Abstract
The innate immune system is present in all animals and is a crucial first line of defence against pathogens. However, animals also harbour large numbers of beneficial microorganisms that can be housed in the digestive tract, in specialized organs or on tissue surfaces. Although invertebrates lack conventional antibody-based immunity, they are capable of eliminating pathogens and, perhaps more importantly, discriminating them from other microorganisms. This Review examines the interactions between the innate immune systems of several model invertebrates and the symbionts of these organisms, and addresses the central question of how these long-lived and specific associations are established and maintained.
Multicellular life arose in a world dominated by microorganisms, and interactions between animals and their microbial companions have profoundly influenced animal evolution. Perhaps the best surveyed habitat for microorganisms in animal hosts is the digestive tract, where the microbial community carries out many crucial functions such as aiding in digestion, providing essential nutrients, protecting against colonization by pathogens and stimulating the immune response1-3. In recognition of the importance of these organisms, the term microbiome was introduced to describe the collective genome of the indigenous microbiota of an animal4. 16S rRNA gene surveys indicate that the specificity of the microorganisms colonizing mammalian digestive tracts is not conserved at the species or strain level, but rather there is conservation at the level of functional genes among diverse microbiota5-8. However, some invertebrate–symbiont model systems that lend themselves to experimental manipulation have revealed a higher degree of specificity that can even be achieved at the species or strain level9-11.
This specificity is multifactorial in origin, and there are some general requirements that quickly reduce the number of organisms capable of establishing a foothold in a particular animal host. Basic physiological requirements include an ability to grow and outcompete other microorganisms at the temperature, redox potential, pH and osmolarity that are found in the host. General nutritional conditions are often set by the host ingesting or providing nutrients at defined intervals and of particular nutritional quality12,13. Many of these factors can also be modified to some degree by the metabolism of the microbial community2,12,14,15. Furthermore, specific organs might help to foster these associations and create microenvironments that are conducive to symbiont growth but inhibit the growth of other microbial competitors. This sequestration at specific sites could serve to protect other environments in the body from colonization.
In addition to these constraints, interactions between symbionts and the host immune system have a crucial role in the establishment and regulation of these microbial communities16-22. For example, in the healthy human gut, immune defence mechanisms are modulated in response to the microbiota, an effect that can be referred to as the development of tolerance21,23. Animal immune systems are often classified based on either the broad and nonspecific innate immune response or the highly specific antibody-based adaptive immune response. Whereas jawed vertebrates use both types of immune response, all invertebrates combat potential pathogens and also foster the formation of mutualistic symbioses in the absence of conventional antibodies. Given that many of these invertebrate host–microorganism partnerships require a high degree of specificity, how can such associations form without obvious mechanisms for distinguishing between specific microorganisms?
In this Review, we explore the interactions between the innate immune system of invertebrates and the symbionts of these organisms, and discuss the role that these interactions have in establishing and maintaining specific microbial communities. This outcome is in stark contrast to the traditional view of the immune system and its role in eliminating pathogenic bacteria, and it is particularly interesting because the innate immune system reportedly lacks robust immunological memory. Given the abundance of model systems, we have selected only a few invertebrate species (FIG. 1) that represent different phylogenetic lineages and for which experimental models have revealed clues about how the innate immune system helps in shaping the microbiota.
Figure 1. Model systems of invertebrate symbioses.
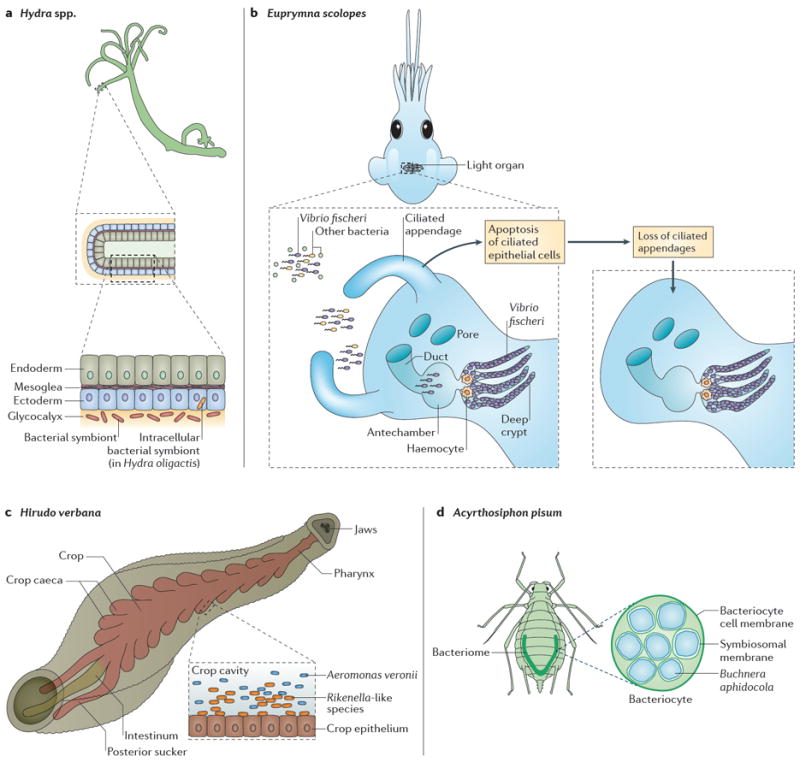
Four model hosts and their symbionts are discussed in this Review. a ∣ Hydra spp. are composed of two epithelial cell layers and house bacterial symbionts on the outermost cell type (the ectoderm), in the glycocalyx. Hydra oligactis also harbors an intracellular symbiont in its ectoderm. b ∣ Euprymna scolopes forms a binary association with the bacterium Vibrio fischeri in the crypts of the light organ. After the squid hatches, V. fischeri is selected from among all other bacteria in the environment and colonizes the juvenile light organ; this colonization initiates a developmental programme that results in morphogenesis of the light organ. c ∣ The leech Hirudo verbana carries two dominant symbionts in the crop (the largest compartment of the digestive tract), Aeromonas veronii and a Rikenella-like species. d ∣ The pea aphid, Acyrthosiphon pisum, houses the obligate intracellular symbiont Buchnera aphidicola in cells known as bacteriocytes, which are located in an organ-like structure known as the bacteriome.
