Abstract
Cytoadherence of red blood cells (RBCs) invaded by Plasmodium falciparum parasites is an important contributor to the sequestration of RBCs causing reduced microcirculatory flow associated with fatal malaria syndromes. The phenomenon involves a parasite–derived variant antigen, the P. falciparum erythrocyte membrane protein 1 (PfEMP1), and several human host receptors, such as chondroitin sulfate A (CSA) which has been explicitly implicated in placental malaria. Elucidating the molecular mechanisms of cytoadherence requires quantitative evaluation, under physiologically relevant conditions, of the specific receptor–ligand interactions associated with pathological states of cell–cell adhesion. Such quantitative studies have not been reported thus far for P. falciparum malaria under conditions of febrile temperatures that accompany malarial infections. In this study, single RBCs infected with P. falciparum parasites (CSA binding phenotype), in trophozoite stage, have been engaged in mechanical contact with the surface of surrogate cells specifically expressing CSA, so as to quantify cytoadherence to human syncytiotrophoblasts in a controlled manner. From these measurements, a mean rupture force of 43 pN was estimated for the CSA–PfEMP1 complex at 37 °C. Experiments carried out at febrile temperature showed a noticeable decrease in CSA–PfEMP1 rupture force (by about 23% at 41 °C and about 20% after a 40 °C heat–treatment), in association with an increased binding frequency. The decrease in rupture force points to a weakened receptor–ligand complex after exposure to febrile temperature, while the rise in binding frequency suggests an additional display of nonspecific binding molecules on the RBC surface. The present work establishes a robust experimental method for the quantitative assessment of cytoadherence of diseased cells with specific molecule–mediated binding.
Keywords: Malaria, cytoadherence, chondroitin sulfate A, force spectroscopy, single–cell probe, diseased cell adhesion, fever
1. Introduction
Life-threatening malaria in humans is known to arise from the Plasmodium falciparum protozoan. The human red blood cells (RBCs), when invaded by the P. falciparum, host a 48–h asexual reproduction cycle of the merozoite, that leads to the clinical symptoms of the disease [1,2]. During this continuous intra–erythrocytic cycle, severe modifications occur in the iRBCs, including (a) an order–of–magnitude decrease in cell stiffness associated with a markedly diminished deformability [35] and (b) the formation of nanoscale protrusions (known as “knobs”) on the extracellular membrane surface that facilitate significant adhesion [6]. A consequence of adhesive knob formation is the sequestration of iRBCs in various organs, such as the brain, lung and placenta, due to the binding of iRBCs to endothelial cells, placental syncytiotrophoblasts, other iRBCs and/or healthy RBCs [7,8]. Collectively these phenomena, termed cytoadherence, deter mature iRBCs from reaching the spleen where they would be cleared from the bloodstream by virtue of their reduced deformability. Cytoadherence thus produces a partial or complete obstruction of blood flow in the microvasculature, which is considered a key contributor to fatal syndromes such as placental malaria.
The nano-scale protrusions form in the later stages of the intra-erythrocytic maturation stage of the merozoites. These protrusions house the knob–associated histidine–rich protein (KAHRP), which provides anchorage for the external display of the adhesive antigen, P. falciparum erythrocyte membrane protein 1 (PfEMP1) (for a review of the ultrastructure of the knob, see [9]). The PfEMP1 variable extracellular cysteine-rich interdomain regions (CIDR) and Duffy-binding-like regions (DBL) modulate adhesion to a variety of host receptors that include: (a) cluster determinant 36 (CD36); (b) inter-cellular adhesion molecule 1 (ICAM1); (c) thrombospondin (TSP); (d) complement receptor 1 (CR1); and (e) chondroitin sulfate A (CSA) [8]. How each individual PfEMP1 polymorph binds to specific host receptors depends on its unique DBL and CIDR. This antigenic variation promotes immune evasion by P. falciparum, and different adhesion phenotypes are correlated with different disease pathologies [10]. For example, CSA has been identified as the main receptor for PfEMP1 attachment to placental cells [11]. This PfEMP1 variant, specifically associated with pregnancy, contributes to severe disease and death of not only the developing fetus but also of the mother carrying her first child; although in subsequent pregnancies, women tend to develop protective antibodies [12].
The mechanical behavior of single biological cells is central to the human malaria pathology and can be investigated by recourse to a variety of independent experimental techniques, such as magnetic twisting cytometry, optical tweezers, micropipette aspiration, microfluidics, and atomic force microscopy, all of which allow direct, real-time testing of single cells [14–17]. In the case of RBCs infected with P. falciparum, quantitative research on mechanical properties has been focused primarily on iRBC elasticity and deformability [5,18]. Recently, an Atomic Force Microscopy (AFM) study, relying on tip functionalization, reported intrinsic kinetic parameters of CD36 and TSP single-molecule interactions with PfEMP1 [19]. In addition, iRBC probes have been used to investigate CD36 clustering occurring on endothelial cell membranes for long duration cell–cell adhesion [20]. However, systematic quantitative details on the cytoadherence involving the receptor implicated in placental malaria have thus far not been reported. Such studies are vital to comprehend the pathogenic basis of malaria occurring in primigravidae, and enable precise diagnostics.
Antimalarial drugs have been mainly directed to the specific metabolic pathways associated with hemoglobin digestion, and act essentially through inhibition of hemozoin crystallization, thereby impairing heme detoxification by the parasite [21]. However, due to the emerging resistance to the artemisinin–based treatments, new antimalarials are an utmost priority [22]. In this context, a fundamental understanding of cytoadherence at physiologically/pathologically relevant temperatures provides valuable information for alternative therapeutic strategies and corresponding drug efficacy assays. Furthermore, quantitative understanding of iRBC cytoadherence mediated by P. falciparum malaria is also necessary to guide and validate recent advances in multi-scale computational simulation methods, such as those involving dissipative particle dynamics (DPD) [23,24]. These techniques enable predictive simulations of the impaired microcirculation arising from reduced deformability and enhanced cytoadherence, that accompany malaria infection. However, the accuracy and validity of these computational models depend heavily on quantitative data for receptor–ligand interactions, which can only be determined through physiologically-relevant experimental assays of cell–cell adhesion.
This paper describes the results of a novel force spectroscopy protocol developed to quantify the adhesion of individual iRBCs to other living cells expressing specific receptors, both at normal physiological temperature and at febrile temperatures symptomatic of malaria. Specifically, RBCs infected with the P. falciparum FCR3–CSA strain (CSA-binding phenotype) have been used to probe Chinese Hamster Ovary (CHO) cells expressing CSA at 37 °C. The influence of febrile temperatures on the adhesion behavior has been assessed by cell–cell probing at 41 °C and, additionally, by incubating iRBCs for 1 h at 40 °C prior to probing CHO cells at 37 °C.
2. Methods
2.1. Cell culture
Parasite culture
P. falciparum FCR3–CSA parasites were maintained in leukocyte-free human RBCs (Research Blood Components, Boston, MA) under an atmosphere of 3% O2, 5% CO2, and 92% N2 in Roswell Park Memorial Institute (RPMI) medium 1640 (Gibco Life Technologies, Rockville, MD) supplemented with 25 mM Hepes (Sigma, St. Louis, MO), 200 mM hypoxanthine (Sigma), 0.209% NaHCO3 (Sigma), and 0.25% albumax I (Gibco Life Technologies). Synchronized cultures were obtained successively by concentration of mature schizonts using plasmagel flotation [25] and sorbitol lysis 2 h after the merozoite invasion [26]. The tests were performed within 24–36 h (trophozoite stage) after merozoite invasion of the erythrocyte.
Counter-cell selection and culture
Selection of the counter-cells involved the following criteria: (i) expression of CSA and (ii) inhibition of non-specific binding. Although CSA has been consistently identified as the dominant placental adhesion receptor [27,28], it has also been shown that ICAM-1 is hyperexpressed on syncytiotrophoblasts of malaria-infected hosts [29]. Therefore, in order to rule out any possible residual binding that may compete with CSA-PfEMP1 when using syncytiotrophoblasts, Chinese Hamster Ovary cells (CHO–K1, CCL–61 American Type Culture Collection) have been chosen as surrogates to these human host cells, and their binding specificity has been demonstrated through static adherence assays. The CHO cells were grown in an incubator at 37 °C with 5% CO2 in a F–12K (ATCC) modified medium containing 10% Fetal Bovine Serum (Gibco, 26140–079) heat inactivated at 56 °C for 0.5 h, and 1% Penicillin / Streptomycin (Biofluids, 303).
