this modification is unique to Toxoplasma (Fig. 3 and S2).
To further validate our data, we showed that several commercial antibodies specific for conserved modified H3 peptides label both human and T. gondii nuclei by an indirect immunofluorescent-antibody assay (IFA) (Fig. S7).
Histone H2A and H2B.
In contrast to H3 and H4, T. gondii histones H2A and H2B show considerable sequence differences from human histones (79% and 75% conservation, respectively) (Fig. S1). These differences are concentrated in the N-terminal regions (especially the first 20 amino acids). Like other organisms, T. gondii encodes H2A variants: H2A.X and H2A.Z. While H2A.X is quite similar to the canonical histone (88% identity), H2A.Z shows only 56% identity to H2A, although it is generally considered the most conserved variant across eukaryotes. H2A is the only histone that has an exposed C-terminal domain (see Fig. S2 for histone variant alignments).
We identified only four modified peptides for the canonical H2A, including one extensively modified peptide with six methyl marks plus one crotonylation mark on peptide K5-K11 (Fig. S5). K96 is conserved in human H2A (K95), but unlike in humans, where this residue is acetylated (H2AK95ac), K96 in TgH2A is methylated (TgH2AK96me2). H2AS126p has a corresponding amino acid in human and Plasmodium H2A (Fig. 2 and Fig. S5). Two H2A.X-modified peptides were identified within the C-terminal region (Fig. S6). H2A.X contains the SQ(E/D) φ motif, where φ is a hydrophobic amino acid and S is a serine that is phosphorylated in response to DNA damage (44). T. gondii has the SQ(E/D) φ motif, where φ is phenylalanine. We identified the phosphorylated S132 (S139p in humans) within this motif, consistent with a conserved function for TgH2A.X, as suggested by Dalmasso et al. (13). In the same peptide, we also identified acetylation of K128 (H2A.XK128ac), a new PTM for this histone variant (Fig. 3 and Fig. S6).
Unlike with canonical H2A and H2A.X, several peptides and their PTM were identified for H2A.Z (Fig. 3 and S6). Unlike in other histones, the first methionine was preserved and the N terminus was acetylated. In other organisms, including T. gondii, H2A.Z deposition correlates with transcriptional activation and H2A.Z is hyperacetylated in the N-terminal region (45). Within the first 40 amino acids, there are 10 lysines (K6, K10, K14, K18, K24, K27, K29, K34, K36, and K37), all of which were acetylated in T. gondii. We also found that lysine K18 can be methylated, creating different PTM combinations with the acetylated residues.
Canonical H2B was also less represented in our data (Fig. 2 and S5). The first methionine was removed, but we found no indications of a PTM at the N terminus. Peptides were identified with acetylation on K4, methylation on K48 (me1), K63 (me1), and K70 (me3), and phosphorylation at S27, S49, S66, and Y74. The N terminus of human H2B has 7 amino acids that are acetylated in association with transcription activation (46). Although several lysines are present, T. gondii H2B has just 1 acetylated mark (K4), suggesting a difference between human and T. gondii H2B, especially at the N-terminal tail.
In contrast, several modifications were identified for H2Bv (also called H2B.Z), the major H2B in T. gondii (13) (Fig. 3 and S6). The first methionine is lost, and there is an acetylation at the N terminus. Additionally, the N-terminal tail is acetylated on lysines 3, 8, 13, 14, and 18. This histone is unique to parasites, and the acetylation PTM are conserved in P. falciparum (Fig. 3).
T. gondii histones are modified by novel PTM not previously seen in protozoa.
Additional modifications on histones, including succinylation, crotonylation, and formylation on lysine residues, have recently been identified (2, 4, 47). Although the role of succinylation sites is not completely elucidated, mutagenesis studies implicated succinylation in nucleosome structure, nucleosome-DNA interaction, telomeric function, and ribosomal DNA (rDNA) silencing (2).
Using different combined proteomic methods, we were able to identify succinylation on peptides from histone H3 (117VTIMPKsuccDIQLAR) (Fig. 6A) and H4 (1acSGRGKGGKGLGKmeGGAKsuccRHRKme2VLR and 24DNIQGITKsuccPAIR) (Fig. 2 and S3). Both H3K122succ and H4K31succ have previously been identified in a variety of eukaryotic species (2), while H4K16succ has not previously been reported. These results were confirmed by IFA nuclear localization of succinyl-lysine (Fig. S8) and immunoblotting (Fig. S8B), where succinyl-lysine antibodies recognized mainly H3. Interestingly, some parasite nuclei that were stained with antibody specific for succinyl-lysine showed stronger staining than with others present in the same vacuole, suggesting possible regulation of this PTM during the cell cycle (Fig. S8A).
N-lysine N-formylation is an additional PTM that emerges from DNA oxidative damage (47). The biological importance of this modification is not clear, but the chemical similarity of formylation with acetylation may suggest interference in acetylation-related pathways (47). We identified several formylated peptides of H3 and H4 (H3K56, H3K122, H4K31, H4K59, H4K67, H4K91). Lysine propionylation was described in 2007 as a new PTM that can be catalyzed by acetyltransferases (48). The function is being elucidated, but there is evidence that suggests a role in coenzyme A (CoA) homeostasis (49). In this study, we detected propionylation on H3 (K115) and H2B (K73). Finally, lysine crotonylation was described in 2011 as a new histone PTM associated with active promoters or enhancers (4). We identified 1 peptide (histone H2A) likely to be lysine crotonylated (Fig. S5). Further study will be necessary to confirm the presence and significance of crotonylation as a histone PTM in T. gondii.