Part b image is modified, with permission, from REF. 30 © (2011) Macmillan Publishers Ltd. All rights reserved.
The immune system of invertebrates
When early multicellular life first associated with microorganisms, hosts would have required sophisticated systems for recognizing and also differentiating beneficial and pathogenic microorganisms. However, unlike vertebrates, invertebrates lack classical antibody-based adaptive immunity, and key molecular and cellular players such as recombination-activating genes (RAGs), B lymphocytes and T lymphocytes are absent24. Although there are indications that some invertebrates have alternative adaptive or anticipatory immune functions and memory-like responses24-27 (BOX 1), to date none of these functions and responses has been shown to be widely evolutionarily conserved among invertebrate phyla. Despite this perceived limitation, invertebrates have mounted efficient defence responses to pathogens for hundreds of millions of years. In addition, growing evidence suggests that the innate immune system interacts differently with symbionts than with pathogens. Although there are many factors associated with invertebrate innate immunity, we focus on three main aspects here: the recognition of microorganisms by pattern recognition receptors (PRRs), cellular immunity in the form of the phagocyte response, and humoral components of the immune system, comprising acellular and biochemical factors (FIG. 2).
Box 1. Alternative anticipatory and memory-like immune mechanisms of invertebrates.
Although invertebrates lack a conventional antibody-based and recombination-activating gene (RAG) protein-mediated adaptive immune system, some invertebrates do possess the ability to generate highly diverse and polymorphic proteins or peptides in response to microorganisms and might have the ability to recognize self versus non-self tissues24-26,145. However, the role of such mechanisms in the establishment of the microbiota has not yet been addressed. These mechanisms, along with the discovery of an additional and alternative adaptive immune system with variable lymphocyte receptors in jawless fish146, suggests that other types of anticipatory and memory-like immunity remain to be discovered. Below are some examples of adaptive immune effectors found in invertebrates.
Fibrinogen-related proteins
Fibrinogen-related proteins (FREPs) contain a fibrinogen and one or two immunoglobulin superfamily domains, and are secreted into the haemolymph of the snail Biomphalaria glabrata in response to trematode parasites25. Although a detailed mechanism of FREP function has not been described, these proteins can bind to parasites and bacteria, and the host can generate a larger FREP repertoire through somatic diversification in response to infection.
Down’s syndrome cell adhesion molecules
Down’s syndrome cell adhesion molecules (DSCAMs) were discovered in Drosophila melanogaster26. By means of differential RNA processing, thousands of protein variants are generated, some of which seem to adhere to and mediate phagocytosis of bacteria.
Immune priming
Examples of memory-like innate immune responses have been described in insects27,42. Hosts that are challenged with specific pathogens will occasionally exhibit a more efficient immune response after a second exposure. In the mosquito Anopheles gambiae, this primed response might be partially mediated by the gut microbiota, as well as by haemocyte differentiation27.
Figure 2. Proposed pathways of innate immune response activation in invertebrates.