2.2. Force spectroscopy
Experimental setup
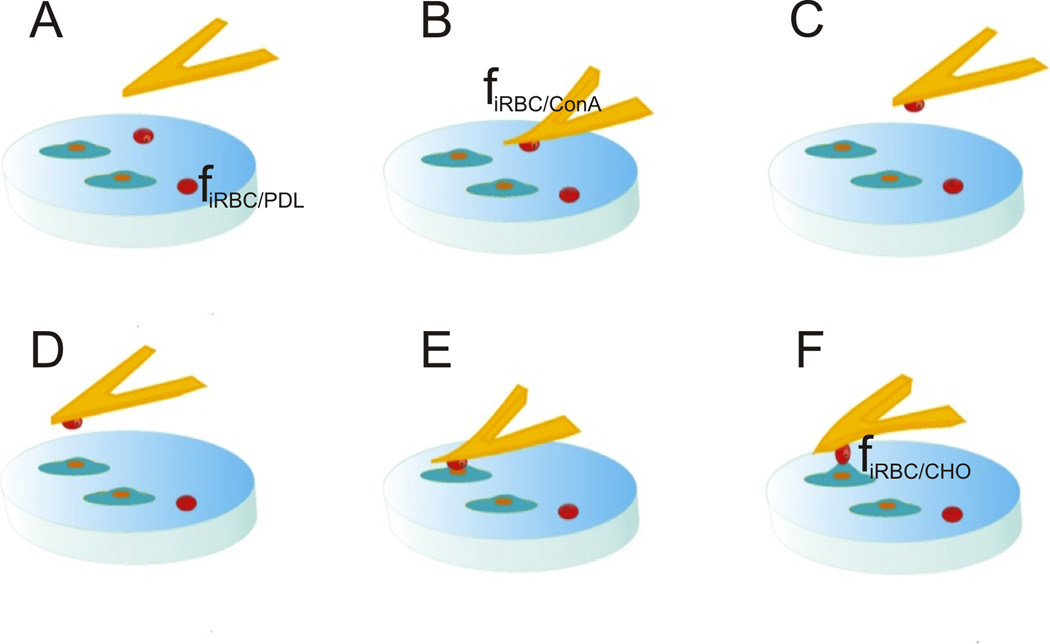
Figure 1 illustrates the experimental setup used for the rupture force measurements. The experiments involved culturing CHO cells on glass slides previously coated with poly–D–lysine (PDL). P. falciparum iRBCs in trophozoite stage were then poured over the CHO cell culture slide and were allowed to weakly bind to the glass substrate through PDL mediation (step A). This gentle immobilization on the glass slide was required for a precise engagement of the tipless cantilever on a chosen iRBC (step B). The prior incubation of the cantilever with concanavalin A (ConA) induced a solid attachment of the iRBC to the cantilever, and the diseased cell could then serve as a contact probe (step C). The attached iRBC was subsequently positioned above a CHO cell (step D) and pressed against this cell until the cantilever deflection reached the value corresponding to a preset trigger force (step E). Precise attachment of the iRBC at the very end of the tipless cantilever prevented the cantilever surface (incubated with ConA) from contacting the counter cell, which would render the adhesion force measurements meaningless. After an appropriately defined contact time (see further discussion to follow), the cantilever was retracted at a set speed until the two cells were completely separated (step F). The cantilever deflection measured during retraction was used to determine the rupture force (f) between the iRBC and the CHO cell. The adhesive mediators (PDL and ConA) required fine–tuning so that fiRBC/substrate < fiRBC/cantilever > fiRBC/CHO. Control experiments were carried out with uninfected RBCs under similar conditions.
Figure 1.
Setup of the force spectroscopy experiments: glass slide pre–coated with PDL and presenting a CHO subconfluent monolayer culture. iRBCs are poured onto the slide and allowed to bind lightly to the substrate (step A); a tipless cantilever previously incubated in a ConA solution is engaged on an iRBC in trophozoite stage (B); the iRBC attached to the tipless cantilever is used as a single–cell probe (D,E,F).
Slide preparation
The glass slides were dipped in 0.1 mg/ml PDL (Sigma) for 10 min, drained and dried overnight at room temperature. Adherent CHO cells growing at 70% confluence were harvested from a cell culture flask after incubation for 5 min with 3 ml of Accutase (Invitrogen, Carlsbad, CA), washed in RPMI medium 1640 (Gibco Life Technologies) and re–suspended to a concentration of 1×106 cells/ml in the CHO culture buffer. A cell suspension drop of 100–150 µl was laid on the PDL pre–coated slide and was incubated for 24–48 h at 37 °C with 5% CO2. The slide with well-spread adherent CHO cells was then gently washed with 1x phosphate-buffered saline (PBS)–Ca –Mg (Invitrogen). P. falciparum iRBCs in trophozoite stage with 2–10% parasitemia were suspended in 1x PBS –Ca –Mg with 5 µg/ml Bovine Serum Albumin (BSA) (Sigma) to 1% hematocryte and were poured over the slide with adherent CHO cells and allowed to stand for 10 min. Non–attached iRBCs were washed with 1x PBS –Ca –Mg with 5 µg/ml BSA and the slide, with adherent CHO cells and lightly attached iRBCs and RBCs, was then immersed in the same buffer and transferred to the microscope liquid cell.
Data acquisition
The force spectroscopy experiments were conducted with an extended-head, three-dimensional molecular force probe (MFP–3D) atomic force microscope (AFM) mounted on an Axiovert Zeiss trans–illuminated microscope (Asylum Research, Santa Barbara, CA). The spring constant (k) of each silicon nitride tipless cantilever (MLCT–O10 Veeco Probes, Camarillo, CA), with a nominal value of 30 mN/m, was calibrated against a glass slide in air using the equipartition method [30]. Each calibrated tipless cantilever was incubated in 1 mg/ml ConA for 30 min prior to the force spectroscopy measurements. The slide containing the cells was loaded into the liquid cell, which was then filled with PBS–Ca –Mg with 5 µg/ml BSA, and was kept at either 37 °C or 41 °C (febrile temperature). The ConA–incubated cantilever was immersed in the heated buffer and the measurements were carried out after allowing for temperature equilibration during only 10–20 min to minimize any deterioration or viability loss of the iRBC(RBC), parasite, and CHO cells. The inverse optical sensitivity of the tipless cantilever was determined by performing an approach/retraction cycle in liquid against the rigid glass slide. The cantilever was subsequently engaged with a contact force of 1 nN for 30 s on a chosen iRBC in trophozoite stage (or a healthy RBC in the control experiments). The cell became strongly attached to the cantilever through ConA mediation and was withdrawn from the substrate upon cantilever retraction. This iRBC (or RBC) was then used to probe CHO cells around the slide.
Each measuring session involved testing one individual iRBC (or RBC) probe, attached to a previously calibrated cantilever, for no more than 150 approach/retraction cycles against different CHO cells. The mechanical tests were performed with a displacement rate (V) of 1.6 µm·s−1, a preset trigger force (F) of ~300 pN, and a dwell time (t) of 0.1 s, under open loop conditions to maximize resolution [31]. The raw data (cantilever deflection and piezo–displacement) were acquired at a sampling rate of 2 kHz. The integrity/viability of the three biological entities involved in the measurements was ascertained by thoroughly monitoring (i) the rotation of the hemozoin crystals inside the parasitophorous vacuole, (ii) the shape preservation and/or any possible echinocyte signs on the iRBC(RBC) probe, and (iii) the spreading of each CHO cell tested. The duration of each measuring session was kept under 1.5 h to avoid the evolution of the iRBC probe out of trophozoite stage. To increase data collection, the maximum separation distances were kept below 12 µm. Since relevant adhesion events occurred for separation distances typically under 5 µm, the 12 µm measurements allowed for the extended baselines in the force versus displacement curves required for correction of hydrodynamic effects.