Ubiquitination and SUMOylation of T. gondii histones.
The histone PTM repertoire is constantly increasing. Ubiquitination and SUMOylation (where SUMO is small ubiquitin-related modifier) are other common histone modifications. Both can be easily distinguished from other modifications by mass spectrometry techniques. The ubiquitin and SUMO small modifiers are 9 kDa and 11 kDa in size, respectively. The attachment of one of these modifications can affect a variety of processes, such as transcriptional activation, DNA damage response, or regulation of histone levels by targeting proteins for degradation. Both PTM were identified on histones of Plasmodium (15, 50), and SUMOylation was identified on Toxoplasma histones by immunologic methods (51). Immunofluorescence confirmed that the parasite nucleus is labeled with antibodies specific for SUMOylation (Fig. S8A). Using our MS approach, we were not able to identify SUMO on T. gondii histones. Although commercial antiubiquitin antibodies did not demonstrate reactivity with T. gondii histones (data not shown), we identified an H3 peptide modified by ubiquitination (Fig. 6B). BLAST search of the AVLALQEAAEAYLVGLFEDTNL sequence matched 100% of the sequence in different strains of T. gondii histone H3 but did not match that in human proteins. The precursor mass error is −2.6 ppm. The strongly matching b and y ions localize the ubiquitination site to K115.
DISCUSSION
Toxoplasma gondii has a complex life cycle that includes sexual and asexual stages in different hosts. To survive, T. gondii requires a refined system to regulate gene expression in response to different stimuli and changes in its environment. Many of these responses to environmental stimuli are achieved via epigenetic gene regulation. Chromatin architecture plays a key role in the regulation of gene expression regulation by controlling the accessibility of genes to the transcriptional machinery. Histones are the fundamental protein unit of chromatin and the main proteins responsible for structural changes in chromatin. PTM of histones provide a platform for recruitment of effector proteins triggering different DNA-related events, including transcription, DNA repair, and DNA replication. The combinations of histone PTM form the histone code, a core mechanism of gene expression regulation in eukaryotic organisms.
Histones are small, basic proteins that arose very early in evolution. There is evidence for the existence of proteins containing histone fold domains in Euryarchaea and histone genes in marine Crenarchaea (52). Histone gene organizations and copy numbers vary considerably across species. Except for TgH2B, for which there are two gene copies, Toxoplasma histones are encoded by single-copy genes on different chromosomes. Some histone genes have introns with introns usually present in histone variants. Toxoplasma histones are highly conserved with mammalian histones. H3 and H4 are the most conserved, and H2A and H2B, though similar, have different modifications, especially at the flexible domains.
We have generated a comprehensive map of histone PTM in Toxoplasma tachyzoites by using a combination of proteomic methodologies. We were able to identify 249 different peptides with 108 modifications in a variety of combinations (see Table S1 and Fig. S8). Peptides for all 4 canonical histones and 5 histone variants were identified. The histone code hypothesis proposes that histone modifications act independently, sequentially, or in collaboration, resulting in a specific event. Here we describe both conserved and unique histone PTM combinations, indicating a specific and unique histone code that exists in T. gondii. Although some combinations are consistent with a transcription activation role, many histone peptides show unique PTM compositions, with a mix of histone PTM associated with gene activation, repression, or unknown functions.
Many of the PTM identified in this study are conserved in humans and Plasmodium, especially those mapped to histone H3 and H4. Garcia et al. (43) showed that although some marks are well conserved, others vary considerably when unicellular eukaryotes are compared with mammalian cells. Toxoplasma displays both activation and silencing marks, although the PTM related with activation are more common.
We analyzed the tachyzoite forms of T. gondii, but it is likely that several histone PTM are conserved in the other life stages of the parasite. Nonetheless, histone PTM are extremely dynamic and change in response to external stimuli, including environmental changes. T. gondii passes through different hosts, and its life cycle forms are distinct morphologically and biochemically. Thus, it is likely that the parasite response to environmental cues involves regulation of histone PTM, which leads to changes in chromatin structure and gene expression. Measuring quantitatively how histone PTM change during developmental transitions is a topic for future investigation.
The Toxoplasma genome is predicted to encode a full repertoire of chromatin remodelers and chromatin-modifying enzymes, including histone deacetylases, histones acetyltransferases, histone demethylases, lysine and arginine methyltransferases, histone kinases, poly-ADP-ribose polymerases, ubiquitin ligases, and SUMO-conjugating enzymes (9, 19). Among the most thoroughly characterized of these histone-modifying enzymes are the histone acetyltransferases TgGCN5-A and TgGCN5B and the histone deacetylase TgHDAC3, which are implicated in regulation of gene expression during the tachyzoite- to bradyzoite-stage conversion (9, 53). In tachyzoites, TgGCN5-A is located in promoter regions of actively transcribed genes that are hyperacetylated (17). In contrast, promoter regions of bradyzoite-specific genes are hypoacetylated and are enriched in TgHDAC3 (17). The importance of these enzymes in gene regulation is supported by the abundance of histone acetylation in T. gondii histones, although broader studies of the T. gondii acetylome suggest that acetylation regulates numerous proteins beyond histones (33). Intriguingly, GCN5 may potentially be involved in propionylation of histones (48), given its documented association with metabolic sensing.