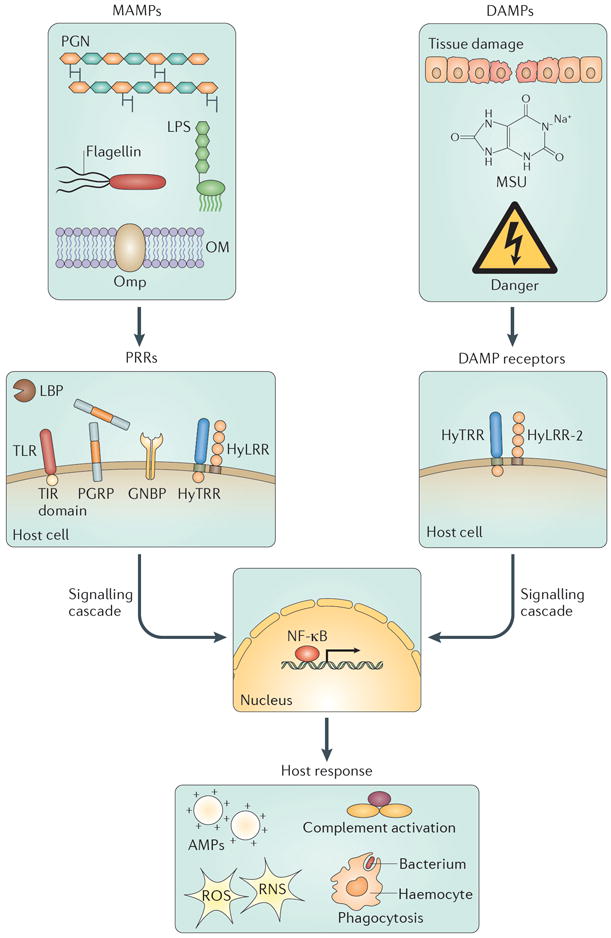
Microorganism-associated molecular patterns (MAMPs) can be recognized as non-self by the host and include compounds such as lipopolysaccharide (LPS), peptidoglycan (PGN), bacterial flagellin and bacterial outer-membrane proteins (Omps). MAMPs can initiate signalling cascades by first interacting with pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and other Toll–interleukin 1 receptor (TIR) domain-containing proteins, peptidoglycan recognition proteins (PGRPs), Gram-negative bacteria-binding proteins (GNBPs) and LPS-binding proteins (LBPs). Hydra spp. also possess Hydra leucine-rich-repeat proteins (HyLRRs), which function with Hydra TIR domain-containing proteins (HyTRRs) to recognize MAMPs (although the specific family members that function as receptors in this pathway have not yet been determined). Other signals that can trigger immune responses are damage-associated molecular patterns (DAMPs), which might result from physical tissue damage and the release of compounds such as monosodium urate (MSU). In Hydra spp., DAMPs can interact with HyLRR-2, which functions with a HyTRR to initiate immune signalling. Both MAMP- and DAMP-mediated signalling can initiate signalling cascades, usually through the nuclear factor-κB (NF-κB) or similar pathway, that lead to the activation of immune responses such as haemocyte trafficking and phagocytosis; induction of the complement pathway; and production of antimicrobial peptides (AMPs), reactive oxygen species (ROS) and reactive nitrogen species (RNS). OM, outer membrane.
Pattern recognition
Cell–cell signalling between microorganisms and their hosts is largely mediated through interactions between host receptors and microorganism-derived ligands23,28-30. Hosts detect specific bacterial components such as peptidoglycan (PGN), lipopolysaccharide (LPS), outer-membrane proteins (Omps), flagellins and other molecules that are expressed by a wide range of bacteria. These molecules were originally termed pathogen-associated molecular patterns (PAMPs) by Janeway in 1989, and this term was popularized by Medzhitov and Janeway in 1997 (REFS 31-33). This concept was based on the idea that the recognition of a molecule such as LPS, which is found in all Gram-negative pathogens but absent in animals, allows receptors of the immune system to recognize the presence of pathogens and launch an immune response. However, PGN, LPS, Omps and flagellin are not pathogen specific, instead being common to many different types of microorganism, including those that form beneficial associations with animals and are generally benign to the host. Indeed, even classical virulence factors such as the type III secretion system (T3SS)34 are required by some beneficial microorganisms for interactions with animal hosts and are therefore not pathogen specific16,35. Thus, the more general term microorganism-associated molecular patterns (MAMPs) was introduced by McFall-Ngai and colleagues36. Some of the best known examples of PRRs are the Toll-like receptors (TLRs) and peptidoglycan recognition proteins (PGRPs), which trigger downstream signalling pathways, often activating cellular and humoral innate immunity effectors such as phagocytes, antimicrobial peptides (AMPs) and reactive oxygen species (ROS) (reviewed in REFS 30,37) (FIG. 2). Because most animal hosts encounter MAMPs from both pathogenic and non-pathogenic microorganisms, additional regulatory levels must be in place that allow for discrimination between beneficial and pathogenic associations.
Phagocytic cells
Phagocytic cells are common to all eukaryotes, and some of the earliest endosymbioses were probably the result of single-celled organisms engulfing bacteria that eventually evolved into organelles such as mitochondria and chloroplasts38,39. As metazoans evolved, circulating phagocytic haemocytes became ubiquitous to most groups except those with only tissue level organization, such as cnidarians; certain organisms, such as the nematode Caenorhabditis elegans, subsequently lost this cell type40,41. Recent evidence suggests that interactions between microorganisms and invertebrate haemocytes can be highly specific and are important for the establishment and maintenance of symbioses in invertebrates18. Furthermore, studies with different fly and mosquito species have shown that haemocytes are likely to be involved with priming the immune system to bacterial challenge27,42. These studies have also shown that haematopoiesis and the development of other components of the innate immune system might be influenced by the presence of bacterial symbionts43. These data, along with the studies of symbioses described below, suggest that haemocytes are capable of highly specific interactions with bacteria.
