Abstract
Primary cilia function as specialized compartments for signal transduction. The stereotyped structure and signaling function of cilia inextricably depend on the selective segregation of molecules in cilia. However, the fundamental principles governing the access of soluble proteins to primary cilia remain unresolved. We developed a methodology termed Chemically-Inducible Diffusion Trap at Cilia (C-IDTc) to visualize the diffusion process of a series of fluorescent proteins ranging in size from 3.2 to 7.9 nm into primary cilia. We found that the interior of the cilium was accessible to proteins as large as 7.9 nm. The kinetics of ciliary accumulation of this panel of proteins was exponentially limited by their Stokes radii. Quantitative modeling suggests that the diffusion barrier operates as a molecular sieve at the base of cilia. Our study presents a set of powerful, generally applicable tools for the quantitative monitoring of ciliary protein diffusion under both physiological and pathological conditions.
INTRODUCTION
A primary cilium is an antenna-like organelle protruding from the apical surface of most cells in a wide variety of organisms1. Primary cilia regulate several signaling systems such as phototransduction, olfaction, and developmental pathways2. To achieve these functions, primary cilia accumulate a specific set of biomolecules including membrane receptors and their downstream soluble effectors. As biosynthetic machinery is not present in primary cilia3, these biomolecules must be transported into primary cilia from the cell body. Thus, understanding protein transport across the physical separation between the site of protein synthesis (the cell body) and the site of protein function (the cilium) is of fundamental significance in the field of ciliary biology and cilia-based diseases4.
The trafficking of membrane proteins into primary cilia has been intensively studied5-7. At the ciliary neck, there is a diffusion barrier regulated by Septin2 and other proteins that limit free lateral diffusion of membrane proteins from the contiguous plasma membrane5, 7. This defining feature maintains a distinct composition of lipids and membrane proteins inside primary cilia4. While the soluble environment in the ciliary lumen is specialized as well, the mechanisms that regulate the transport of soluble molecules into or out of primary cilia remain poorly understood.
The transport of soluble molecules between the cytosol and the ciliary lumen has been most rigorously examined in rod photoreceptor cells, which possess a connecting cilium (CC) that provides a conduit between the cell body and the outer segment (OS)8. The massive vectorial transport of arrestin, a soluble protein, across the CC into the OS was shown to occur by simple diffusion driven by a concentration gradient generated when photons expose arrestin-binding sites on rhodopsin, which is highly concentrated in the OS9. In this system, monomers, dimers and trimers of green fluorescent protein (GFP) freely diffused across the CC, suggesting that there is no fixed pore that limits the diffusion of soluble proteins at least up to 80 kDa10.
In contrast to these studies of photoreceptor cilia, a recent study proposed that molecules above 67 kDa are excluded from primary cilia in epithelial cells by a fixed pore at the ciliary base that has similar properties and molecular composition to the nuclear pore complex (NPC)11. In this study the kinetics of accumulation in cilia was not directly monitored; instead the ratio of fluorescence between the cilium and the cytosol at a given time point after microinjection of a labeled tracer was measured. Since diffusion is a kinetic process, such end-point assays cannot distinguish between differences in the rate of diffusion as well as differences in the propensity of molecules to occupy the cytoplasm and the cilium. A striking example of this is found in dark-adapted photoreceptors, where arrestin is excluded from the OS because of molecular crowding effects and not because its diffusion is restricted across the CC10.
These considerations and the divergent conclusions reached by studies in rods and cultured cells prompted us to deploy chemically-inducible dimerization to visualize ciliary diffusion in living cells. Our technique bypasses the need for perturbations such as microinjection or detergent permeabilization commonly used to introduce diffusion probes into cells. With both high sensitivity and fine temporal resolution, we explored the ciliary diffusion barrier faced by soluble cytoplasmic proteins ranging in Stokes radius from 3.2 to 7.9 nm (molecular weight: 40 to 650 kDa). While the rate of ciliary influx of this series of probes was strongly dependent on their size, the ciliary diffusion barrier allowed the entry of soluble proteins with a Stokes radius as large as 7.9 nm. Our kinetic data using a large series of diffusion probes was most consistent with a sieve-like barrier at cilia whose mesh radius is larger than 7.9 nm. The present study highlights a powerful technique that enables quantitative characterization of the ciliary diffusion barrier for soluble proteins and substantially revises our physical model of this barrier.
RESULTS
A chemically-inducible technique for trapping soluble proteins inside cilia of living cells
The measurement of diffusion between two compartments requires the presence of a concentration gradient. We begin by describing a new technique generating such a gradient between the cytoplasm and the cilium by using a high-affinity interaction to trap soluble proteins that diffuse into primary cilia. This approach is conceptually identical to the arrestin gradient generated across the photoreceptor CC when light generates arrestin binding sites on rhodopsin in the OS8. The basis of our technology is chemically-inducible dimerization12, where a chemical dimerizer such as rapamycin induces the dimerization of FKBP-rapamycin binding domain (FRB) and FK506 binding protein (FKBP) (Fig. 1a). By localizing the FRB partner protein in cilia, we hypothesized that rapamycin addition could be used to trap any FKBP fusion protein that diffused into cilia, thus generating a sink that would set up a concentration gradient of the FKBP fusion protein across the putative ciliary diffusion barrier.