Since the parasite-derived antigens are expected to remain expressed on the iRBC membrane upon returning to 37 °C after exposure to fever, any spurious effects occurring on the CHO cells during the measurements at 41 °C could be controlled by probing CHO cells kept at 37 °C with iRBCs previously exposed to heat. Relatively short exposures to 41 °C resulted in a high destruction rate of the parasites, and this rendered excessively time-consuming to spot iRBCs with viable parasites suited for attachment to the cantilever (see Figure 1). However, heat-treatments at 40 °C for 1 hour proved to be an appropriate compromise, and 37 °C/40 °C(1h) experiments have been implemented to demonstrate that the differences observed in adhesion behavior as a result of short-term exposure to febrile temperatures are related to iRBC changes, and that they do not reflect a CHO response.
Data analysis
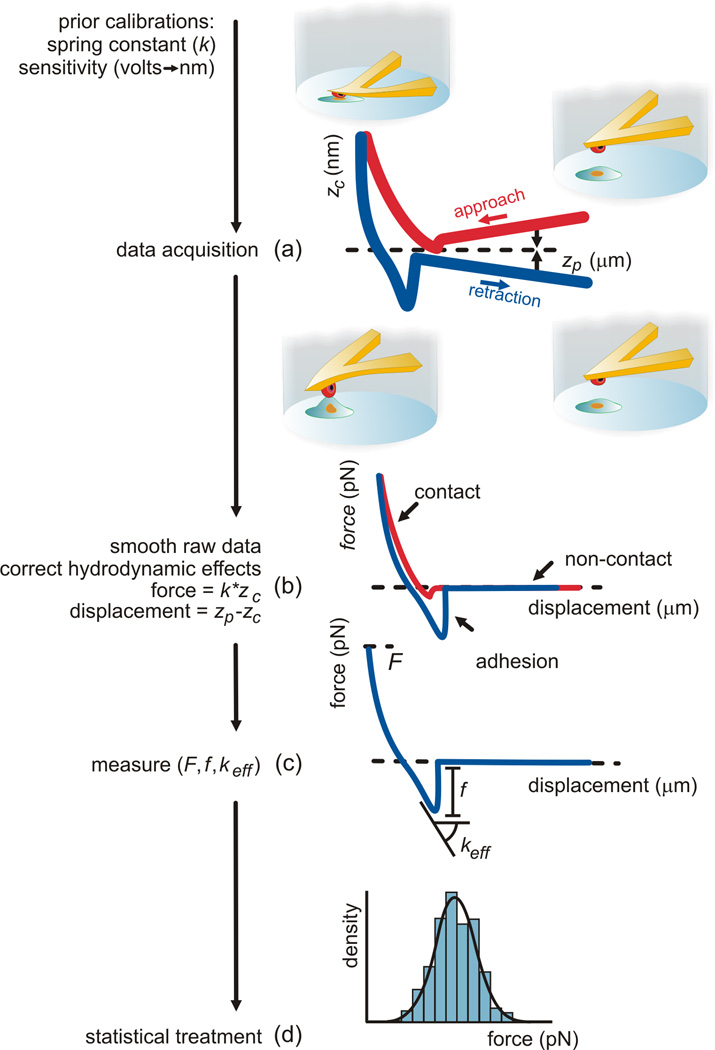
Figure 2 describes the sequence of steps involved in data analysis [31,32]. The cantilever deflection versus piezo–displacement curves (Figure 2(a)) were smoothed with a 100-point moving average. Any tilt and/or curvature of the baselines resulting from hydrodynamic effects and local thermal imbalances were corrected with a polynomial function not higher than 3rd order (Figure 2(b)). Since the trigger force is imposed as a deflection difference relative to the initial value, the hydrodynamic effects produced some scatter in the effective trigger force, F, which has been taken into account in interpreting the data. Force versus displacement curves were obtained by converting the measured cantilever deflection, zc, into force using the cantilever spring constant, k, calibrated previously, and converting the measured piezo–displacement, zp, into probe/sample separation (true displacement) through subtraction of the cantilever deflection (as described in Figure 2(b)). The offset observed at rupture in the retraction curve was used to quantify the rupture force f associated with each approach/retraction cycle (Figure 2(c)). The effective spring constant, keff, of the adhesion system was determined from the slope of a line fitted to the region preceding rupture strictly for iRBC/CHO retraction curves exhibiting single–rupture events (Figure 2(c)). The curve analysis was performed using custom routines written for MATLAB (The MathWorks, Natick, MA) with close monitoring for each individual curve. The adhesion force values resulting from each series of force versus displacement curves are used for the statistical analysis (d).
Figure 2.
Schematic drawings of the force spectroscopy data analysis procedure (adapted from [31,32]). The larger thickness of the curves in (a) represents the noise removed subsequently with the smoothing operation (b). Correction of hydrodynamic effects and conversion of deflection to force and piezo-displacement to (true) displacement are carried out for each curve (b) before measurement of the adhesion force f, contact force applied F and effective stiffness of the binding complex keff (c). The adhesion data from a series of approach/retraction curves are used for the statistical analysis (d).
In general, the fraction of multiple receptor–ligand ruptures tends to increase with the binding frequency and can be estimated using Poisson distribution statistics. When only 25% of the retraction curves show rupture events, 86% of them are expected to correspond to unbinding of a single receptor–ligand pair, 12% to double bindings and < 2% can be assigned to other multiple interactions. For a binding frequency of 75% those fractions are, respectively, 46%, 32%, < 22% (see [33] for a detailed explanation). Thus the study of single receptor-ligand binding requires low binding frequency, and minimization strategies are usually based on readjusting the contact conditions. However, in the present study low binding frequency conditions could not be attained even for the lowest values of contact time and contact force recommended for single–molecule experiments (100–400 ms and 100–300 pN, respectively, for single-cell probes against functionalized surfaces [31]). Nevertheless, the relatively delicate contact conditions imposed have led to multimodal force distributions, which could be interpreted in terms of single and multiple receptor–ligand ruptures and allowed the determination of the single-molecule binding force.
Statistical treatment
Assuming the rupture force f as a positive variable, the following Gaussian mixture probability density function is proposed to account for the multimodality of the rupture force distributions observed experimentally:
| (1) |
The following conditions are imposed to the mixture weights: ai ≥ 0 (i = 0,1,2,…,n) and a0 + a1 + … + an = 1. The positive integer n is determined from both the distribution of experimental data and the likelihood ratio test (LRT) used for model comparison. Each bi (i = 1,…, n–1) value corresponds to the mean rupture force of i receptor–ligand complexes, and bn corresponds to the mean rupture force of ≥ n molecules binding. The standard deviation of the corresponding rupture force is denoted by ci (i = 1,…, n). In each of the individual density functions, Φ(.) represents the cumulative distribution function of the standard normal distribution. The maximum likelihood (ML) approach [34] has been adopted to estimate the parameters, ai, bi and ci, of the Gaussian mixture density function. The 95% confidence intervals have been constructed based on the asymptotic normality of the estimators. The differences in mean rupture force between the cases of 37 °C, 41 °C and 37 °C/40 °C(1h) have been evaluated by comparing the estimated bi values under different scenarios, with statistical significance and 95% confidence intervals of the differences provided by two–sided Z–tests. All the above statistical computing has been conducted using version 2.12.0 of R (The R Project for Statistical Computing, free software for the GNU operating system [35]).
2.3. CSA and PfEMP1 expression and binding specificity
Immunofluorescence following force spectroscopy
Immediately after the force spectroscopy experiments the presence of CSA on the membranes of the CHO cells was tested by immunofluorescence. The test slides were washed with PBS and the CHO cells were fixed with 2% paraformaldehyde in 1x PBS. The fixed cells were permeabilized with 0.1% triton in 1x PBS and stained for 30 min at 4 °C with purified mouse IgG2A anti–CSA mAb (BD Pharmingen, San Diego, CA). This specific antibody was detected with Alexafluor 488 goat anti–mouse (Molecular Probes, Eugene, OR) diluted 1/1000. CHO cell nuclei were stained with 4',6–diamidino–2–phenylindole (DAPI) 5 µg/mL (Invitrogen). Negative controls were produced without the specific anti-CSA antibody.