As in other organisms, TgH3 is extensively modified. However, the high homology between H3 and H3.3 does not allow us to assign specific PTM to the individual proteins; therefore, our analysis cannot establish whether the H3 PTM are specific to either subtype or shared by both. We identified only 2 unique peptides from CenH3, consistent with the low abundance of this variant and its very specific function in the centromeric region (12).
H2A and H2B were identified by a limited number of peptides (4 and 10, respectively), with more peptides being found for the variants H2A.Z and H2Bv. H2A.Z is the most divergent between H2A subspecies; however, it is considered the most conserved H2 variant between various eukaryotes (45). H2A.Z has several lysines at the N terminus that can be acetylated. In P. falciparum, H2A.Z is located in intergenic regions around the transcription start sites with H3K9ac and H3K4me3 (54). We found 10 acetylated lysines at the N terminus of H2A.Z. Most of those marks are conserved in Plasmodium but not in humans or yeast, which have an H2A.Z with a shorter N-terminal tail (Fig. 3 and S2). The parallels with P. falciparum have yet to be investigated, but differences are likely to exist, as centromeres in P. falciparum are enriched in H2A.Z but not H3K9me2/3 (55) whereas centromeres in T. gondii are the only chromosomal regions enriched in H3K9me2/3 (12) but are not enriched in H2A.Z (S. C. Nardelli and K. Kim, unpublished).
H2A and H2A.X are highly similar and share the SQ motif at the C terminus, although only H2A.X has the complete SQ(E/D) ϕ motif that is phosphorylated in response to DNA double-stranded breaks (13). Finally, H2Bv is a parasite-specific variant that interacts with H2A.Z at promoter regions of active genes in Toxoplasma (13), as well as at transcriptions start sites (TSS) in Trypanosoma brucei, together with H3K4me3 and H3K76me3 (corresponding to H3K79 in yeast and humans) (56). The role of H2Bv in transcription activation is consistent with the abundant acetylation in the N-terminal tail of H2Bv as well as H2A.Z. TgH2Bv showed 5 acetylated lysines that are conserved in Plasmodium (15). Since these variant histones localize at actively transcribed genetic loci, the greater abundance of those variants than of canonical histones may reflect the greater constitutive activation of most genes in T. gondii than of mammalian genes (18). It is also possible that the low abundance of peptides corresponding to H2A and H2B was an artifact of sample preparation; e.g., the accessibility of these proteins was altered during acid extraction, due to their enrichment in highly compacted chromatin.
Many of the histone PTM identified here are conserved in humans and other species, as is expected due to the high sequence similarity among histones, which may reflect an underlying shared eukaryotic histone code. Modifications associated with transcription activation, including H3K9ac, H3K14ac, H3R17me, H3K18ac, H3K23ac, and H3K27ac, as well as H4K5ac, H4K8ac, H4K12ac, and H4K16ac, are among the most abundant PTM and peptides found in our analysis. In Plasmodium, most histone PTM identified were associated with transcription activation. About 60% of Plasmodium genes are constitutively expressed (15), and the repressive mark H3K9me3 is predominantly associated with antigenically variant genes, such as var genes and subtelomeric regions (57). The lack of detection of repressive marks in Plasmodium likely reflects the substoichiometric abundance of many histone PTM. Recently, Jeffers and Sullivan identified several acetylation marks in histones of Toxoplasma using an acetyl-lysine enrichment strategy (33). Although most of those PTM were identified in this study, some are missing from our survey (H4K31ac, H2BK33/34, H2BK98/99, and H2BK110/111). Thus, for complete characterization of low-abundance histone PTM, the use of several complementary approaches is likely to yield the most comprehensive catalogue of PTM.
We were able to identify PTM associated with classic repressive marks, including H3K9me, H3K36me, and H3K79me, although peptides with these PTM were less abundant than peptides with PTM associated with active marks. We also observed that residues, like H3K9, that can be either methylated or acetylated were most commonly detected in the acetylated form. Collectively, these data suggest that most T. gondii tachyzoite chromatin is marked with PTM associated with the open active state.
During the entire process, we were mindful of the potential for contamination of our histone preparations with human histones. Since H3 and H4 proteins are highly conserved, many peptides cannot be conclusively assigned to only T. gondii histones. We attempted to minimize contamination by purifying the parasites from human host cells by filtration, prior to processing them for MS. In support of our methodology, we identified numerous modified peptides unique to T. gondii histone sequences, enabling us to validate T. gondii histone PTM conclusively. To further confirm the identification of highly conserved PTM across T. gondii and human chromatin, we performed immunofluorescence assays with commercial antibodies to characterized histone PTM. These antibodies labeled both human and Toxoplasma nuclei, supporting the identification of the particular histone PTM in both host and parasite nuclei (Fig. S7 and S8).
Finally, we found by MS that several recently described histone modifications, lysine succinylation, propionylation, formylation, and probably crotonylation, are present on T. gondii histones. The functions of these modifications have not been elucidated. Using commercial antibodies, we confirmed by immunofluorescence that succinylation is detected in parasite nuclei. Antibody studies also suggest that T. gondii histones are modified by SUMOylation, as previously described for T. gondii (51). Ubiquitination, a conserved PTM, was identified in Plasmodium (15) and is also present on T. gondii histone H3. Although commercial ubiquitin antibodies did not react to T. gondii histones by immunoblotting or IFA, we were able to identify a unique ubiquitinated lysine on H3 (K115), suggesting that ubiquitinylated T. gondii histones are substoichiometric and not highly abundant. Enrichment procedures for ubiquitination may be required to identify other ubiquitinated histones in T. gondii.