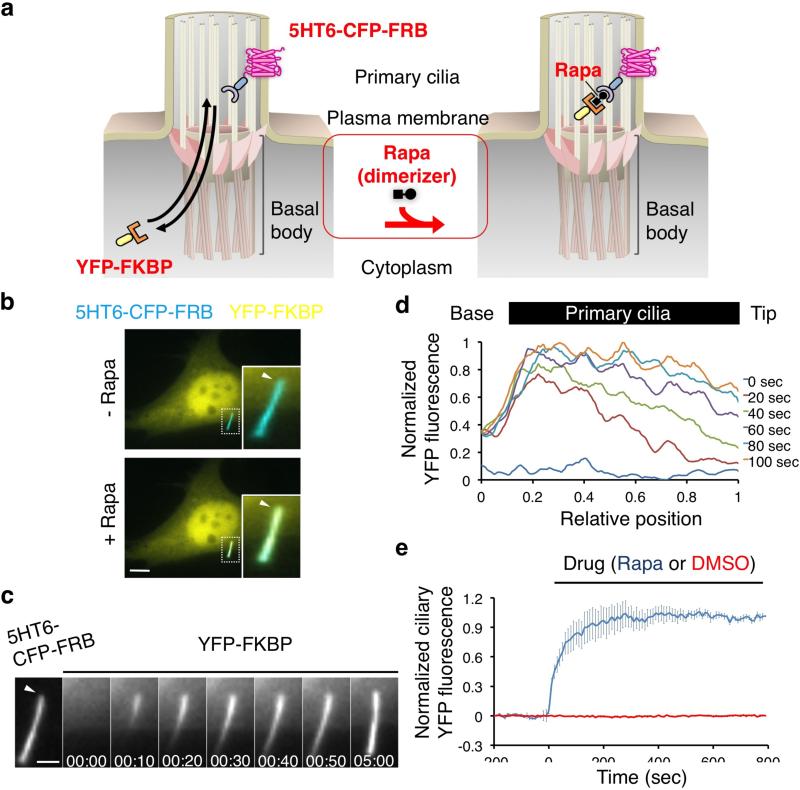
Figure 1.
Chemically-inducible diffusion trap of soluble proteins inside primary cilia. (a) Schematic diagram of ciliary influx of cytosolic YFP-FKBP proteins using a chemically-induced dimerization system. Available cytosolic YFP-FKBP in primary cilia can be trapped by rapamycin to a primary cilia membrane marker, 5HT6-CFP-FRB. (b) Fluorescence image of NIH 3T3 cells expressing 5HT6-CFP-FRB (false-colored red) and YFP-FKBP (green) before and 5 min after addition of 100 nM rapamycin, which induced accumulation of YFP-FKBP in the 5HT6-CFP-FRB-labeled primary cilia. Insets (enlarged views of the boxed regions) show cilia visualized as overlays of the color channels. Scale bars, 10 μm. (c) Video frames illustrating the translocation of YFP-FKBP into 5HT6-CFP-FRB-labeled primary cilia before and after addition of 100 nM rapamycin. Scale bars, 3 μm. (d) A line-scan analysis of the YFP signal intensity along the primary cilia after addition of 100 nM rapamycin for the indicated time. Cilium is indicated perpendicularly with its tip on the right (n = 3 cells from three independent experiments). (e) Time course of YFP fluorescence intensity in primary cilia of the cells treated with 100 nM rapamycin (blue) or 0.1% dimethyl sulfoxide (DMSO) (red). Error bars represent S.E.M. (n = 16 cells from three independent experiments).
We employed the 5-hydroxytryptamine receptor 6 (5HT6)13 to target a fusion protein of cyan fluorescent protein (CFP) and FRB to the ciliary membrane (Fig. 1a). Immunofluorescence images confirmed that the resulting fusion protein, 5HT6-CFP-FRB was highly enriched in primary cilia (Supplementary Fig. 1). When we co-transfected NIH 3T3 cells with 5HT6-CFP-FRB and yellow fluorescent protein (YFP)-tagged FKBP (YFP-FKBP), the former was highly enriched in cilia and the latter was distributed throughout the cytoplasm (Fig. 1b, left panels). Addition of rapamycin for 5 min led to a striking increase in YFP fluorescence in primary cilia (Fig. 1b, right panels). Time-lapse, dual-color fluorescence imaging revealed spatial dynamics and kinetics of the process (Supplementary Video1). More specifically, time series of YFP fluorescence in primary cilia and line scan analysis demonstrated that YFP-FKBP gradually accumulated in cilia from the base to tip until saturation was reached, presumably because all the 5HT6-CFP-FRB binding sites were occupied and thus the concentration gradient was abolished (Fig. 1c,d). Lateral diffusion of YFP-FKBP, 5HT6-CFP-FRB, and rapamycin complexes within the cilia membrane could also contribute to the spatial temporal evolution of the YFP fluorescence profiles. The time required for half-maximal (t1/2) accumulation in cilia for YFP-FKBP was 57 ± 5 s (Fig. 1e).
To rule out the possibility that our results were cell type specific, we tested the influx assay in another cell type, IMCD3, commonly used as a model in cilia biology. No significant difference was found in the influx rate of YFP-FKBP into primary cilia between NIH3T3 and IMCD3 cells (p=0.749, 57 ± 5 s and 68 ± 32 s, respectively) (Supplementary Fig. 2).