Fluorescence-activated cell sorting (FACS)
PfEMP1 expression on trophozoite-stage cells after a transient exposure to 40 °C for 1 h was assessed by flow cytometry and compared with the results obtained with cells kept at 37 °C. After incubation at the different temperatures, RBCs were washed and stained for 30 min at 4 °C with a rabbit polyclonal antibody anti-PfEMP1 (gift from Michael F. Duffy, University of Melbourne, Australia), followed by an Alexa Fluor 594 goat anti-rabbit IgG specific antibody diluted 1/1000. An isotype–matched rabbit antibody was used as negative control. The parasite nuclei were stained with SYTO16 from molecular probes (Invitrogen). Analysis was performed using FACSCalibur and Cell Quest software (BD Biosciences).
Static adherence assays
The CSA–PfEMP1 binding specificity was controlled by using CSA in solution as a PfEMP1 blocking agent. The assays were performed by incubating the iRBC suspension in culture flasks with CHO monolayers (70 % confluence) for 1 h with gentle agitation every 15 min. A control condition was prepared with a pre–incubation of 100 µg/ml CSA (Sigma). Adherence was assessed microscopically after washing away unbound RBCs, fixing with methanol (Sigma) and staining with Giemsa Blood Staining Solution (J.T.Baker, Center Valley, PA).
3. Results
3.1 Force spectroscopy
Force versus displacement curves
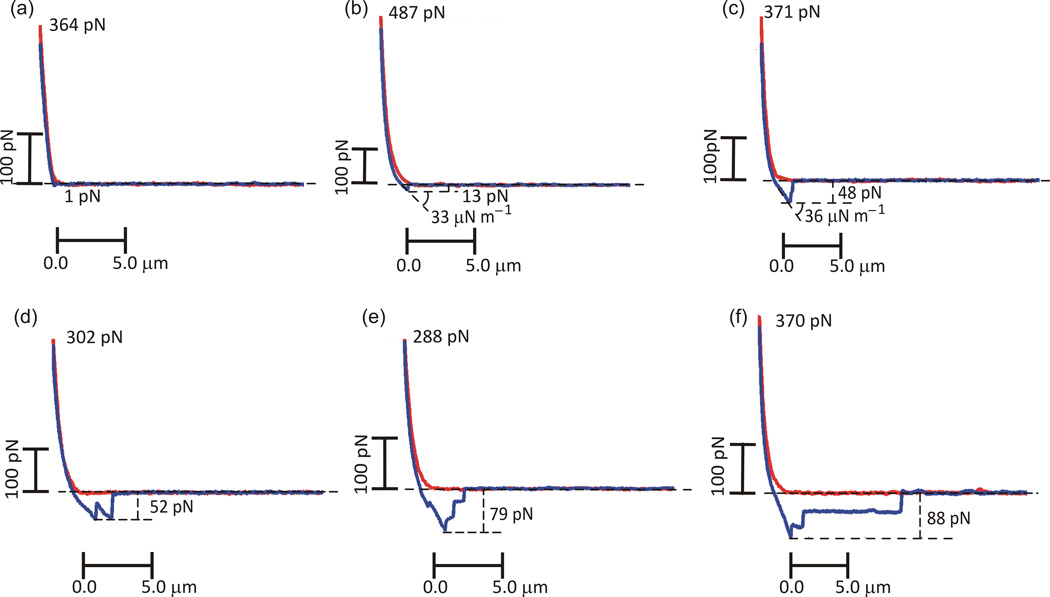
Figure 3 shows representative force versus displacement curves obtained with the cell–cell adhesion experiments. The gentle contact conditions imposed resulted in extremely small binding forces (typically tens of pN). The highest frequency for all tested conditions corresponded in fact to curves displaying no detectable adhesion or very low adhesion (Figure 3(a)), although a relatively high number of curves exhibted single rupture events (Figure 3(b) and (c)). The experiments also resulted in force versus displacement curves compatible with molecule unfolding [14] (Figure 3(d)), multiple rupture events (Figure 3(e) and subsequent discussion) and tethering plateaux (Figure 3(f)). Numerous such results, along with a rigorous statistical analysis, were used to characterize the cell-cell adhesion behavior in a systematic and quantitative manner.
Figure 3.
Representative force versus displacement curves: (nearly) no detectable adhesion, f < 5 pN (a), single rupture events (b,c), molecule unfolding-type (d), multiple rupture events (e), thethering plateau (f). The corresponding rupture forces, f, and effective spring constant, keff, are indicated, respectively in pN in µN/m.
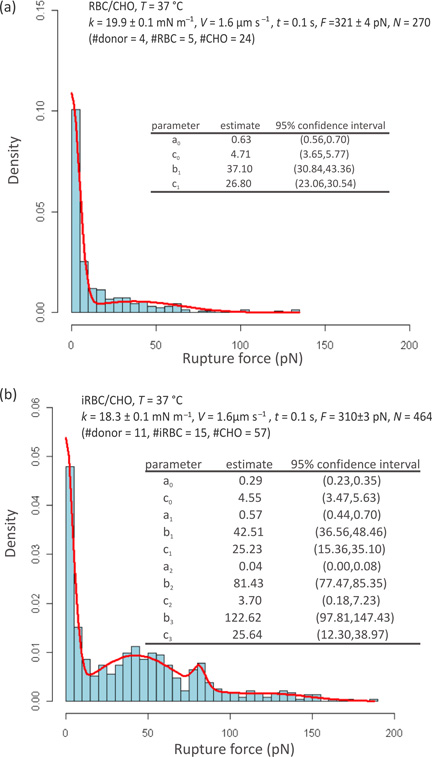
Cell adhesion
Figure 4 shows the results obtained at 37 °C using individual RBCs (a) and iRBCs (b) to probe CHO cells. Figure 4(a) presents the rupture force distribution determined from 270 curves obtained with 5 RBCs from 4 healthy donors that were used to probe 24 CHO cells with k = 19.9 ± 0.1 mN/m and F = 321 ± 4 pN. Figure 4(b) presents the rupture force distribution determined from 464 curves with 15 iRBCs, cultured from blood samples from 11 healthy donors, which were used to probe 57 CHO cells with k = 18.3 ± 0.1 mN/m and F = 310 ± 3 pN. Estimated Gaussian mixture density functions p(f) are presented as solid lines and their mean values and standard deviations are listed in the inset tables.
Figure 4.
Rupture force distributions obtained from measurements carried out at 37°C using RBC (a) and iRBC (b) probes. Estimated Gaussian mixture density functions are presented as solid lines. The estimated parameters, where bi corresponds to the mean value and ci to the standard deviation, are included in the inset tables together with the respective 95% confidence intervals.
As expected, the binding frequency was generally low for uninfected probes, with the rupture force being essentially zero as seen in Figure 4(a). The mean rupture force for non-specific binding (b1 in Figure 4(a)) was estimated to be 37 pN. The considerably higher binding frequency observed for infected cells in Figure 4(b) is consistent with the higher cytoadherence associated with mature iRBCs. Although some non-specific binding is expected, as inferred from Figure 4(a), the maxima at 43 pN and 81 pN in Figure 4(b) are postulated to be associated with the rupture of, respectively, single and double receptor–ligand complexes, with the subsequent maximum representing all other multiple bindings.