In conclusion, we provide here a comprehensive survey of T. gondii tachyzoite histone PTM. Many of these PTM have not been previously reported. The histone peptides identified show complex combinations of modifications, providing an important step for understanding how the histone code contributes to epigenetic regulation in this organism. Elucidation of the regulatory pathways for histone PTM should provide insight into T. gondii differentiation and gene regulation and may lead to therapeutic approaches to treat latent and active infection with this pathogen.
MATERIALS AND METHODS
Chemicals and reagents were high-purity grade and obtained from the sources cited in Text S1 (methods) in the supplemental material.
Cell culture and parasite purification.
Human foreskin fibroblasts (HFF) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin and maintained at 37°C with 5% CO2. Confluent HFF were infected with RH Δhxgprt tachyzoites (58) at a multiplicity of infection of 3 to 5 parasites per cell. After 40 h of infection, infected cells were washed and monolayers were scraped from the flasks and passed through 20-, 23-, and 25-gauge needles. Tachyzoites were purified from host cell debris with a 3.0-μm Nuclepore filter (Whatman GE Healthcare). Once no intact human cells were visible, the lysate containing intact tachyzoites was washed twice with phosphate-buffered saline (PBS) to remove host cell contaminants, and cell pellets were frozen in liquid nitrogen.
Immunoblots and immunofluorescence.
Immunoblot assays were performed using histones resolved by 15% SDS-PAGE. Nitrocellulose membranes were blocked with 5% nonfat milk in PBS and incubated with anti-succinyl lysine antibodies (1:400; PTM Biolabs Inc.) overnight. Membranes were washed with PBS containing 0.05% Tween 20 and incubated with secondary antibody coupled to horseradish peroxidase (Amersham) and chemiluminescence substrate for detection (Immobilon; Millipore).
For indirect-immunofluorescence experiments, HFF were grown on coverslips until confluent. Tachyzoites (106) were used for overnight infection followed by fixation with 4% p-formaldehyde in PBS for 20 min. Fixed cells were washed three times with PBS, permeabilized with 0.25% Triton X-100 in PBS for 10 min, blocked for 30 min with PBS containing 1% bovine serum albumin (BSA), and incubated with antibodies specific for succinylated lysine (1:200; PTM Biolabs Inc.), SUMO (1:500; gift of M. A. Hakimi [51]), H3K4me1, H3K4me2, H3K4me3, H3K9ac (all from Millipore), H3R17me2 (Abcam), and H3K27ac (LP Bio). After being incubated for 1 h, the slides were washed three times with PBS and incubated with secondary antibodies (Molecular Probes). Slides were stained with 0.01 mM 4′,6-diamidino-2-phenylindole (DAPI) to detect nucleic acids. Slides were mounted with Vectashield (Vector Laboratories) and analyzed with a DeltaVision Core microscope (inverted Olympus model IX71; Albert Einstein College of Medicine Analytical Imaging Facility).
Histone sample preparation.
Histone samples were prepared using an acid extraction protocol (59) with modifications (see Text S1 in the supplemental material). Approximately 5 × 108 fresh or frozen parasites were lysed in 1 ml of lysis solution composed of 0.25 M sucrose, 1 mM EDTA, 3 mM CaCl2, 0.01 M Tris HCl (pH 7.4), and 0.5% saponin and protease inhibitors. Histones were solubilized by the addition of 1 ml of 0.4 N HCl. Acid-soluble proteins were recovered by adding 8 volumes of acetone to the supernatant and precipitating them overnight at −20°C. The resultant histone-enriched pellet was resuspended in 30 to 50 µl of ultrapure water.
Proteomic analysis of histone preparations.
Full details of the proteomics sample preparation and analysis are in the supplemental material. Four complementary proteomic approaches were used to analyze the histone PTM (Fig. 1 and Text S1 in the supplemental material) using the high-resolution LTQ Orbitrap Velos mass spectrometer. These included (i) in-solution digestion with trypsin followed by 2D LC-MS/MS analysis by higher-energy collisional dissociation (HCD), electron transfer dissociation (ETD), or collision-induced dissociation (CID); (ii) separation of histone acidic extracts by SDS-PAGE and in-gel trypsin digestion followed by 2D LC-MS/MS; (iii) in-solution Asn-N digestion followed by 1-dimensional (1D) LC-MS/MS with ETD analysis; and (iv) middle-down fractionation of histone acidic extract by off-line HPLC and in-solution Asp-N digestion, followed by 1D LC-MS/MS with ETD analysis.
Data analysis.
MS/MS raw data were converted to text files (Mascot generic files [mgf]) with Proteome Discoverer 1.10 (Thermo Fisher Scientific Inc.). MS/MS searches were routinely performed against a human protein database as well as the T. gondii protein database (60). Since searches with large MS/MS data files against both T. gondii and human protein databases for the identification of all possible varieties of histone posttranslational modifications are time- and CPU-intensive, we used a two-step search strategy to accelerate and facilitate the searches. Mascot searches were first performed against a limited protein database composed of sequences of T. gondii histones H3, H4, H2A, and H2B, as well as the variant histones H3.3, H2Bv, H2A.Z, H2A.X, and CenH3 and a potential H1 (TgME49_115570). Mascot searches were repeated using different combinations of PTM (see Text S1 in the supplemental material for details). The peptide mass tolerance used was 20 or 40 ppm, and the product mass tolerance was ±0.1 Da for product ions (for HCD or ETD, when Orbitrap was used for MS/MS) and ±0.25 Da (for CID, when an ion trap was used for MS/MS).