Humoral defences
In addition to the cellular immune response, animals possess acellular, chemical-based mechanisms that constitute the humoral response and also shape the microbial community. This response includes the production of AMPs, which lyse cells, and ROS, which oxidize lipids and proteins and also damage DNA. Both pathogens and symbionts must overcome these insults to thrive in the specific niche of the host. AMPs and ROS are among the most widely used immune effector mechanisms among both vertebrates and invertebrates, and they appear to have been conserved throughout evolution44-46. In addition, many invertebrates possess components of the complement system, which assists in the detection of microorganisms and MAMPs using opsins and lectin–ligand interactions47.
Invertebrate models of symbioses
Below, we describe four model invertebrates, from diverse taxa, that form associations ranging in complexity from involving a single symbiotic species to diverse consortia (FIG. 1). These symbionts are localized in or on specialized cells, tissues or organs. The first of these model systems is cnidarians of the genus Hydra, which represent basal metazoans that have a simple tissue organization48-50. Owing to their basal position in the animal lineage, these organisms provide an important model for understanding microbial interactions with hosts. This basal position also means that the immune system processes which these cnidarians use to shape the composition of their microbial community are likely to be present in more complex animals. Two lophotrochozoan models are also discussed: Euprymna scolopes (the Hawaiian bobtail squid), which houses a single bacterial species, Vibrio fischeri, in a specialized light organ51-53; and Hirudo verbana (a medicinal leech), which has a simple microbial community in its gut that is dominated by two bacterial species, Aeromonas veronii and a Rikenella-like bacterium54-56. Of the ecdysozoan insects, Drosophila melanogaster has a moderately complex gut microbiota comprising 5–20 species and is the best characterized invertebrate model system57. However, the features of the innate immune response of D. melanogaster have been reviewed elsewhere recently30,58,59. Instead of this system, for our fourth model we discuss the association between Acyrthosiphon pisum (the pea aphid) and its primary endosymbiont, Buchnera aphidicola60,61. We explore these systems with regard to, first, whether the symbionts possess specific capabilities to escape or neutralize the immune response; second, common mechanisms by which the innate immune system of the host detects and responds to bacterial symbionts (TABLE 1) and third, how the immune response is influenced by spatial and temporal factors, and whether additional signals such as tissue damage are also required for immune stimulation.
Table 1.
Innate immune effectors in selected invertebrates that harbour associations with bacterial symbionts
| Organisms | PRR–MAMP signalling
|
Humoral components
|
||||
|---|---|---|---|---|---|---|
| PRRs | NF-κB and/or other immune signalling pathways | Haemocyte response | AMPs | Complement- like factors | ROS and RNS | |
| Hydra spp. | HyTRRs and HyLRRs | Present (NF-κB pathway) | Absent | Present | Some present | Present |
|
| ||||||
| Euprymna scolopes | PGRPs, TLR and LBPs | Present (NF-κB pathway) | Present | Unknown | Present | Present |
|
| ||||||
| Hirudo verbana | Predicted PRRs | Mostly present (NF-κB pathway) | Present | Present | Present | Present |
|
| ||||||
| Acyrthosiphon pisum | Toll and GNBPs, but no PGRPs | Present (Toll and JAK–STAT pathways; reduced and incomplete Imd pathway) | Present | Reduced | Present | Present |
AMPs, antimicrobial peptides; GNBPs, Gram-negative bacteria-binding proteins; HyLRRs, Hydra leucine-rich repeat proteins; HyTRRs, Hydra Toll–interleukin-1 receptor domain-containing proteins; Imd, immune deficiency; JAK–STAT, janus kinase–signal transducer and activator of transcription; LBPs, lipopolysaccharide-binding proteins; MAMP, microorganism-associated molecular pattern; NF-κB, nuclear factor-κB; PGRPs, peptidoglycan recognition proteins; PRR, pattern recognition receptor; RNS, reactive nitrogen species; ROS, reactive oxygen species; TLR, Toll-like receptor.
Bacterial symbiosis in Hydra spp
Hydra vulgaris and related species of the phylum Cnidaria consist of two main cell layers, the ectoderm and the endoderm50 (FIG. 1a). A feature that distinguishes the members of this phylum from the other animal models described in this Review is the lack of motile phagocytic cells. Thus, Hydra spp. provide the opportunity to study the immune response of ectodermal and endodermal cells62. General features of the cnidarian immune response include AMPs, PRRs and a signalling cascade related to the nuclear factor-κB pathway (NF-κB pathway)63. Another generic protective feature of cnidarians that is also widespread among animals is a mucus layer known as the glycocalyx, which covers the ectoderm (FIG. 1a).