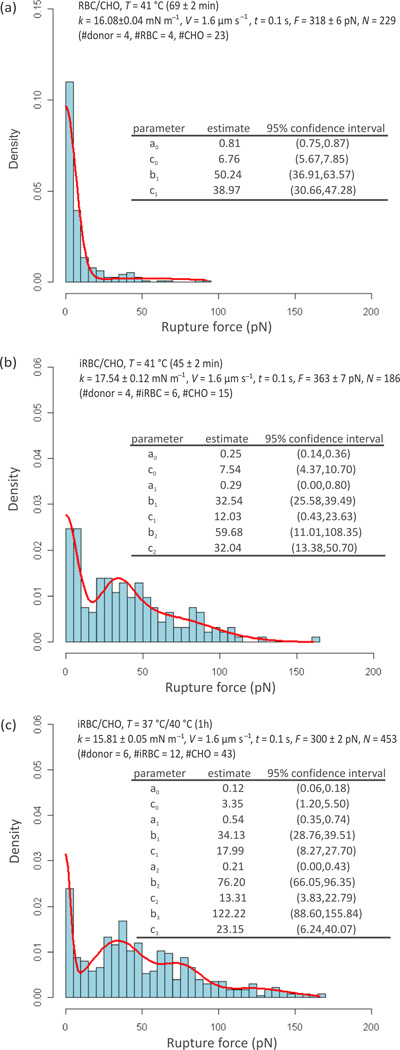
Figure 5 shows the results obtained at 41 °C using individual RBCs (a) and iRBCs (b) to probe CHO cells, together with the results produced at 37 °C using iRBC probes previously incubated for 1 h at 40 °C (c). Figure 5(a) represents the rupture force distribution determined from 229 curves obtained with 4 RBCs from 4 healthy donors that were used to probe 23 CHO cells with k = 16.1 ± 0.1 mN/m and F = 318 ± 6 pN. Figure 5(b) shows the rupture force distribution determined from 186 curves obtained with 6 iRBCs, cultured from blood obtained from 4 healthy donors, that were used to probe 15 CHO cells with k = 17.5 ± 0.1 mN/m and F = 363 ± 7 pN. Due to the intrinsic characteristics of the experimental setup, the period of exposure to 41 °C increased for the successive approach/retraction cycles taken with each iRBC(RBC). The exposure of the iRBCs(RBCs) to 41 °C was on average 1.15 ± 0.03 h and 0.75 ± 0.03 h for (a) and (b), respectively. Figure 5(c) represents the rupture force distribution determined from 451 curves obtained at 37 °C with 12 iRBCs, cultured from 6 healthy donors (independent from the donors of the previous experiments) and exposed to 40 °C for 1 h immediately before contact–probing against 43 different CHO cells with k = 15.8 ± 0.1 mN/m and F = 300 ± 2 pN.
Figure 5.
Rupture force distributions obtained at 41 °C with RBC (a) and iRBC (b) probes, and at 37 °C after a transient exposure of iRBC probes to 40 °C for 1 h (c). Estimated Gaussian mixture density functions are presented as solid lines. The estimated parameters, where bi corresponds to the mean value and ci to the standard deviation, are included in the inset tables together with the respective 95% confidence intervals.
As seen in Figure 5(a), the adhesion behavior of uninfected cells exposed to fever temperature is similar to that observed at 37 °C (Figure 4(a)): essentially no rupture force is noticed without any other pronounced maxima. The mean rupture force for non–specific binding (b1 in Figure 5(a)) was estimated to be 50 pN. As shown in Figure 5(b), experiments at 41 °C with iRBC probes also resulted in a higher binding frequency than with uninfected probes (Figure 5(a)). In the multimodal rupture force distribution obtained at 41 °C, the maximum at 33 pN is likely associated with the rupture of single receptor–ligand complexes, while the subsequent maximum corresponds to multiple bindings. Comparison with the results obtained at 37 °C (Figure 4(b)) shows a decrease from 43 to 33 pN in single receptor–ligand binding force, which occurred in association with an increase in binding frequency (76% of the curves evidenced f > 5 pN at 37 °C against 88% at 41°C).
A relatively low number of curves was obtained at 41 °C since these experiments were particularly difficult to perform: (i) a large proportion of the CHO cells tended to recoil with exposure to the febrile temperature, and as such could not be tested; and (ii) equilibration of temperature in the AFM liquid cell was harder to attain at 41 °C than at 37 °C, which led to a challenging control of the trigger force applied (compare the F values in the inset legends of Figures 4(b) and 5(b)). In addition, due to the experimental setup, the time of exposure to the febrile temperature increased along the sequence of force versus displacement curves obtained with each iRBC(RBC) probe. Moreover, the effect of the febrile temperature on the CHO cells that remained well spread may not suitably represent the syncytiotrophoblasts adhesion behavior at 41 °C. These shortcomings were absent from the 37°C/40°C(1h) control experiments.
Similarly to the experiments carried out at 37 °C and 41 °C with iRBC, the rupture forces at 37°C/40°C(1h) present a multimodal distribution with maxima at 34, 76 and 122 pN, which are postulated to be associated with the rupture of single, double and all other multiple receptor–ligand complexes, respectively. A comparison of the 37°C and 37°C/40°C(1h) data between Figures 4(b) and 5(c) shows a decrease in single receptor–ligand binding force (43 versus 34 pN, respectively) and an increased binding frequency (76% of the curves evidenced f > 5 pN at 37 °C against 88% after exposure to 40 °C(1h)).
The statistical significance of the mean rupture force differences can be inferred from Table 1. The estimated parameters for the various iRBC/CHO scenarios show that b1 varies –23% when the measurement temperature changes from 37 °C to 41 °C, which is statistically significant as indicated by the p values provided for each comparison; and that b1 varies −20% after a transient exposure of the probes to 40 °C for 1 h, which is also statistically significant.
Table 1.
Statistical significance of the difference between the mean values estimated from the full datasets for single and double binding at different temperatures, respectively Δb1 and Δb2.
| Conditions | Δbi(pN) | 95% Confidence interval | p–value |
|---|---|---|---|
| 41 °C versus 37 °C1 | Δb1:−10.0 | (−19.1, −0.8) | 0.03 |
| 37°C/40°C(1h) versus 37 °C | Δb1: −8.4 | (−16.4, −0.4) | 0.04 |
| Δb2: −5.2 | (−16.1, −5.7) | 0.35 |
Due to the reduced dataset for 41°C the b2 maximum is expected to correspond to all multiple binding and not only to double binding. This impaired a meaningful comparison of the b2 values at 41 °C and 37 °C.
The average effective spring constant, keff, of the adhesion complex has been determined empirically from force versus displacement curves that strictly presented a discrete single rupture event, as illustrated in Figure 3(b) and (c), and whose rupture force contributed to the 43 pN (37 °C) and 34 pN (37°C/40°C(1h)) maxima, i.e., for rupture forces in the 15–75 pN range at 37 °C and in the 10–60 pN range at 41°C. The results so obtained are presented in Table 2. The two sets of keff values are statistically different from each other with P = 0.007, and can be translated into effective loading rates (r = V.keff) of 72 ± 4 and 54 ± 5 pN/s, respectively.
Table 2.
Effective spring constant measured from single–rupture event curves showing rupture forces around the 43pN (37°C) and 34 pN (37°C/40°C(1h)) means.