All MS/MS spectra matching those of posttranslationally modified histone peptides were extracted from the original mgf and combined into a new mgf for a second search using the same search parameters as the first search (described in Text S1 in the supplemental material) against a combined protein database comprised of all protein sequences of T. gondii and humans.
All spectra for the possible hits of T. gondii histone peptides containing posttranslational modifications were inspected manually. To accept the identification of an MS/MS spectrum as a modified T. gondii histone peptide, the following criteria were used: (i) the MS/MS spectrum was identified as a histone peptide of T. gondii in the second database search, (ii) the observed peptide mass error was within ±7 ppm of the theoretical peptide mass (the observed MS/MS fragment mass error was within ±0.1 Da [when Orbitrap was used for MS/MS] or 0.25 Da [when the ion trap was used for MS/MS]), (iii) the MS/MS spectrum had 5 or more consecutive N-terminal or C-terminal fragment ions that matched the theoretical fragment ions, and (iv) more than 80% of major fragment ions matched N-terminal or C-terminal fragment ions.
The overall peptide false-discovery rate was 3.8% (based upon results of a Mascot decoy database search), and a list of the peptide masses, including their errors, is provided in Table S1 in the supplemental material.
MS/MS spectra of the identified modified histone peptides for each histone (H2A, H2B, H3, and H4) and histone variant (H2Bv, CenH3, H2A.Z, and H2A.X) are available for download at http://fiserlab.org/biodefense/ (EPICdB) under Downloads. The peptide sequence and observed m/z and charge state of a precursor ion are shown for each modified peptide.
SUPPLEMENTAL MATERIAL
Alignment of canonical histones from Toxoplasma gondii, Plasmodium falciparum, Homo sapiens, and Saccharomyces cerevisiae. The genes for T. gondii histones have single copies, except that for T. gondii H2B. The accession numbers of the genes for the following histones are as indicated: H3 (T. gondii, TGME49_061240; P. falciparum, PFF0510w; H. sapiens, AAA52651.1), H4 (T. gondii, TGME49_039260; P. falciparum, PF11_0061; H. sapiens, AAA52652.1; and S. cerevisiae, EDV12280.1), H2A (T. gondii, TGME49_061250; P. falciparum, PFF0860c; H. sapiens, AAN59974.1), and H2B (T. gondii, TGME49_105160 and TGME49_051870; P. falciparum, PF11_0062; H. sapiens, CAA41051.1; and S. cerevisiae, CAA81268.1). S. cerevisiae does not have a canonical H3 or H2A. Instead S. cerevisiae H3 (ScH3) and ScH2A more closely resemble H3.3 and H2A.Z but act as canonical histones. Yellow amino acids represent the identical amino acids between all species. Download
Alignment of histone variants from Toxoplasma gondii, Plasmodium falciparum, Homo sapiens, and Saccharomyces cerevisiae, namely, H3.3 (T. gondii, TGME49_018260; P. falciparum, PFF0865w; H. sapiens, CAA29288.1; and S. cerevisiae, AAS64349.1), CenH3 (T. gondii, TGME49_025410; P. falciparum, PF13_0185; H. sapiens, AAH02703.1; and S. cerevisiae, CAA81884.1), H2AZ (T. gondii, TGME49_100200; P. falciparum, PFC0920w; H. sapiens, AAA35984.1; and S. cerevisiae, EDV10597.1), H2A.X (T. gondii, TGME49_061580; H. sapiens, NP_002096.1; and S. cerevisiae, CAA81267.1), and H2Bv (T. gondii, TGME49_009910; P. falciparum, PF07_0054). In P. falciparum, H2AX is not present. H2Bv is a variant specific for parasites. Download
Combinations of PTM identified on histone H4. The different combinations of modified H4 peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), succinylation (succ), formylation (form), and oxidation (ox). The unique peptide sequences (U) that distinguish TgH4 from HsH4 (I54T, I65V, K67R, S69A, R77K, I86V, and S89A [highlighted in yellow]) are well represented among the H4 peptides. The brackets show the region where the indicated modifications occur (1.2 or 3 methyl groups are present at the selected areas), but the exact location of the modifications could not be determined. Download
Combinations of PTM identified on histone H3. The different combinations of modified H3 peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), succinylation (succ), formylation (form), oxidation (ox), phosphorylation (p), and ubiquitination (ub). The unique peptide sequences (U) that distinguish TgH3 from HsH3 (S22T, M31A, S32T, I35V, D59E, L90M, A96C, R134A [highlighted in yellow]) are well represented among the H3 peptides. The brackets show the region where the indicated modifications occur (1, 2, 3, or 4 methyl groups are present on the selected areas), but the exact locations of the modifications could not be determined. Download
Combinations of PTM identified on histone H2A and H2B. The different combinations of modified peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), propionylation (prop), formylation (form), oxidation (ox), and phosphorylation (p). The brackets show the region where the indicated modification occurs. On H2A histone, one crotonylation mark and 3 methyl groups were found distributed on residues 5 to 10. The specific combination of the modifications could not be determined. On H2B, 4 methyl groups and 2 oxidations are present on the selected areas. Download
Combinations of PTM identified on T. gondii histone variants. The different combinations of modified peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), oxidation (ox), and phosphorylation (p). The bracket shows an acetylation mark at the selected region for H2Bv. Download
Indirect immunofluorescence using antibodies specific for conserved histone PTM. Commercial antibodies for the indicated histone modification were tested by IFA on HFF infected with tachyzoites. Labeling is observed for human nuclei and tachyzoite nuclei. The boxed area indicates the higher-magnification (zoom) detail. Download
Uncommon and novel modifications identified on T. gondii histones. (A) Indirect immunofluorescence was performed on HFF infected with RH strain tachyzoites using antibodies specific for succinyl-lysine (PTM Biolabs Inc.) and TgSUMO (gift of M. A. Hakimi). Nuclei were stained with DAPI. In both cases, we localized the modifications inside the parasite nucleus. (B) Western blot assays of histone acid-enriched samples. Positions of histones are indicated next to the Coomassie blue-stained gel. Antibody specific for succinylated-lysine recognized H3 as well as H2B and H2A. Download
List of modified histone peptides. The table lists the sequences of the identified histone peptides and peptide scores of the Mascot database search against the combined protein database of H. sapiens and Toxoplasma gondii. The observed m/z, theoretical m/z, peptide ion charge state, observed peptide mass, theoretical peptide mass, and the measurement error of peptide m/z are shown for every peptide in this table. Note that those modifications that could not be unambiguously localized to specific amino acids are highlighted with colors and parentheses. For example, for an H3 peptide, ARTKmeQTARKSTGGKAPRK(3me) QLASKAARKSAPMSGGIKKPHR(4me) YRPGTVALR, the three methyl groups (3me) could potentially be localized at K9, K14, R17, and K18, and the four methyl groups (4me) could be exactly localized to K37 or R40.