The microbial consortia of Hydra spp
A ground-breaking study revealed that H. vulgaris and the closely related species Hydra oligactis are capable of shaping their epidermal microbiota64, which might aid in the defence against pathogenic fungi (T. Bosch, personal communication). A comparison of H. vulgaris and H. oligactis isolates collected from two lakes — including some isolates of both species from the same lake —with strains that had been maintained for more than 30 years in a laboratory revealed two interesting results64. One intracellular symbiont, an alphaproteobacterium related to Rickettsia and Ehrlichia spp., had been retained in laboratory cultures of H. oligactis for more than 30 years but was absent in the H. vulgaris laboratory strain. Furthermore, the diversity and composition of the extracellular bacteria associated with the epithelium differed between the two species. Both laboratory and wild isolates of H. vulgaris harboured substantially more species than H. oligactis from either source, suggesting that H. vulgaris is permissive to a wider range of microorganisms than H. oligactis. Analysis of 16S rRNA gene sequence data showed that although several bacterial species are common to both the wild H. vulgaris and the wild H. oligactis isolates, as a whole each species possesses a distinct microbiota and might thus use different mechanisms to shape these microbial communities. Presumably, these mechanisms must include recognizing specific microorganisms and/or fostering an environment that leads to the establishment of the community.
A key follow-up study discovered that Hydra spp. can mount a powerful immune response in the absence of motile phagocytic cells65. This response is contingent on two independent components: the presence of bacterial flagellin (a classic example of a MAMP) and the secretion of a tissue damage or danger signal, known as a damage-associated molecular pattern (DAMP) (FIG. 2). Interestingly, Hydra spp. do not appear to encode typical TLR-like proteins. Instead, two proteins are required for MAMP recognition and signal transduction: a Hydra Toll–interleukin-1 receptor domain (TIR domain)-containing protein (HyTRR) and Hydra leucine-rich-repeat protein 2 (HyLRR-2), containing a transmembrane domain. HyLRR-2 interacts with the HyTRR protein in response to flagellin. When exposed to a DAMP, such as monosodium urate (which is released from injured cells), an even stronger induction of defence genes is observed. For example, there is increased production of AMPs such as periculin-1. It is tempting to speculate that the requirement for both a MAMP and a DAMP to launch an effective immune response constitutes a mechanism that allows Hydra spp. to distinguish pathogens from symbionts.
This study has several important implications65. First, it demonstrates that an ancient lineage of multicellular life can recognize broad categories of microorganisms based on their MAMPs and can integrate DAMPs (which are induced only by organisms that harm the host) to induce an immune response (FIG. 2). This ability to integrate the two signals (MAMPs and DAMPs) may contribute to shaping of the microbiota, but further mechanistic studies are needed to clarify this. A further study also demonstrated that periculins are not only important in mounting an immune response in developing H. vulgaris embryos but also instrumental in establishing the bacterial community, suggesting that the innate immune system is crucial for both the establishment and long-term maintenance of host-associated bacterial communities in Hydra spp.20.
The Euprymna scolopes–Vibrio fischeri symbiosis
Cephalopods are among the most complex invertebrates. They have a closed circulatory system with a highly developed vasculature that allows circulating phagocytic haemocytes to migrate between tissues and interact with both symbiotic and non-symbiotic microorganisms18,66-68. These invertebrates also express PRRs that can recognize MAMPs in a number of tissues, and they are capable of mounting potent antimicrobial responses using ROS as well as other biochemical mechanisms19,68,69.
The association between the Hawaiian bobtail squid, E. scolopes, and the bioluminescent bacterium V. fischeri has served as a model for understanding how beneficial bacteria influence animal development, colonize host epithelia and interact with the host innate immune system19,51-53,70-72. In contrast to the other examples discussed in this Review, this association is binary, involving a single host and a single symbiont. Thus, this system offers an opportunity to study the involvement of the immune system in establishing and maintaining such high specificity.
Within the squid, V. fischeri is housed in a vascularized light organ that is responsible for a behaviour known as counter-illumination; the squid uses light produced by the symbiont to mask itself from predators when it is foraging at night73. The bacteria are contained within epithelium-lined crypt spaces that are directly connected to the environment via ducts which terminate in the host’s mantle cavity (FIG. 1b). The association is transmitted environmentally: after hatching, the juvenile squid obtain bacteria from the surrounding sea water, selecting V. fischeri, which constitutes less than 0.1% of the 106 bacteria that are found per ml of sea water74,75. There are many mechanisms by which the partners ensure this specificity, and the innate immune system of the host seems to play a central part in this process18,19,44,52,70,72,76-78.
Pattern recognition in the squid
Host recognition of bacterial MAMPs is crucial for many colonization events in the E. scolopes–V. fischeri association. For example, one of the earliest host responses is copious secretion of mucus following exposure to the external environment when the juvenile squid emerges from the safety of its egg case79,80. The presence of bacterial PGN is detected, and in response, the superficial ciliated fields of the juvenile light organ secrete mucus, which aids in the development of a V. fischeri biofilm outside the nascent light organ. Interestingly, V. fischeri dominates this biofilm over other environmental bacteria81. The nature of this dominance is unknown, but this aggregation serves as one of the first sites of specificity in this association. During the first days of the association, V. fischeri MAMPs, including LPS, PGN and PGN derivatives, play an important part in mediating specificity and initiating the onset of colonization36,82. For example, LPS and the PGN derivative trachaeal cytotoxin (TCT) work synergistically to induce normal development of the light organ by initiating apoptosis and regression of the juvenile ciliated fields within days of colonization by V. fischeri36 (FIG. 1b). Although MAMPS such as PGN, LPS and TCT are often implicated as virulence factors among pathogens, in this symbiotic relationship these MAMPs actually carry out a beneficial function by inducing proper tissue development in the host.