| iRBC/CHO 37 °C |
iRBC/CHO 37°C/40°C(1h) |
|
|---|---|---|
| Gaussian mixture mean and standard deviation: b1c1 (pN) |
42.5, 25.2 | 34.1, 18.0 |
| Rupture force range around the mean [pN] |
15 – 75 | 10 – 60 |
| Fraction of single rupture event curves in the force range |
58/211 | 59/217 |
| Effective spring constant (keff) (strictly single rupture event curves) [µN/m] |
44.8 ± 2.6 | 33.7 ± 3.0 |
3.2 CSA and PfEMP1 expression and binding specificity
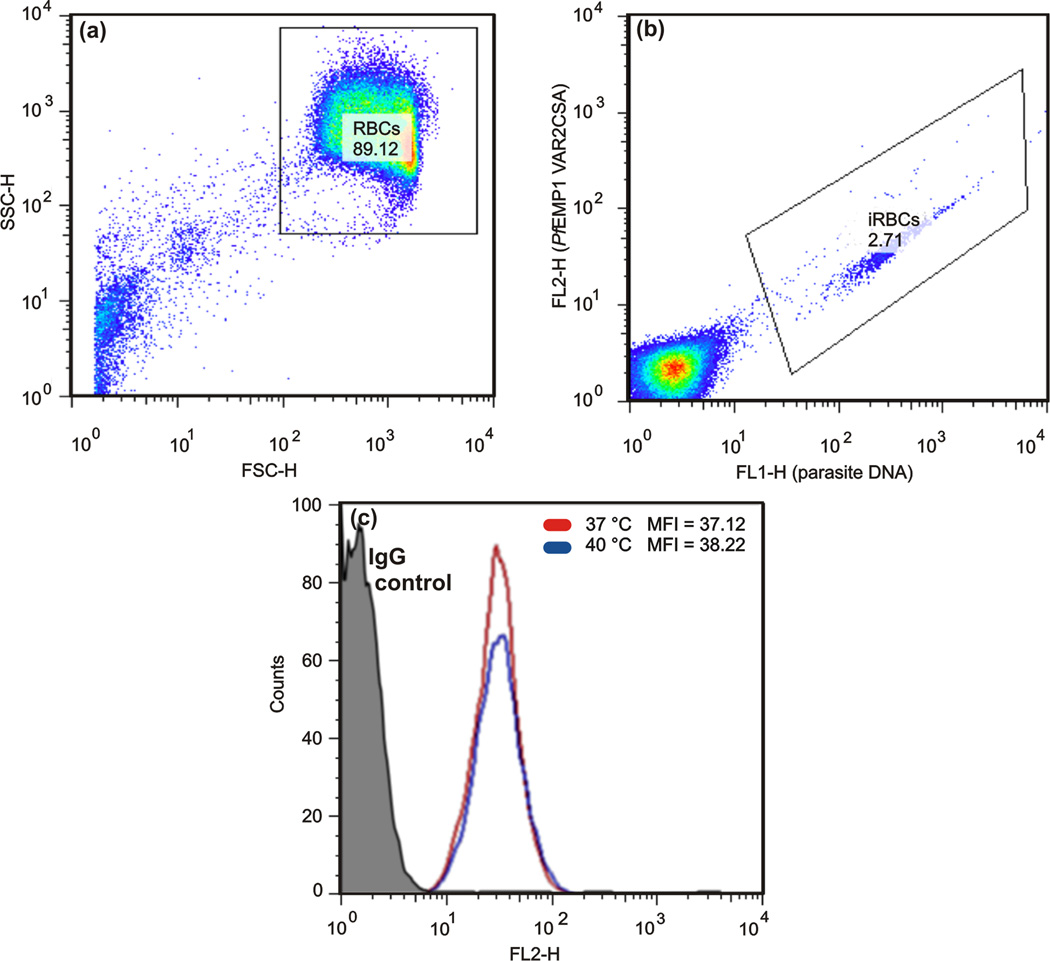
Immunofluorescence investigations carried out on slides used for force spectroscopy experiments attested the presence of CSA on the membranes of the CHO cells (results included as supporting material). FACS demonstrated that the mean level of PfEMP1 expression after a transient exposure to 40 °C (1 h) is similar to that observed at 37 °C (Figure 6).
Figure 6.
FACS data pointing to similar mean fluorescent intensity (MFI) for cells exposed and non–exposed to febrile temperatures. (a) RBCs were gated on a side scatter/forwardscatter(SSC-H/FSC-H).(b) Trophozoite–stage iRBCs were identified using SYTO16 and the PfEMP1 expression level was detected with a rabbit polyclonal antibody (mAb) anti-PfEMP1, followed by an Alexa Fluor 594 anti-rabbit IgG specific antibody. (c) Overlay of histograms analyzing PfEMP1 expression on iRBCs at 37°C (red line) or exposed to 40°C (blue line). The corresponding labeled isotype was used as negative control (black line)
The CSA–binding specificity of the FCR3–CSA parasite strain was confirmed through static adherence assays that demonstrated the blocking effect of CSA in solution on the adhesion of iRBCs to CHO cells (results included as supporting material).
4. Discussion
Force spectroscopy has been used to evaluate quantitatively the CSA–PfEMP1 adhesion complex at 37 °C, as well as to investigate the PfEMP1 response upon exposure to febrile temperature. The results indicate that, when all other experimental parameters are held equal, febrile temperature decreases both the CSA–PfEMP1 binding force (f) and the stiffness of the adhesion complex (keff), while enhancing the binding frequency.
Possible effects on the adhesion behavior, in terms of rupture force and stiffness of the receptor-ligand complex, arising from the experimental conditions require consideration. Specifically:
PDL influence on the subsequent adhesion behavior of the cell – Due to the cell-cell force spectroscopy setup, the gentle immobilization of the iRBC on the slide through PDL mediation is required for precise attachment of the cell at the end of the cantilever. The control experiments carried out with RBCs demonstrate that healthy cells pulled off from PDL show residual adhesion to CHO cells (Figure 4(a) and Figure 5(a)) in contrast to the behavior seen with iRBCs. This indicates that the weak binding to the substrate through PDL mediation is not likely to have a strong influence on the rupture force distribution differences observed.
Stiffness of the cantilevers employed – The adhesive properties of each iRBC have been assessed with a specific cantilever calibrated individually. Although cantilevers with outlier stiffness values have been discarded, some experimental scatter in the k values was inevitable (see inset legends in Figures 4 and 5). Nevertheless, this is not expected to have affected the experimental results since the cantilever stiffness variations have been taken into account in the determination of the force.
Contact force effectively applied (F) – The force applied affects the cell–cell contact conditions and lower trigger forces are known to result in decreased binding frequency [31]. However, the comparison of the binding frequency obtained at 37°C/40°C(1h) for iRBC/CHO with F = 300 ± 7 N (88% of the curves presented f > 5 pN) with the one attained at 37°C for iRBC/CHO with F = 310 ± 3 N (76% of the curves presented f > 5 pN), shows that the slightly lower trigger force at 37°C/40°C(1h) did not have a determining influence on the experimental results.
Stiffness of the intermediate structures included in the force transducer, i.e., of the iRBC itself and of any linkers between the iRBC cytoskeleton and PfEMP1 – The mechanical response of the total force transducer, translated into keff, includes the properties of the cell, of the receptor–ligand complex and of any biological linkers, which are three orders of magnitude more compliant than the cantilevers used and thus dictate the system spring constant [36]. The intermediate biological structures in the force transducer system are expected to have stiffness values comparable to the ones of the CSA–PfEMP1 receptor–ligand complex. Indeed, the stiffness of iRBCs in trophozoite stage has been estimated from optical tweezers experiments carried out at 25°C to be ~20 µN/m [13]. Nevertheless, magnetic twisting cytometry experiments have shown that the stiffness of iRBCs in mature stages increases acutely within minutes of exposure to febrile temperature [18]. Therefore, the decrease in stiffness upon exposure to febrile temperature, observed in the current study, is probably related to changes in the CSA–PfEMP1 complex (and/or in any molecular linker associated with PfEMP1 anchorage to the knob structural elements), and not to the overall cell mechanical behavior which is dominated by the cytoskeleton deformation.
Binding frequency impact on the ability to discriminate single/multiple rupture events – Rupture force distributions obtained with functionalized tips frequently show a pronounced peak at lower forces, corresponding to single molecule binding, followed by lower peaks or shoulders at higher forces, attributed to multiple ruptures [37–39]. In the present study the multimodal distributions obtained with iRBCs, and the level of mean rupture forces estimated from the Gaussian mixture probability density functions, indicate that the data acquired can be discussed in terms of single, double and multiple ruptures of adhesion complexes. The estimated mean rupture force of 43 pN together with the empirical 72 ± 4 pN/s loading rate, attributed to single CSA–PfEMP1 complexes, are in fact in agreement with the TSP–PfEMP1 and CD36–PfEMP1 data obtained with functionalized tips tested against mature iRBCs at 37°C under low binding frequency conditions [19].