Experimental procedures. Download
ACKNOWLEDGMENTS
We thank Mohammed Ali Hakimi for the gift of TgSUMO antibodies.
This research was supported by National Institutes of Health grants R01AI087625 (K.K.), RC4AI092801 (K.K., W.J.S.), R01AI095094 (L.M.W.), 5T32AI070117 (S.C.N.), and R01AI083162 (S.O.A., W.J.S.), as well as NIH instrument grant 1S10RR029398 for the LTQ Orbitrap Velos mass spectrometer system. We also acknowledge the support of the Einstein-Montefiore Center for AIDS Research, funded by grant P30AI051519.
Footnotes
Citation Nardelli SC, Che F-Y, Silmon de Monerri NC, Xiao H, Nieves E, Madrid-Aliste C, Angel SO, Sullivan WJ, Jr, Angeletti RH, Kim K, Weiss LM. 2013. The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. mBio 4(6):e00922-13. doi:10.1128/mBio.00922-13.
REFERENCES
- 1. Khorasanizadeh S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259–272 [DOI] [PubMed] [Google Scholar]
- 2. Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. 2012. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11:100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanover JA. 2010. Epigenetics gets sweeter: O-GlcNAc joins the “histone code.” Chem. Biol. 17:1272–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146:1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 6. Dubey JP, Lindsay DS, Speer CA. 1998. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11:267–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. 2005. The transcriptome of Toxoplasma gondii. BMC Biol. 3:26. 10.1186/1741-7007-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darkin-Rattray SJ, Gurnett AM, Myers RW, Dulski PM, Crumley TM, Allocco JJ, Cannova C, Meinke PT, Colletti SL, Bednarek MA, Singh SB, Goetz MA, Dombrowski AW, Polishook JD, Schmatz DM. 1996. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U. S. A. 93:13143–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dixon SE, Stilger KL, Elias EV, Naguleswaran A, Sullivan WJ., Jr. 2010. A decade of epigenetic research in Toxoplasma gondii. Mol. Biochem. Parasitol. 173:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ, Jr, Wang H, Brunk BP. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36:D553–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dalmasso MC, Echeverria PC, Zappia MP, Hellman U, Dubremetz JF, Angel SO. 2006. Toxoplasma gondii has two lineages of histones 2b (H2B) with different expression profiles. Mol. Biochem. Parasitol. 148:103–107 [DOI] [PubMed] [Google Scholar]
- 12. Brooks CF, Francia ME, Gissot M, Croken MM, Kim K, Striepen B. 2011. Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 108:3767–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalmasso MC, Onyango DO, Naguleswaran A, Sullivan WJ, Jr, Angel SO. 2009. Toxoplasma H2A variants reveal novel insights into nucleosome composition and functions for this histone family. J. Mol. Biol. 392:33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miao J, Fan Q, Cui L, Li J, Li J, Cui L. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369:53–65 [DOI] [PubMed] [Google Scholar]
- 15. Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. 2009. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome Res. 8:3439–3450 [DOI] [PubMed] [Google Scholar]
- 16. Cui L, Miao J. 2010. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot. Cell 9:1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr, Cesbron-Delauw MF, Hakimi MA. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25:10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLOS Pathog. 3:e77. 10.1371/journal.ppat.0030077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bougdour A, Braun L, Cannella D, Hakimi MA. 2010. Chromatin modifications: implications in the regulation of gene expression in Toxoplasma gondii. Cell. Microbiol. 12:413–423 [DOI] [PubMed] [Google Scholar]
- 20. Shvartsburg AA, Zheng Y, Smith RD, Kelleher NL. 2012. Separation of variant methylated histone tails by differential ion mobility. Anal. Chem. 84:6317–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eliuk SM, Maltby D, Panning B, Burlingame AL. 2010. High resolution electron transfer dissociation studies of unfractionated intact histones from murine embryonic stem cells using on-line capillary LC separation: determination of abundant histone isoforms and translational modifications. Mol. Cell. Proteomics 9:824–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chu F, Nusinow DA, Chalkley RJ, Plath K, Panning B, Burlingame AL. 2006. Mapping post-translational modifications of the histone variant MacroH2A1 using tandem mass spectrometry. Mol. Cell. Proteomics 5:194–203 [DOI] [PubMed] [Google Scholar]
- 23. Lee AY, Paweletz CP, Pollock RM, Settlage RE, Cruz JC, Secrist JP, Miller TA, Stanton MG, Kral AM, Ozerova ND, Meng F, Yates NA, Richon V, Hendrickson RC. 2008. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J. Proteome Res. 7:5177–5186 [DOI] [PubMed] [Google Scholar]
- 24. Garcia BA, Shabanowitz J, Hunt DF. 2007. Characterization of histones and their post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 11:66–73 [DOI] [PubMed] [Google Scholar]
- 25. Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. 2009. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8:2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia BA. 2009. Mass spectrometric analysis of histone variants and post-translational modifications. Front. Biosci. (Schol. Ed.) 1:142–153 [DOI] [PubMed] [Google Scholar]