Several E. scolopes PRRs have been characterized, including a TLR, five PGRPs and three LPS-binding proteins (LBPs)44,70,76-78. The E. scolopes PGRPs (EsPGRPs) have cell- and tissue-specific expression patterns, which may confer a tissue-specific ability to respond to PGN and TCT depending on the MAMPs that are encountered in different microenvironments. For example, EsPGRP1 is expressed in the nuclei of host epithelial cells but is lost from late-stage apoptotic cells during V. fischeri-induced morphogenesis78. A V. fischeri mutant that is deficient in lytic transglycosylase activity (and is therefore defective for TCT release)83 prevents this loss of EsPGRP1 (REF. 78), suggesting that V. fischeri influences the expression of host PRRs. EsPGRP2 is secreted in host-derived mucus during colonization and is also localized in the light organ crypts after the onset of V. fischeri colonization76. EsPGRP2 has also been shown to have TCT-degrading amidase activity76 that might ameliorate the deleterious effect of this powerful toxin, which in pathogenic associations is cytotoxic to host mucosal and epithelial surfaces84,85. The ability to degrade TCT might be especially important during periods when symbiont concentrations, and presumably TCT concentrations, are high. The roles of EsPGRP3, EsPGRP4 and EsPGRP5 are currently unknown, but these proteins have been detected in haemocytes70 (B. Rader and S.V.N., unpublished observations). Although the three E. scolopes LBPs (EsLBPs) have yet to be characterized in detail, EsLBP1 has been detected in the crypt spaces, and its expression is upregulated in symbiotic hosts44,86.
Phagocytosis in the squid
Host haemocytes migrate from the vascular system through the epithelium of the light organ crypt to interact with the symbionts66,67,87, and recent work has focused on understanding whether these cells can differentiate between symbiotic and non-symbiotic bacteria18. Haemocytes have been shown to recognize and bind members of the family Vibrionaceae to different degrees, and V. fischeri has low binding activity18. When the symbionts are eliminated from the light organ of the adult squid (using antibiotics for a period of 5 days), haemocytes from these cured hosts bind significantly more V. fischeri cells than haemocytes obtained from untreated or fully colonized animals. The increase in adhesion is specific to V. fischeri cells, as binding of non-symbiotic bacteria remains unchanged. Thus, variation among related bacterial species in their ability to avoid adhesion to haemocytes, coupled with an altered haemocyte response to colonization, might contribute to the specificity of this symbiotic interaction. As the haemocytes migrate into the crypts and probably sample these microenvironments, the ability of V. fischeri to avoid adherence could contribute to long-term persistence of the organism at these sites. This change in the binding affinity of haemocytes is a striking example of symbiont-induced host tolerance. The mechanism (or mechanisms) by which V. fischeri mediates this immune tolerance in the host remains unknown, but a bacterial Omp might be involved. Deletion of the V. fischeri gene encoding OmpU leads to a significant increase in binding of the bacterium to squid haemocytes, suggesting that this protein helps V. fischeri to avoid phagocytosis18. Ongoing research is focusing on an analysis of the transcriptomes and proteomes of haemocytes from symbiotic and cured hosts, and a recent study has shown that EsPGRP5, nitric oxide synthase (NOS) and a squid orthologue of complement component C3 are all differentially expressed in these cells depending on the colonization state70.
These findings in the adult squid host raise questions about how the immune system of the juvenile host develops in response to colonization. In many animals, the immune system undergoes a maturation and development period in response to microbial and environmental challenges, and this often leads to tolerance or homeostasis43,88-90. Although haemocytes with internalized bacteria have been observed within the crypts of colonized juvenile hosts, no engulfed bacteria have been observed in the light organ haemocytes of adults, despite the fact that these cells are exposed to high concentrations of V. fischeri67. Another study also revealed that there is an infiltration of haemocytes in the superficial ciliated field of the nascent light organ within 2 hours of exposure to V. fischeri66. These observations suggest that the cellular immune response of the host is altered by the symbionts and might mature in response to the persistent colonization by V. fischeri. Although the development of the immune system in E. scolopes has not been characterized in detail, these studies suggest that symbiosis with V. fischeri is one factor that contributes to immune system maturation.
Chemical factors
Chemical components of the innate immune system have also been implicated in the E. scolopes–V. fischeri symbiosis, and several studies have shown that ROS and reactive nitrogen species (RNS) have an important role in this association91-93. E. scolopes produces squid halide peroxidase (sHPO), a protein similar to vertebrate myeloperoxidase. This enzyme converts H2O2 into hypohalous acids (for example, hypochlorous acid), which are microbiocidal compounds91. sHPO is expressed in tissues that directly contact bacteria, including the light organ, gills and the accessory nidamental gland (a female reproductive organ that harbours a consortium of bacteria)94. In the light organ, V. fischeri can presumably overcome this challenge owing to its expression of a periplasmic catalase that degrades H2O2 (REF. 95).