Several controls were performed to establish that YFP-FKBP accumulation was caused by the trapping of YFP-FKBP that diffused into cilia. During the experimental time period, we did not observe an increase in the levels of the 5HT6-CFP-FRB bait in primary cilia (Supplementary Fig. 3). Second, trapping required a freely diffusing form of YFP-FKBP because a variant of YFP-FKBP anchored to the cytoplasmic face of the endoplasmic reticulum (YFP-FKBP-Cb5) by a single transmembrane domain could not be recruited to cilia by rapamycin14 (Supplementary Fig. 4). Finally, we sought to exclude the important possibility that the observed accumulation of YFP-FKBP was not due to trapping within cilia but rather due to the transport of complexes between 5HT6-CFP-FRB, YFP-FKBP and rapamycin that formed outside cilia. Previous reports have shown that the diffusion of 5HT6 across the ciliary membrane protein barrier is slow (hours)7 compared to our assay timescale (min). We confirmed the kinetics of 5HT6 by performing a fluorescence recovery after photobleaching (FRAP) assay. 5HT6-GFP and 5HT6-GFP-FRB fluorescence did not recover inside primary cilia for 90 min after photobleaching (Supplementary Fig. 5), suggesting that there is negligible flux of new molecules of 5HT6-GFP-FRB into the cilium during our experiments.
Taken together, these studies indicate that the addition of rapamycin produces a diffusion trap in primary cilia, acutely generating a concentration gradient that can be used to follow the diffusion of any protein fused to YFP-FKBP.
Chemically-inducible trapping using orthogonal dimerizer systems
We also constructed a ciliary diffusion trap using an orthogonal chemical dimerizer system which uses a gibberellin analog (GA3-AM) and two completely different protein domains15 instead of FRB and FKBP. Upon addition of GA3-AM, YFP-labeled Gibberellin insensitive DWARF1 (YFP-GID1) accumulated in primary cilia carrying 5HT6 fused to the N-terminal 92 amino acids of Gibberellin insensitive (5HT6-GAI(S)) (Supplementary Fig. 6). The influx rate of YFP-GID1 into primary cilia was roughly 3 times slower than that of YFP-FKBP (185 ± 29 s vs. 57 ± 4.6 s). This difference could be due to the larger size of YFP-GID1 (616 amino acids) compared to YFP-FKBP (392 amino acids), differences in the permeability of the chemical dimerizers, or dimerization affinity. These results suggest that the FKBP-FRB and GID1-GAI(S) systems can be used together in cells to induce two separate dimerization driven signaling manipulations.
Probing the diffusion barrier of primary cilia using chemically-inducible trapping
We then performed ciliary trapping experiments using YFP-FKBP proteins that were fused to a series of diffusion probe proteins whose native molecular weights ranged from 40-650 kDa (Table 1). These test proteins were chosen according to the following criteria: (a) uniform distribution in the cytosol (Supplementary Fig. 7), (b) lack of effect on the localization of 5HT6-CFP-FRB (Supplementary Fig. 7), and (c) lack of effect on the length of cilia (Supplementary Fig. 8). To determine the Stokes radii (Rs) of these proteins, extracts made from cells over-expressing each YFP-tagged protein were fractionated by gel filtration chromatography and the Rs of each protein was calculated using a set of standards (Supplementary Fig. 9). The YFP-FKBP fusion proteins varied in size between 32 and 79 Å (Table 1). Of note, YFP-FKBP fusion proteins of full-length β-galactosidase (YFP-FKBP-β-Gal) and its N-terminally truncated variant (YFP-FKBP-ΔN β-Gal) formed a tetramer (Rs: 79 Å) and dimer (Rs: 62 Å), respectively, as described previously16, 17. We then measured the rate at which these proteins accumulated in primary cilia after the addition of rapamycin. Our results indicated a decrease in accumulation rates as protein size increased (Fig. 2a and Supplementary Fig. 7). A single exponential curve fit the data (R2 = 0.85), suggesting the existence of a diffusion barrier that is dependent on protein size (Fig. 2a). Fig. 2b illustrates representative fluorescence images of the ciliary accumulation of three proteins (YFP-FKBP, YFP-FKBP-Luciferase, and YFP-FKBP-β-Gal) after rapamycin addition. One concern in the above experiment was that we were detecting the translocation of smaller degradation products of the probe proteins and not the full-length proteins. To address this possibility, we constructed two additional fusion proteins, YFP-Luciferase-FKBP and YFP-β-Gal-FKBP, in which the YFP used for detection by fluorescence microscopy was separated from the FKBP used for dimerization by the probe protein. In this arrangement, any degradation product would separate the YFP from the FKBP, rendering it undetectable. Only the full length protein would carry both domains and thus be detected in our assays. There was no significant difference in the accumulation rate between YFP-Luciferase-FKBP and YFP-FKBP-Luciferase (p=0.85, 229 ± 100 s vs. 197 ± 83 s), or YFP-β-Gal-FKBP and YFP-FKBP-β-Gal (p=0.65, 813 ± 171 s vs. 997 ± 334 s) (Fig. 2a), strongly suggesting that the observed accumulation rates reflect those of full length proteins. Table 1 summarizes the accumulation rates together with other size-related parameters. We also investigated whether the chemical dimerizers themselves could affect primary cilia function. The addition of rapamycin or GA3-AM alone did not affect cilia length after one hour (Supplementary Fig. S10). This result indicates that non-specific effects on cilia due to the addition of chemical dimerizers are unlikely to occur over the time scale of our diffusion assays, which were typically complete within 30 minutes.
Table 1.