- 27. Sidoli S, Cheng L, Jensen ON. 2012. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J. Proteomics. 75:3419–3433 [DOI] [PubMed] [Google Scholar]
- 28. Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3:1154–1169 [DOI] [PubMed] [Google Scholar]
- 29. Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. 2007. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Green EM, Morrison AJ, Gozani O. 2012. New marks on the block: Set5 methylates H4 lysines 5, 8 and 12. Nucleus 3:335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reinberg D, Chuikov S, Farnham P, Karachentsev D, Kirmizis A, Kuzmichev A, Margueron R, Nishioka K, Preissner TS, Sarma K, Abate-Shen C, Steward R, Vaquero A. 2004. Steps toward understanding the inheritance of repressive methyl-lysine marks in histones. Cold Spring Harb. Symp. Quant. Biol. 69:171–182 [DOI] [PubMed] [Google Scholar]
- 32. Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, Tardieux I, Belrhali H, Hakimi MA. 2007. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol. Cell. Biol. 27:5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeffers V, Sullivan WJ., Jr. 2012. Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 11:735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Côté J. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28:1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748 [DOI] [PubMed] [Google Scholar]
- 36. Shahbazian MD, Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100 [DOI] [PubMed] [Google Scholar]
- 37. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 38. Wu J, Cui N, Wang R, Li J, Wong J. 2012. A role for CARM1-mediated histone H3 arginine methylation in protecting histone acetylation by releasing corepressors from chromatin. PLoS One 7:e34692. 10.1371/journal.pone.0034692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309–25317 [DOI] [PubMed] [Google Scholar]
- 40. Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. 2011. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 108:540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dorn ES, Cook JG. 2011. Nucleosomes in the neighborhood: new roles for chromatin modifications in replication origin control. Epigenetics 6:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Cadahia B, Drobic B, Davie JR. 2010. 298:H3–H4 phosphorylation: dual role in mitosis and interphase. Biochem. Cell Biol. 87:695–709 [DOI] [PubMed] [Google Scholar]
- 43. Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. 2007. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282:7641–7655 [DOI] [PubMed] [Google Scholar]
- 44. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- 45. Talbert PB, Henikoff S. 2010. Histone variants—ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11:264–275 [DOI] [PubMed] [Google Scholar]
- 46. Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189–1192 [DOI] [PubMed] [Google Scholar]
- 47. Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. 2007. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. 2007. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. 2007. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282:30239–30245 [DOI] [PubMed] [Google Scholar]
- 50. Issar N, Roux E, Mattei D, Scherf A. 2008. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell. Microbiol. 10:1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Braun L, Cannella D, Pinheiro AM, Kieffer S, Belrhali H, Garin J, Hakimi MA. 2009. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 39:81–90 [DOI] [PubMed] [Google Scholar]
- 52. Eirín-López JM, Dryhurst D, Méndez J, Jusio J. 2009. Long-term evolution of histone families: old notions and new insights into their mechanisms of diversification across eukaryotes, p 139–162 In Pontarotti P, Evolutionary biology: concept, modeling, and application. Springer-Verlag, Berlin, Germany [Google Scholar]
- 53. Bhatti MM, Livingston M, Mullapudi N, Sullivan WJ., Jr. 2006. Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 5:62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bártfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Françoijs KJ, Stunnenberg HG. 2010. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 6:e1001223. 10.1371/journal.ppat.1001223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoeijmakers WA, Stunnenberg HG, Bártfai R. 2012. Placing the Plasmodium falciparum epigenome on the map. Trends Parasitol. 28:486–495 [DOI] [PubMed] [Google Scholar]
- 56. Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. 2009. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 23:1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lopez-Rubio JJ, Mancio-Silva L, Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190 [DOI] [PubMed] [Google Scholar]
- 58. Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 [DOI] [PubMed] [Google Scholar]
- 59. Toro GC, Galanti N. 1990. Trypanosoma cruzi histones. Further characterization and comparison with higher eukaryotes. Biochem. Int. 21:481–490 [PubMed] [Google Scholar]