The enzyme NOS is necessary for the production of nitric oxide, which can react with oxygen or superoxide anions to generate antimicrobial RNS such as peroxynitirite and dinitrogen trioxide. Previous studies have shown that both NOS and nitric oxide are active during initiation of the E. scolopes–V. fischeri symbiosis92,96,97. Specifically, NOS and nitric oxide have been detected in host-derived vesicles found within the mucous secretions in which V. fischeri aggregates before entry into the light organ, and they have also been reported in the ciliated epithelium, ducts and antechambers (FIG. 1b). Successful colonization leads to an irreversible attenuation of the NOS and nitric oxide levels in the light organ, and LPS and TCT seem to be required to induce this response, suggesting that the symbiont and/or host can adjust the innate immune response to facilitate colonization92,97. V. fischeri also contains a haem nitric oxide–oxygen (H-NOX)-encoding gene, which probably acts as a nitric oxide sensor, along with a number of potential nitric oxide detoxification genes, including hmp (encoding flavohaemoprotein), norVW (encoding the anaerobic nitric oxide reductase flavorubredoxin (NorV) and the associated NorV oxidoreductase (NorW)) and the nrf operon (encoding cytochrome c nitrite reductase) (reviewed in REF. 96).
Other innate immune factors
Recent analyses of the E. scolopes–V. fischeri system have also identified a number of putative members of the complement cascade that might play a part in modulating symbiosis19,98. An orthologue of complement component C3, one of the crucial components of the cascade, has been localized to the apical surfaces of the crypt epithelium, although the role of this orthologue in the symbiosis has yet to be analysed in detail98. Quantitative PCR was carried out to compare expression of the C3-encoding gene in haemocytes from symbiotic and cured hosts, revealing that transcript levels in haemocytes from naive or cured squid are significantly higher than the levels in haemocytes isolated from a fully colonized light organ, suggesting that V. fischeri dampens the complement response in these cells70. Recent analyses of the light organ and haemocyte proteomes also found several thioester-containing proteins70,93. In invertebrates, these proteins have an important role in the innate immune response as members of a complement-like system or as protease inhibitors; future studies will focus on investigating the role of C3 and other members of the complement response in this symbiosis99-101.
The leech–bacteria symbiosis
Compared to vertebrates, some invertebrates house relatively simple microbial communities in their gut. Although in some cases this is probably due to the extremely alkaline conditions in the invertebrate gut13, other invertebrates lack such an obvious barrier to colonization. One example of these other invertebrates is the Hungarian medicinal leech, Hirudo verbana, in which the microbial community in the largest compartment of the digestive tract, the crop, is dominated by two bacterial species, Aeromonas veronii and a Rikenella-like bacterium54-56,102 (FIG. 1c). The digestive-tract symbionts are thought to provide the host with vitamin B, which is in low abundance in blood103. So how is this simple microbial community established and maintained, and what is the role of the leech immune system in these processes104?
Immunity in leeches is mediated by haemocytes (also known as amoebocytes) that circulate in the haemolymph and in the lumen of the digestive tract105. In addition, several AMPs have been identified in leech species, including theromacin, theromyzin, lumbricin and neuromacin. These peptides are found in large fat cells, neurons and microglia, but their presence in other regions of the leech body has not been excluded106. Expressed sequence tag (EST) data also indicate that proteins with some similarity to PRRs might also be present in Hirudo medicinalis and H. verbana107.
Phagocytosis in the leech
The crop is a storage compartment for the ingested blood meal and, analogous to the human colon, the site where water and osmolytes are absorbed105. In a single feeding, a leech can consume more than five times its body weight in blood. The ingested erythrocytes are concentrated by the leech absorbing water from the blood meal, and the removal of osmolytes makes the intraluminal fluid isosmotic with the leech haemolymph105. Interestingly, the complement system of ingested vertebrate blood remains active for one or two days and kills sensitive bacteria in the gut108. Although this is an important barrier to microbial colonization of the gut, it is only transitory and not sufficient to explain the low diversity of microorganisms.
Another mechanism that contributes to specificity in the leech is phagocytosis of sensitive bacteria by haemocytes that patrol the crop16,104. The importance of haemocytes was discovered by a signature-tagged-transposon mutagenesis screen, which obtained A. veronii mutants that had lost the ability to colonize the medicinal leech gut16,109. One of these mutants had a defective T3SS and was found to be phagocytosed by haemocytes, whereas the parent strain adhered to haemocytes but was not phagocytosed. These data suggest that the leech haemocytes did not actively differentiate between mutant and parental A. veronii cells but that the parental cells possess protective mechanisms that inhibit phagocytosis. When animals were colonized with a mixture of the T3SS mutant and the parent strain, the mutant was still cleared from the leech, indicating that the T3SS of the wild-type strain does not interfere with phagocytosis of other bacteria (such as the mutant strain). This lack of cross-protection for cells that are susceptible to phagocytosis (in this case, the T3SS mutant) is likely to be important for maintaining the specificity of the association. Taken together, these data suggest that specific features of the bacterial symbiont promote avoidance of the innate immune system in the digestive tract of the leech.