Size and influx rate of FKBP probe proteins
| Test Proteins | Experimen tal Rs (nm)1 | Experiment al MW (kDa)2 | Estimate d MW (kDa)3 | T1/2 accumulatio n time (sec) | Slope of integrated fluorescenc e in cilia (a. u.)4 | Diffusion coefficients inside cilia (μm2 /s) |
|---|---|---|---|---|---|---|
| YFP-FKBP | 3.2 | 40 | 43 | 57 ± 5 | 130 ± 25 | 5.58 ± 0.52 |
| YFP-FKBP-PKIM | 4.1 | 39 | 49 | 60 ± 2 | 248 ± 65 | 5.33 ± 1.5 |
| YFP-FKBP-Grp1(229-772) | 4.1 | 57 | 62 | 101 ± 33 | 69 ± 11 | 3.99 ± 0.54 |
| YFP-FKBP-Grp1(229-1200) | 4.4 | 72 | 78 | 95 ± 21 | 100 ± 29 | 2.57 ± 0.61 |
| YFP-Luciferase-FKBP | 4.4 | 100 | 101 | 229 ± 101 | 119 ±17 | 1.44 ± 0.19 |
| YFP-FKBP-Luciferase | 4.5 | 100 | 102 | 197 ± 83 | 30 ± 7 | 0.91 ± 0.23 |
| YFP-FKBP-Grp1(1-772) | 4.9 | 69 | 71 | 130 ± 22 | 134 ± 85 | 1.97 ± 0.74 |
| YFP-FKBP-Grp1(1-1200) | 5.0 | 88 | 87 | 266 ± 59 | 52 ± 13 | 0.69 ± 0.37 |
| YFP-FKBP-Tiam1 | 5.1 | 124 | 107 | 193 ± 64 | 29 ± 9 | 0.90 ± 0.25 |
| YFP-FKBP-ΔN β-Gal | 6.3 | 322 | 305 | 466 ± 83 | 45 ± 27 | 0.69 ± 0.26 |
| YFP-FKBP-β-Gal | 7.6 | 659 | 622 | 813 ± 171 | 18 ± 13 | 0.32 ± 0.15 |
| YFP-β-Gal-FKBP | 7.9 | 651 | 626 | 997 ± 334 | 14 ± 5 | 0.28 ± 0.25 |
Measured and calculated from a native gel filtration assay with internal standards.
Calculated from a western blot analysis.
Estimated based on amino acid sequences.
Corresponds to J in equation (1).
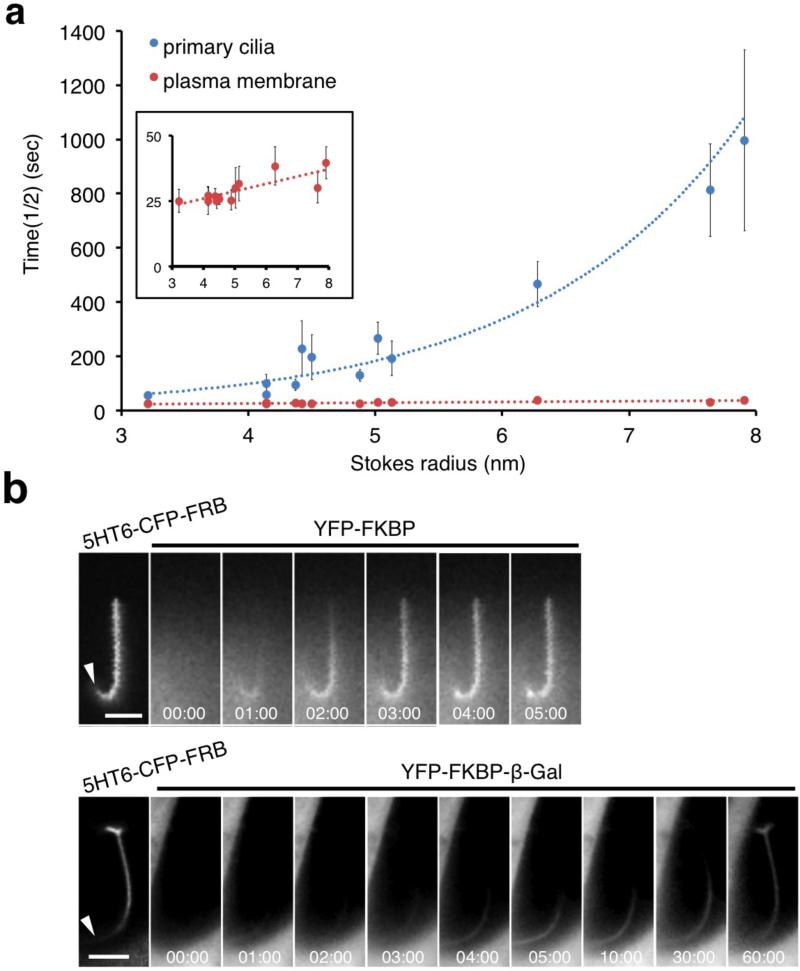
Figure 2.