Non-specific binding – The rupture force distributions obtained for uninfected cells (Figure 4(a) and Figure 5(a)) revealed that, even in the absence of PfEMP1, some non-specific binding existed between RBC probes and CHO cells. This behavior may be responsible for an overestimation of the bi values for the iRBC probes, which is also suggested by the average rupture forces determined strictly from single rupture event curves in the ranges presented in Table 2, i.e, 37 ± 1.9 pN (37°C) and 30 ± 1.6 pN (37°C/40°C(1h)) with p = 0.005, which compare, respectively, with b1 = 43 pN at 37°C and b1 = 34 pN at 37°C/40°C(1h) in Figures 4(b) and 5(c). Nonetheless, non-specific binding is expected to increase with febrile temperature exposure [40] and cannot justify the observed decrease in the single-molecule binding force (Δb1 in Table 1).
PfEMP1 and CSA levels of expression – Each knob, with a diameter of 80–130 nm, has been estimated to exhibit about 10 PfEMP1 molecules by considering a globular shape for the antigen and the expected molecular packing for mature iRBC [41]. Assuming an average inter-knob distance of 200 nm [42] and a square array for the knob spatial distribution, the 10 PfEMP1 molecules/knob translate into a density of about 250 molecules per µm2 of the iRBC membrane. The videos acquired during the experiments (some of which are included as supporting material) show that the contact area between the iRBCs and the CHO cells was around 20 µm2 (roughly the projected area of the iRBC probes). This indicates that, under the current force spectroscopy experimental conditions, approximately 5000 PfEMP1 molecules are available for binding. Given the binding frequencies of 76% at 37 °C and 88% at 37°C/40°C(1h), the probability of CSA–PfEMP1 interaction can roughly be estimated as 0.02% in both conditions. This probability depends critically not only on the contact conditions imposed but also on the level of expression of CSA on the particular cell type used as a surrogate to the human syncytiotrophoblast. Nevertheless, although the cell-cell adhesion force is dependent on the density of interacting ligand-receptor pairs, the latter parameter is not expected to critically affect the behavior of single ligand-receptor binding.
The previous analysis indicates that the shift to lower values consistently and significantly observed for the mean rupture force (b1) after exposure to febrile temperature (see Table 1) is not an artifact of the experimental methods employed. Instead, it may reflect a weakened CSA–PfEMP1 binding, which occurs in association with a lower stiffness of the adhesion system and an increased binding frequency. The physical meaning of these results is discussed next.
The binding force of any ligand–receptor complex is a dynamic property dependent on the loading rate (r = keff.V) employed during forced unbinding and rupture force data acquired at different loading rates can be used to reveal thermodynamic and kinetic details governing adhesion processes. The Bell–Evans model predicts that the most probable rupture force (b1) increases linearly with the natural logarithm of r [43,44]:
| (2) |
where T is the absolute temperature, kB is the Boltzmann constant, xb is the rate of unstressed spontaneous unbinding (driven by thermodynamics) and xb can be understood as the length over which the ligand and receptor disengage from each other upon unbinding. In the case of CD36–PfEMP1 the rupture force decreases 2–3 pN when the loading rate varies from 72 to 54 pN/s at 37 °C (where xb and koff are, respectively, 0.76 nm and 0.001 s−1 [19]), and the same is observed for TSP–PfEMP1 (where xb and Koff are, respectively, 0.36 nm and 0.144 s−1 [19]). This suggests that the reduction in loading rate with heat exposure (Table 2) may not fully justify the b1 variation observed for CSA–PfEMP1 binding at 37°C/40°C(1h) versus 37 °C (Δb1 in Table 1). This suggests that the decrease in binding force resulting from exposure to febrile temperatures, observed for the biological system used to model the adhesion behavior of iRBC/syncytiotrophoblasts, may have resulted from conformational changes in the CSA–PfEMP1 complex and/or kinetic changes due to an altered mobility of the molecules on the iRBC surfaces.
The role of febrile temperatures on the adhesion of iRBC to recombinant 5CD36 and ICAM-1 has been previously investigated [45]. In this study, heating ring-stage iRBCs to 40 °C for 2 h enhanced the number of adherent cells and was associated with an increase in trafficking of PfEMP1 to the cell surface.However, this effect seems absent in mature stages since the mean level of PfEMP1 expression has not been altered by the transient exposure to 40 °C (Figure 6). On the other hand, febrile temperatures are known to promote phosphatidylserine expression on iRBCs membranes during parasite maturation [40]. This additional display of non–specific receptors may justify the increase in binding frequency observed for iRBC after exposure to febrile temperatures. In fact, in terms of overall cell adhesion, the weakening effect of single and multiple CSA-PfEMP1 binding may be balanced or superseded by increased non-specific binding.
Quantifying cell-cell cytoadherence poses challenging constraints and requires additional parameter control when compared with force spectroscopy involving functionalized tips. Yet the opportunity to investigate receptor–ligand interactions under physiologically relevant conditions, and with mediation of biologic linkers, supplants the additional experimental complexity. Such quantitative understanding is critical for the development of computational models of blood rheology in health and disease. Furthermore, in the context of disease diagnostics, measuring cell-cell adhesion at the molecular level can be used to evaluate quantitatively the parasite phenotypes in terms of binding to receptors associated with specific pathologies. In addition, the present experimental method affords new possibilities for drug screening protocols that could potentially target cytoadherence as a means to therapeutically modulate blood flow dynamics.
5. Concluding remarks
This paper presents new experimental protocols as well as novel statistical techniques to analyze experimentally-derived multimodal probability density functions. Quantitative details of cytoadherence influenced by molecular level mechanisms for P. falciparum iRBCs have been presented under physiologically/pathologically relevant conditions. The results obtained here have established that the binding force of the CSA–PfEMP1 complex decreases significantly with exposure to febrile temperature. These trends accompany a higher compliance of the adhesion complex and an increased binding frequency for the iRBC/CHO model system. The quantitative analysis of cytoadherence in terms of the distinct binding phenotypes can open new pathways for screening drugs that target cytoadherence.
Supplementary Material
Acknowledgements
The research work was supported by the Infectious Diseases Interdisciplinary Research Group of the Singapore–MIT Alliance for Research and Technology (SMART). PAC was additionally supported by the MIT–Portugal program and the Fulbright Commission. The authors thank Dr. J. Smith (SBRI, Seattle, Washington, USA) for providing FCR3–CSA parasites.