- 60. Madrid-Aliste CJ, Dybas JM, Angeletti RH, Weiss LM, Kim K, Simon I, Fiser A. 2009. EPIC-DB: a proteomics database for studying apicomplexan organisms. BMC Genomics 10:38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Khare SP, Habib F, Sharma R, Gadewal N, Gupta S, Galande S. 2012. HIstome—a relational knowledgebase of human histone proteins and histone modifying enzymes. Nucleic Acids Res. 40:D337–D342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of canonical histones from Toxoplasma gondii, Plasmodium falciparum, Homo sapiens, and Saccharomyces cerevisiae. The genes for T. gondii histones have single copies, except that for T. gondii H2B. The accession numbers of the genes for the following histones are as indicated: H3 (T. gondii, TGME49_061240; P. falciparum, PFF0510w; H. sapiens, AAA52651.1), H4 (T. gondii, TGME49_039260; P. falciparum, PF11_0061; H. sapiens, AAA52652.1; and S. cerevisiae, EDV12280.1), H2A (T. gondii, TGME49_061250; P. falciparum, PFF0860c; H. sapiens, AAN59974.1), and H2B (T. gondii, TGME49_105160 and TGME49_051870; P. falciparum, PF11_0062; H. sapiens, CAA41051.1; and S. cerevisiae, CAA81268.1). S. cerevisiae does not have a canonical H3 or H2A. Instead S. cerevisiae H3 (ScH3) and ScH2A more closely resemble H3.3 and H2A.Z but act as canonical histones. Yellow amino acids represent the identical amino acids between all species. Download
Alignment of histone variants from Toxoplasma gondii, Plasmodium falciparum, Homo sapiens, and Saccharomyces cerevisiae, namely, H3.3 (T. gondii, TGME49_018260; P. falciparum, PFF0865w; H. sapiens, CAA29288.1; and S. cerevisiae, AAS64349.1), CenH3 (T. gondii, TGME49_025410; P. falciparum, PF13_0185; H. sapiens, AAH02703.1; and S. cerevisiae, CAA81884.1), H2AZ (T. gondii, TGME49_100200; P. falciparum, PFC0920w; H. sapiens, AAA35984.1; and S. cerevisiae, EDV10597.1), H2A.X (T. gondii, TGME49_061580; H. sapiens, NP_002096.1; and S. cerevisiae, CAA81267.1), and H2Bv (T. gondii, TGME49_009910; P. falciparum, PF07_0054). In P. falciparum, H2AX is not present. H2Bv is a variant specific for parasites. Download
Combinations of PTM identified on histone H4. The different combinations of modified H4 peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), succinylation (succ), formylation (form), and oxidation (ox). The unique peptide sequences (U) that distinguish TgH4 from HsH4 (I54T, I65V, K67R, S69A, R77K, I86V, and S89A [highlighted in yellow]) are well represented among the H4 peptides. The brackets show the region where the indicated modifications occur (1.2 or 3 methyl groups are present at the selected areas), but the exact location of the modifications could not be determined. Download
Combinations of PTM identified on histone H3. The different combinations of modified H3 peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), succinylation (succ), formylation (form), oxidation (ox), phosphorylation (p), and ubiquitination (ub). The unique peptide sequences (U) that distinguish TgH3 from HsH3 (S22T, M31A, S32T, I35V, D59E, L90M, A96C, R134A [highlighted in yellow]) are well represented among the H3 peptides. The brackets show the region where the indicated modifications occur (1, 2, 3, or 4 methyl groups are present on the selected areas), but the exact locations of the modifications could not be determined. Download
Combinations of PTM identified on histone H2A and H2B. The different combinations of modified peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), propionylation (prop), formylation (form), oxidation (ox), and phosphorylation (p). The brackets show the region where the indicated modification occurs. On H2A histone, one crotonylation mark and 3 methyl groups were found distributed on residues 5 to 10. The specific combination of the modifications could not be determined. On H2B, 4 methyl groups and 2 oxidations are present on the selected areas. Download
Combinations of PTM identified on T. gondii histone variants. The different combinations of modified peptides are shown with the positions of PTM. Abbreviations include acetylation (ac), methylation (me), oxidation (ox), and phosphorylation (p). The bracket shows an acetylation mark at the selected region for H2Bv. Download
Indirect immunofluorescence using antibodies specific for conserved histone PTM. Commercial antibodies for the indicated histone modification were tested by IFA on HFF infected with tachyzoites. Labeling is observed for human nuclei and tachyzoite nuclei. The boxed area indicates the higher-magnification (zoom) detail. Download
Uncommon and novel modifications identified on T. gondii histones. (A) Indirect immunofluorescence was performed on HFF infected with RH strain tachyzoites using antibodies specific for succinyl-lysine (PTM Biolabs Inc.) and TgSUMO (gift of M. A. Hakimi). Nuclei were stained with DAPI. In both cases, we localized the modifications inside the parasite nucleus. (B) Western blot assays of histone acid-enriched samples. Positions of histones are indicated next to the Coomassie blue-stained gel. Antibody specific for succinylated-lysine recognized H3 as well as H2B and H2A. Download
List of modified histone peptides. The table lists the sequences of the identified histone peptides and peptide scores of the Mascot database search against the combined protein database of H. sapiens and Toxoplasma gondii. The observed m/z, theoretical m/z, peptide ion charge state, observed peptide mass, theoretical peptide mass, and the measurement error of peptide m/z are shown for every peptide in this table. Note that those modifications that could not be unambiguously localized to specific amino acids are highlighted with colors and parentheses. For example, for an H3 peptide, ARTKmeQTARKSTGGKAPRK(3me) QLASKAARKSAPMSGGIKKPHR(4me) YRPGTVALR, the three methyl groups (3me) could potentially be localized at K9, K14, R17, and K18, and the four methyl groups (4me) could be exactly localized to K37 or R40.
Experimental procedures. Download