Exponential dependence of influx rate on size. (a) The influx rate of FKBP probe proteins of various sizes into primary cilia. Cells were transfected with one of 13 YFP-FKBP probe proteins with either 5HT6-CFP-FRB (primary cilia) or Lyn-CFP-FRB-GAI(S) (plasma membrane). The graph shows the half time (t1/2) of influx kinetics of each FKBP probe protein to primary cilia (blue) or plasma membrane (red) as a function of its Stokes radius (n ≥ 10 cells from three independent experiments). The inset graph indicates a close up view of t1/2 for the plasma membrane trapping. The YFP-FKBP probe proteins tested are the following (in order of size): YFP-FKBP, YFP-FKBP-PKIM, YFP-FKBP-Grp1(229-772), YFP-FKBP-Grp1(229-1200), YFP-Luciferase-FKBP, YFP-FKBP-Luciferase, YFP-FKBP-Grp1(1-772), YFP-FKBP-Grp1(1-1200), YFP-FKBP-Tiam1, YFP-FKBP-ΔN β-Gal, YFP-β-Gal-FKBP, and YFP-FKBP-β-Gal. (b) Time-series fluorescence images of YFP-FKBP, YFP-FKBP-Luciferase, and YFP-FKBP-β-Gal inside primary cilia upon addition of 100 nM rapamycin. Left images indicate 5HT6-CFP-FRB anchored to primary cilia. Scale bars, 3 μm.
Chemically-inducible trapping at the plasma membrane does not exhibit any diffusion barrier
The Stokes-Einstein diffusion equation predicts a linear relationship between diffusion coefficients and the inverse of the Stokes radii (i.e., Rs−1) for freely diffusing molecules without any barrier. This was exactly what was observed when rapamycin was used to trap YFP-FKBP fusion proteins at the plasma membrane where a membrane-anchored FRB was expressed (Fig. 2a and Supplementary Fig. 11). The kinetics of plasma membrane recruitment of our diffusion probes is consistent with the previous observation that diffusion of macromolecules in the cytoplasm is unhindered by barriers18. The difference in size dependency on accumulation kinetics (linear for recruitment to the plasma membrane but exponential for recruitment to the cilium) strongly supports the existence of a size-dependent barrier at primary cilia.
Localization of diffusion limiting components in cilia
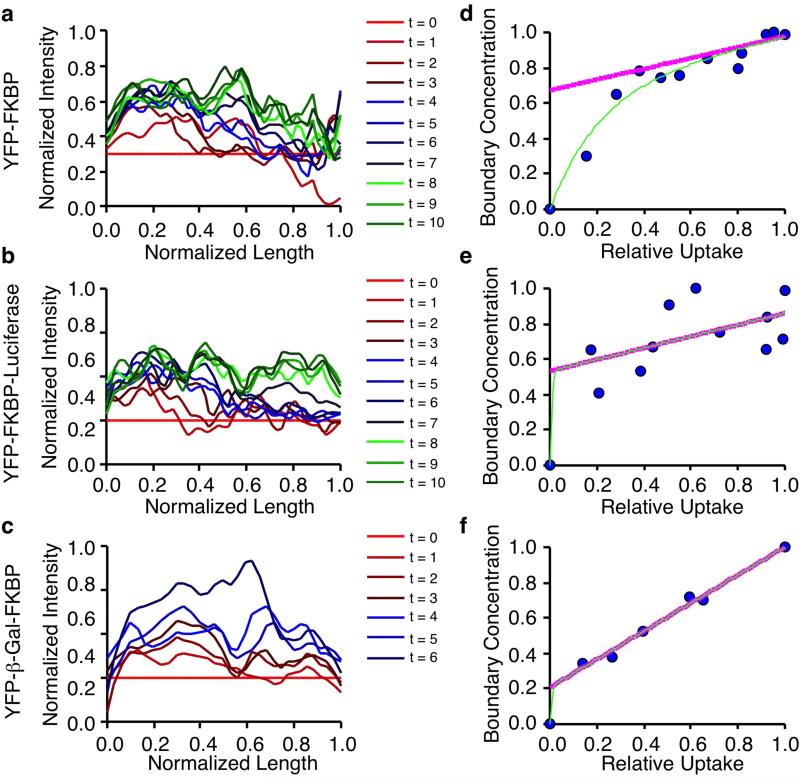
Our kinetic data supported the notion of hindered diffusion into primary cilia, but it was not apparent whether diffusion into cilia was limited by a putative barrier at the entrance and/or structures within cilia and how these relationships would evolve for different sized constructs. We reasoned that the time evolved YFP fluorescence profiles of different FKBP constructs within cilia, as shown in Fig. 1D, could provide clues to the localization of diffusion limiting components in cilia. We capitalized on theory derived from crystal diffusion19, where analysis of boundary concentrations (analogous to FKBP construct concentrations at the base of cilia) in relation to uptake (accumulated FKBP constructs in cilia) could be used to determine whether the surface permeability (a barrier at the base of cilia) or intracrystalline diffusion (intracilia diffusion) was limiting. Using this analysis, we calculated line scan data from three constructs which spanned the spectrum of our tested size range (3.2 - 7.9 nm) (Fig. 3a-c) and derived the corresponding boundary concentration vs. relative uptake plots (Fig. 3d-f). Details on how these values were determined can be found in the supplementary information. Based on the plots, we calculated a factor ‘W’, which estimates the influence of a permeability barrier versus intracilia factors in regulating uptake into cilia19. W values close to 0 indicate that entry into primary cilia is rate determining, while W values close to 1 indicate diffusion within cilia is rate determining. Intermediate W values indicate that both factors contribute to governing ciliary diffusion. We found a gradation of W values from high to low as FKBP construct size increased (Fig. 3d-f), suggesting that as protein size increases, diffusion into cilia becomes increasingly hindered by a barrier at the base of primary cilia.
Figure 3.