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Cooke BM, Mohandas, Coppel RL. The malaria-infected red blood cell: Structural and functional changes. Adv Paraistol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nash GB, O'Brien E, Gordon–Smith EC, Dormandy JA. Abnormalities in the mechanical properties of red blood cells caused by Plasmodium falciparum. Blood. 1989;74:855–861. [PubMed] [Google Scholar]
- 4.Suresh S, Spatz J, Mills JP, Micoulet A, Dao M, Lim CT, Beil M, Sefferlein T. Connections between single-cell biomechanics and human disease states: gastrointestinal cancer and malaria. Acta Biomater. 2005;1:16–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Mills JP, Diez–Silva M, Quinn DJ, Dao M, Lang MJ, Tan KSW, Lim CT, Milon G, David PH, Mercereau–Puijalon O, Bonnefoy S, Suresh S. Effect of plasmodial RESA protein on deformability of human red blood cells harboring Plasmodium falciparum . PNAS. 2007;104:9213–9217. doi: 10.1073/pnas.0703433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deitsch KW, Wellems TE. Membrane modifications in erythrocytes parasitized by Plasmodium falciparum . Mol Biochem Parasitol. 1996;76:1–10. doi: 10.1016/0166-6851(95)02575-8. [DOI] [PubMed] [Google Scholar]
- 7.Ho M, White NJ. Molecular mechanisms of cytoadherence in malaria. Cell Physiology. 1999;276:C1231–C1242. doi: 10.1152/ajpcell.1999.276.6.C1231. [DOI] [PubMed] [Google Scholar]
- 8.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Normark J. Surface antigens and virulence in Plasmodium falciparum malaria. Stockholm, Sweden: PhD thesis Karolinska Institutet; 2008. [Google Scholar]
- 10.Pasternak ND, Dzikowski R. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum . Int J Biochem Cell Biol. 2009;41 doi: 10.1016/j.biocel.2008.12.012. 1463-1436. [DOI] [PubMed] [Google Scholar]
- 11.Fried M, Duffy PE. Adherence ofPlasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272 doi: 10.1126/science.272.5267.1502. 1502-1404. [DOI] [PubMed] [Google Scholar]
- 12.Bentley GA, Gamain B. How does Plasmodium falciparum stick to CSA? Let's see in the crystal. Nature Struct Mol Biol. 2008;15:895–897. doi: 10.1038/nsmb0908-895. [DOI] [PubMed] [Google Scholar]
- 13.Suresh S. Mechanical response of human red blood cells in health and disease: Some structure-property-function relationships. J Mater Res. 2006;21:1871–1877. [Google Scholar]
- 14.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nature Materials. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 15.Lim CT, Zhou EH, Quek ST. Mechanical models for living cells - a review. Journal of Biomechanics. 2006;39:195–216. doi: 10.1016/j.jbiomech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Helenius J, Heisenberg C–P, Gaub HE, Muller DJ. Single-cell force spectroscopy. J Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 17.Vedula SRK, Lim TS, Kausalya PJ, Lane EB, Rajagopal G, Hunziker W, Lim CT. Quantifying Forces Mediated by Integral Tight Junction Proteins in Cell–Cell Adhesion. Experim Mech. 2009;49:3–9. [Google Scholar]
- 18.Marinkovic M, Diez–Silva M, Pantic I, Fredberg JJ, Suresh S, Butler JP. Febrile temperature leads to significant stiffening of Plasmodium falciparum parasitized erythrocytes. Am J Physiol Cell Physiol. 2009;296:C59–C64. doi: 10.1152/ajpcell.00105.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ang L, Lim TS, Shi H, Yin J, Tan SJ, Li Z, Low BC, Tan KSW, Lim CT. Molecular Mechanistic insights into the endothelial receptor mediated cytoadherence of Plasmodium falciparum-infected erythrocytes. PLoS One. 2011;6:e16929. doi: 10.1371/journal.pone.0016929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis SP, Amrein M, Gillrie MR, Kristine Lee K, Muruve DA, May Ho M. Plasmodium falciparum-induced CD36 clustering rapidly strengthens cytoadherence via p130CAS-mediated actin cytoskeletal rearrangement. FASEB J. 2012;26:1119–1130. doi: 10.1096/fj.11-196923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fidock DA, Eastman RT, Ward SA, Meshnick SR. Recent highlights in antimalarial drug resistance and chemotherapy research. Trends Parasitol. 2008;24:537–544. doi: 10.1016/j.pt.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien C, Henrich PP, N Passi, Fidock DA. Recent clinical and molecular insights into emerging artemisinin resistance in Plasmodium falciparum . Curr Opin Infect Dis. 2011;24:570–577. doi: 10.1097/QCO.0b013e32834cd3ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedosov DA, Caswell B, Suresh S, Karniadakis GE. Quantifying the biophysical characterics of Plasmodium-falciparum-parasitized red blood cells in microcirculation. PNAS. 2011;108:35–39. doi: 10.1073/pnas.1009492108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedosov DA, Lei H, Caswell B, Suresh S, Karniadakis GE. Multiscale modeling of red blood cell mechanics and blood flow in malaria. PLoS Comput Biol. 2011;7:e1002270. doi: 10.1371/journal.pcbi.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasvol G, Wilson RJ, Smalley ME. Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood. J Ann Trop Med Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- 26.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 27.Doritchamou J, Bertin G, Moussiliou A, Bigey P, Viwami F, Ezinmegnon S, Fievet N, Massougbodji A, Deloron P, Ndam NT. First-trimester Plasmodium falciparum infections display a typical “Placental” Phenotype. J Infect Dis. 2012;206:1911–1919. doi: 10.1093/infdis/jis629. [DOI] [PubMed] [Google Scholar]
- 28.Rogerson SJ, Hviid L, Duffy PE, Leke RFG, Taylor DW. Malaria in pregnancy: pathogenesis and immunity. Lancet Infect Dis. 2007;7:105–117. doi: 10.1016/S1473-3099(07)70022-1. [DOI] [PubMed] [Google Scholar]
- 29.Sartelet H, Garraud O, Rogier C, Milko-Sartelet I, Kaboret Y, Michel G, Roussilhon C, Huerre M, Gaillard D. Hyperexpression of ICAM-1 and CD36 in placentas infected with Plasmodium falciparum: a possible role of these molecules in sequestration of infected red blood cells in placentas. Histopathology. 2000;36:62–68. doi: 10.1046/j.1365-2559.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 30.Butt H, Jaschke M. Calculation of thermal noise in atomic force microscopy. Nanotech. 1995;6:1–7. [Google Scholar]
- 31.Franz CM, Taubenberger A, Puech P–H, Muller DJ. Studying integrin-mediated cell adhesion at the single-molecule level using AFM force spectroscopy. Sci STKE. 2007;406:1–16. doi: 10.1126/stke.4062007pl5. [DOI] [PubMed] [Google Scholar]
- 32.Colaço R, Carvalho PA. Atomic Force Microscopy in Bioengineering Applications, in Scanning Probe Microscopy in Nanoscience and Nanotechnology. In: Bhushan B, editor. NanoScience and Technology series. Vol. 3. Springer-Verlag; 2012. pp. 397–430. [Google Scholar]
- 33.Tees DF, Waugh RE, Hammer DA. A microcantilever device to assess the effect of force on the lifetime of selectin-carbohydrate bonds. Byophys J. 2001;80:668–682. doi: 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casella G, Berger RL. Statistical Inference. Pacific Grove, CA: Duxbury; 2002. [Google Scholar]
- 35. http://www.gnu.org/
- 36.Yuan C, Chen A, Kolb P, Moy VT VT. Energy landscape of avidin–biotin complexes measured by atomic force microscopy. Biochemistry. 2000;39:10219–10223. doi: 10.1021/bi992715o. [DOI] [PubMed] [Google Scholar]
- 37.Dupres V, Menozzi FD, Locht C, Clare BH, Abbott NL, Cuenot S, Bompard C, Rae D, Dfrene Y. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nature Methods. 2005;2:515–520. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 38.Guo S, Ray C, Kirkpatrick A, Lad N, Akhremitchev BB. Effects of multiple-bond ruptures on kinetic parameters extracted from force spectroscopy measurements: revisiting biotin-streptavidin interactions. Biophys J. 2008;95:3964–3976. doi: 10.1529/biophysj.108.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu C, Kirkpatrick A, Ray C, Guo S, Akhremitchev BB. Effects of Multiple-Bond Ruptures in Force Spectroscopy Measurements of Interactions between Fullerene C60 Molecules in Water. J Phys Chem C. 2008;112:5085–5092. [Google Scholar]
- 40.Pattanapanyasat K, Sratongno P, Chimma P, Chitjamnongchai S, Polsrila K, Chotivanich K. Febrile temperature but not proinflammatory cytokines promotes phosphatidylserine expression on Plasmodium falciparum malaria-infected red blood cells during parasite maturation. Cytometry A 77. 2010:515–523. doi: 10.1002/cyto.a.20879. [DOI] [PubMed] [Google Scholar]
- 41.Joergensen L, Salanti A, Dobrilovic T, Barfod L, Hassenkam T, Theander TG, Hviid L, Arnot DE. The kinetics of antibody binding to Plasmodium falciparum VAR2CSA PfEMP1 antigen and modelling of PfEMP1 antigen packing on the membrane knobs. Malar J. 2010;9:100. doi: 10.1186/1475-2875-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rug M, Prescott SW, Fernandez KM, Cooke BM, Cowman AF. The role of KAHRP domains in knob formation and cytoadherence of P falciparum-infected human erythrocytes. Blood. 2006;108:370–378. doi: 10.1182/blood-2005-11-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 44.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udomsangpetch R, Pipitaporn B, Silamut K, Pinches R, Kyes S, Looareesuwan S, Newbold C, White NJ. Febrile temperatures induce cytoadherence of ring-stage Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci USA. 2002;99:11825–11829. doi: 10.1073/pnas.172398999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.