Localization of ciliary diffusion barriers. (a-c) Representative examples of YFP fluorescence line scans of indicated constructs through time from the base to tip of cilia. Data is normalized to the line scan at t = 0. (d-f) Analysis of boundary concentration vs. relative uptake of the corresponding FKBP constructs in a-c. The green line indicates the fit of the following equation- y = a(1 – ecx) + bx (1). The purple line indicates the fit of y = a + bx (2) using coefficients derived from the fit of (1). W denotes the average Y intercept of (2) from multiple cells along with the standard error of the mean (S.E.M). Number of cells are indicated.
A molecular sieve acts as a passive barrier at the base primary cilia
Our analysis above suggested that a diffusion barrier at the base of cilia regulates the entry of proteins in a size dependent manner. To shed light on the physical nature of the barrier, we calculated the diffusion coefficients (D) that characterize the movement of each of the FKBP constructs into primary cilia and then compared them to the D's predicted by various models. We reasoned that the very initial flux of FKBP complexes across a proposed diffusion barrier into the cilium could be described by Fick's first law
| (1) |
where [Fcy] and [Fci] are concentrations of the FKBP construct in the cytoplasm and cilia, respectively, D is the diffusion coefficient of the FKBP construct, and x is the length of the diffusion barrier. We used equation (1) to solve for D for each construct using parameters derived from our experiments ([Fcy] and J), and structural details of primary cilia from our experiments and the literature4, 20-23 (Fig. 4a). Full details of the measurements and assumptions used can be found in the supplementary information.
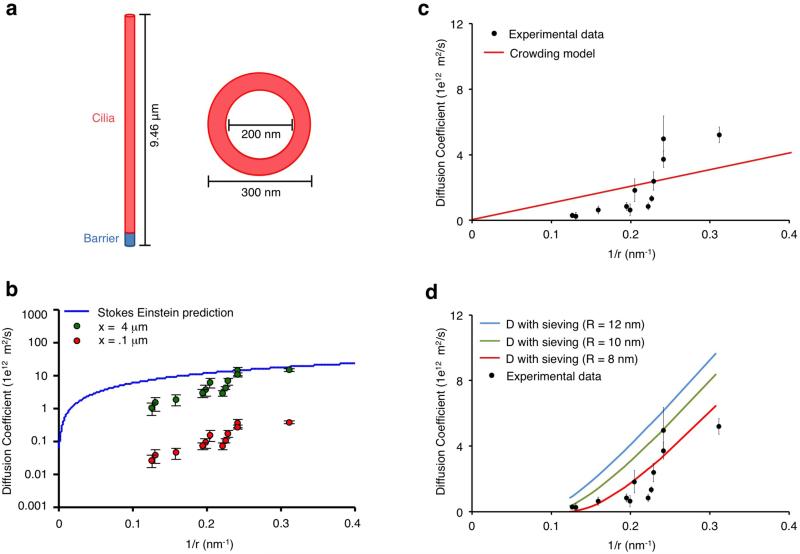
Figure 4.
Analysis of the diffusion barrier at primary cilia. (a) Structural depiction of the geometry of the primary cilia used to calculate diffusion coefficients. (b) Variation of barrier length and its effect on diffusion coefficients. Increases in the barrier length cause an upward shift in diffusion coefficients. The Stokes Einstein equation curve is included as an upper boundary to the diffusion coefficients. (c) Experimentally derived diffusion coefficients are plotted as a function of the inverse of their Stokes radii in comparison to a molecular crowding model shown in red. (d) Experimentally derived diffusion coefficients are plotted as a function of the inverse of their Stokes radii in comparison to a molecular sieving model. The resulting curves for different mesh sizes are included in the plot with an optimal fit at R = 8 nm. The YFP-FKBP probe proteins plotted are the followings: (in order of size): YFP-FKBP, YFP-FKBP-PKIM, YFP-FKBP-Grp1(229-772), YFP-FKBP-Grp1(229-1200), YFP-Luciferase-FKBP, YFP-FKBP-Luciferase, YFP-FKBP-Grp1(1-772), YFP-FKBP-Grp1(1-1200), YFP-FKBP-Tiam1, YFP-FKBP-ΔN β-Gal, YFP-β-Gal-FKBP, and YFP-FKBP-β-Gal.
As a first step, given that the length of the barrier is unknown, we assessed the effect of varying barrier length on our calculated D's (Fig. 4b). We used the Stokes Einstein equation as an upper boundary for D's to confine the range of possible barrier lengths (range of 0 to 4 μm) (Fig. 4b). Our calculated D's displayed a linear relationship with barrier length and for subsequent analysis we fixed the barrier length at 1.5 μm. Interestingly, the relationship between the experimentally determined D's and the inverse of Stokes radii of the probe proteins was non-linear, a hallmark of a size-dependent barrier (Fig. 4c). This size dependency was independently confirmed by performing FRAP experiments using the smallest and largest probe proteins at primary cilia. The measured diffusion coefficients were 4.3 ± 2.3 μm2/s for GFP-FKBP and 0.3 ± 0.1 μm2/s for GFP-FKBP-β-Gal (Supplementary Fig. 12). As the photobleaching was targeted to a ciliary subregion, these values reflect the diffusion both inside and into the primary cilia.
We next assessed the ability of different models of hindered diffusion to account for the discrepancy between predicted and experimentally measured D's. First, we investigated whether molecular crowding in the primary cilia could explain our data by implementing a Stokes radius independent molecular crowding term24,
| (2) |
where Dc is the D in a molecularly crowded environment, Dsc is the D predicted by the Stokes-Einstein equation in cytoplasm and C is a constant crowding term. We compared Dc for each FKBP construct to our experimental D‘s and found that the closest fit C = 0.19 was a poor fit to our data (R2 = 0.43, RMSE = 1.31, Fig. 4c).
Next, we investigated molecular sieving as a mechanism to explain our observed hindered diffusion. NPCs have been shown to contain a molecular sieve in the lumen which impedes passive diffusion into the nucleus in a size dependent manner25. We hypothesized that a similar sieve may exist at primary cilia and modeled the sieve with the following equation25-
| (3) |
where Dm is the diffusion coefficient in the mesh, Dsc is the diffusion coefficient from the Stokes-Einstein equation in cytoplasm, r is the Stokes radius of the FKBP construct, and R is the estimated mesh radius of the sieve. When we compared Dm for each probe protein with the experimentally derived D's, we found that molecular sieving could indeed provide a reasonable explanation for our data (Fig. 4d). The best fit for Dm from our standard cilia geometry (Fig. 4a) was obtained with a mean mesh radius of 8 nm (RMSE = .90, Fig. 4d and Supplementary Fig. 13a). One caveat to our analysis is that the calculated mesh radius will vary depending on the assumed barrier length (Supplementary Fig. 13b). When we fit the two parameters (mesh radius and barrier length) to our data, the overall best fit waswith a mesh radius of 8 nm and a barrier length of 1.4 μm (Supplementary Fig. 13c).
DISCUSSION
CID techniques have been used extensively to manipulate the level and activity of signaling molecules12. In this study, we adapted this technique to probe passive permeability barriers in cells. Specifically, we performed kinetic measurements of the ciliary accumulation of a series of diffusion probes into cilia, and suggest that the ciliary barrier for soluble proteins consists of a molecular sieve which hinders the entry of proteins in a size dependent manner. As more than 90% of the soluble proteins in mammalian cells are smaller than 650 kDa26, the size of our largest diffusion probe, our observation implies that most of the cytoplasmic proteins have the potential to freely enter cilia, albeit with kinetics that are determined by protein size. These results have two important implications for our view of how ciliary protein composition is regulated. Over long time scales, the steady state distribution of proteins in cilia is unlikely to be regulated by a barrier that excludes specific proteins, but rather simply by the selective retention of proteins in cilia due to binding interactions with other cilia-resident molecules and by molecular crowding10. However, for kinetically-controlled, rapid processes, such as signaling reactions, the sieve-like barrier could present a significant factor in limiting protein entry, with active or facilitated transport processes playing important regulatory roles.
One of the most well-characterized soluble diffusion barriers in cells are NPCs, which control the passage of biomolecules to and from the nucleus by excluding proteins larger than 60 kDa, slowing down the diffusion of proteins 30-60 kDa in size, and allowing anything smaller than 30 kDa to enter27. A recent study demonstrated that soluble proteins above a specific size threshold of 67 kDa are restricted from passively entering the primary cilia of epithelial cells in a manner analogous to the NPC11, in contrast to our result. This study used an end-point assay in which the ciliary distribution of soluble probe proteins was measured 5 minutes after microinjection into the cytoplasm. Thus, this study could not determine the rates of diffusion of the various test probes, which could easily lead to errors in visualizing the ciliary accumulation of larger, slow-diffusing probes. In contrast, the present C-IDTc technique not only provides diffusion kinetics, but also bypasses the need for perturbations such as microinjection or detergent permeabilization, preserving the physiological intracellular environment.
Our modeling suggests that the ciliary permeability barrier behaves like a mesh with sieving properties reminiscent of NPCs25, but with much larger pore radii. The largest pore radius proposed to exist at the NPC is approximately 4.3 nm25. However, in our experiments, inert proteins as large as 7.9 nm were able to enter cilia, suggesting a population of pores larger than 7.9 nm must exist at the cilia base. Our modeling further revealed that a pore radius of 8 nm gave the best fit to our data, but we are unable to rule out the possibility of a heterogeneous distribution of pore radii. The diffusion barrier seen at the base of dendritic spines in neurons28 could be more analogous to primary cilia than the nucleus, as neither structure is entirely compartmentalized by lipid membranes. Rather, both compartments are contiguous with the plasma membrane and decorated by septin molecules at their necks7, 29. The exponential increase in diffusivities we observed has been similarly noted for large molecules in porous media30. This further suggests that porous material at the base of cilia might thus form a molecular sieve which exponentially affects the entry of proteins into cilia in a size-dependent manner.
Unfixed pores for passive diffusion and preferential retention are both mechanisms observed in the photoreceptor connecting cilium (CC)10. Of note, ultrastructural studies on photoreceptor CC have not detected any structural features that deviate from primary cilia4, with some exceptions such as the specialized cilia of chondrocytes31. In CC photoreceptors, steric volume exclusion is one primary driving force that limits protein accumulation in the OS. However, our kinetic analysis revealed that the molecular flux across the cilia barrier significantly decreased with protein size in the presence of comparable concentration gradients. We suggest that volume exclusion may not be a major defining feature of the ciliary barrier in fibroblasts.
What is the molecular identity of the sieve-like barrier at the primary cilia? Ultrastructural studies have yet to find evidence for such meshwork-like structure at the base of primary cilia. However, these studies have revealed an electron-dense region around the ciliary subcompartment known as the transition zone (TZ). Indeed, the TZ is known to accumulate a series of proteins including nephrocystin complexes (NPHP1, NPHP4, etc.), BBSomes, nucleoporins, Cep290 and Cep16411, 32-37. Septin2, a barrier molecule for membrane receptors, also accumulates at the TZ, which suggests the TZ may serve as a molecular sieve for soluble proteins. As nucleoporins form hydrophobic hydrogels at NPCs, they are reasonable candidates for the ciliary barrier11. However, given that much larger proteins can passively enter primary cilia, we suggest that the organization of the nucleoporin FG-repeats may be different between these two compartments.
We have used the C-IDTc technique to answer fundamental questions about soluble protein diffusion into primary cilia. This method should be useful in future studies aimed at defining the molecular composition of this passive barrier. In principle, this technique should allow the time-resolved, inducible recruitment of any two proteins to primary cilia using the orthogonal recruitment modules we have developed. We believe that this methodology will allow perturbation of ciliary signaling reactions in ways that have previously been impossible, for instance by the recruitment of signaling proteins such as small GTPases or enzymes that can modulate the levels of second messengers such as phosphoinositide lipids.
Methods
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Acknowledgement
We thank Akiko Seki (Stanford University) for the 5HT6 construct, Sen Takeda (University of Yamanashi) for the GFP-IFT88 construct, and Michael Wolfgang (Johns Hopkins University) for β-Gal and Luciferase constructs. We also thank D.N.R., T.K., H.I. and S.T. for helpful discussions. This study was supported in part by the National Institute of Health (NIH) (GM092930 and DK090868 to Takanari I., and R00CA129174 and R21NS074091 to R.R.) and other grants (Grant-in-Aid for Challenging Exploratory Research 23650197 to H.N. and Pew Foundation to R.R.). Y.C.L is a postdoctoral fellow supported by the National Science Council in Taiwan. S.C.P. is a recipient of the National Science Scholarship from A*STAR in Singapore.
Footnotes
Author Contribution
Y.C.L., S.C.P. and Takanari I. generated DNA constructs, and Y.C.L., S.C.P., and J.J. performed cell biology experiments. B.L. analyzed data with R.R., P.N., A.L. and Takanari I., while P.N. performed biochemical experiments with R.R. H.N. performed FRAP experiments with Takafumi I. Y.C.L., B.L., R.R. and Takanari I. wrote the paper.
Competing Financial Interest
There is no competing financial interest associated with the present study.
References
- 1.Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 2.Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 2009;111:p39–53. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 4.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chih B, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 6.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Q, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr., Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Nair KS, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci U S A. 2012;109:203–208. doi: 10.1073/pnas.1115109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kee HL, et al. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012 doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derose R, Miyamoto T, Inoue T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013;465:409–417. doi: 10.1007/s00424-012-1208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsu T, et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto T, et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat Chem Biol. 2012 doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celada F, Zabin I. A dimer--dimer binding region in beta-galactosidase. Biochemistry. 1979;18:404–406. doi: 10.1021/bi00570a002. [DOI] [PubMed] [Google Scholar]
- 17.Matthews BW. The structure of E. coli beta-galactosidase. C R Biol. 2005;328:549–556. doi: 10.1016/j.crvi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Seksek O, Biwersi J, Verkman AS. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kärger J, Chmelik C, Heinke L, Valiullin R. A new view of diffusion in nanoporous materials. Chemie Ingenieur Technik. 2010;82:779–804. [Google Scholar]
- 20.Czlapinski JL, et al. Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. J Am Chem Soc. 2008;130:13186–13187. doi: 10.1021/ja8037728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narita K, Kawate T, Kakinuma N, Takeda S. Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic. 2010;11:287–301. doi: 10.1111/j.1600-0854.2009.01016.x. [DOI] [PubMed] [Google Scholar]
- 22.Okada Y, Takeda S, Tanaka Y, Izpisua Belmonte JC, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura K, Kawate T, Takeda S. Signaling through the primary cilium affects glial cell survival under a stressed environment. Glia. 2011;59:333–344. doi: 10.1002/glia.21105. [DOI] [PubMed] [Google Scholar]
- 24.Dauty E, Verkman AS. Molecular crowding reduces to a similar extent the diffusion of small solutes and macromolecules: measurement by fluorescence correlation spectroscopy. Journal of Molecular Recognition. 2004;17:441–447. doi: 10.1002/jmr.709. [DOI] [PubMed] [Google Scholar]
- 25.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT). Mol Cell Proteomics. 2006;5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 29.Tada T, et al. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahimi M, Jue VL. Diffusion of large molecules in porous media. Phys Rev Lett. 1989;62:629–632. doi: 10.1103/PhysRevLett.62.629. [DOI] [PubMed] [Google Scholar]
- 31.Jensen CG, et al. Ultrastructural, tomographic and confocal imaging of the chondrocyte primary cilium in situ. Cell Biol Int. 2004;28:101–110. doi: 10.1016/j.cellbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 34.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 35.Salomon R, Saunier S, Niaudet P. Nephronophthisis. Pediatr Nephrol. 2009;24:2333–2344. doi: 10.1007/s00467-008-0840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szymanska K, Johnson CA. The transition zone: an essential functional compartment of cilia. Cilia. 2012;1:10. doi: 10.1186/2046-2530-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Schnakenburg C, Fliegauf M, Omran H. Nephrocystin and ciliary defects not only in the kidney? Pediatr Nephrol. 2007;22:765–769. doi: 10.1007/s00467-007-0434-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.