Because A. veronii is also a pathogen of mammals, it was possible to assess whether the same T3SS mutant could cause disease in mice, and to test for cytotoxicity: the T3SS mutant had reduced virulence in mice and was significantly less cytotoxic to mouse macrophages16. A. veronii possesses at least two type III effectors, AexT and AexU110,111. AexT had previously been identified in Aeromonas salmonicida, an important fish pathogen112,113, and AexU is a novel toxin that was discovered in an Aeromonas hydrophila diarrhoeal isolate, as well as in the leech symbiont A. veronii110,111. The role of the T3SS in causing cytotoxicity to mouse macrophages while protecting the bacterium against phagocytosis in the leech suggests that this system has two very different roles in these two animals.
Analysis of the genome of the second dominant leech symbiont, the Rikenella-like bacterium, did not reveal the presence of a T3SS, but these bacteria are either exclusively associated with the host mucus or present in microcolonies that are surrounded by a polysaccharide matrix, which could serve as a protective barrier against phagocytosis102. Thus, both of these H. verbana symbionts (A. veronii and the Rikenella-like bacterium) possess protective mechanisms that do not directly interfere with the activity of haemocytes. These data suggest that symbiosis in H. verbana is at least partially mediated by the ability of the symbionts to avoid phagocytosis, in contrast to symbiosis in E. scolopes, for which both symbiont avoidance of phagocytosis by haemocytes and the ability of haemocytes to discriminate between symbionts and non-symbionts might be important for specificity.
Chemical factors and pattern recognition
In vivo transcriptomes of the digestive-tract symbionts, in combination with analyses of A. veronii mutants, suggest that the bacteria encounter membrane stress inside the leech crop109,114. A potential source of this stress is AMPs; for example, lumbricin and neuromacin were found in the central nervous system of the closely related leech H. medicinalis106. In the distantly related leech Theromyzon tessulatum, AMPs were also detected in the leech intestinal epithelium115-117, raising the possibility that AMPs are active in the leech crop. How the expression of AMPs is regulated in leeches is currently unknown. Furthermore, 12 different transcripts that are predicted to encode PRRs have also been detected in EST libraries from H. medicinalis and H. verbana107. Some of the predicted PRRs are only detected in libraries constructed from either embryos or the nervous system, which suggests that the leech exhibits tissue-specific expression of PRRs, although functional studies have not yet been carried out.
Acyrthosiphon pisum–Buchnera aphidicola
The pea aphid, A. pisum, and its symbiotic bacterium, Buchnera aphidicola, have co-evolved to form an association that is both highly specific and essential for viability of both the host and the symbiont118. Aphids are dependent on B. aphidicola for essential amino acids, and the vertically transmitted bacterium is an obligate endosymbiont that has an extremely reduced genome. B. aphidicola resides within specific host cells (bacteriocytes) and obtains essential nutrients from the host119 (FIG. 1d). A number of studies have described the genes that are expressed in host bacteriocytes, and in addition to bacterial proteins involved in the delivery of essential amino acids and vitamins to insect hosts, A. pisum innate immunity factors have also been detected in these cells, suggesting that components of the innate immune system are important in interfacing with B. aphidicola120-122.
However, unlike other insects, the innate immune system of A. pisum lacks PGRPs and defensins, and has a non-functioning immune deficiency pathway (Imd pathway)123. It has been suggested that the symbiosis with B. aphidicola contributed to the evolution of reduced immune capabilities in the aphid host, and this hypothesis was recently examined122. Specifically, when D. melanogaster Schneider 2 cells were challenged with B. aphidicola, an increase in the expression of a PGRP as well as components of the Imd pathway was observed, and there was a subsequent production of AMPs, which led to the clearance of B. aphidicola within 2 days. These data suggest that B. aphidicola would face a serious threat from a fully functional immune system in aphids and/or that a reduced immune response facilitated the establishment of the B. aphidicola symbiosis.
One interesting question is whether, and how, hosts adapt differently to bacterial endosymbionts that have reduced genomes and are vertically transmitted (such as B. aphidicola), as opposed to bacterial endosymbionts that have large genomes and are environmentally transmitted, and whether obligate endosymbionts lead to reduced immune capabilities in other systems. Aphids are not, however, defenceless; for example, they produce lysozyme, and their haemolymph contains up to five different types of immune cell, at least two of which are capable of phagocytosing bacteria120,124,125. Pea aphids often harbour facultative secondary bacterial symbionts126 (Serratia symbiotica, ‘Candidatus Hamiltonella defensa’ or ‘Candidatus Regiella insecticola’), and aphids appear to have differing numbers of haemocytes depending on the type of secondary symbiont that they carry, suggesting that colonization by specific symbionts can influence host cellular immunity124. Haemocytes from aphids also seem to be capable of phagocytosing both B. aphidicola and secondary symbionts, but the role of these cells in mediating symbioses remains to be determined. Pea aphids possess components of the Toll and janus kinase–signal transducer and activator of transcription (JAK–STAT) signalling pathways, as well as prophenoloxidase, a key component of the insect melanization defence response, and nitric oxide synthase123. Secondary symbionts such as ‘Ca. Hamiltonella defensa’ and ‘Ca. Regiella insecticola’ also seem to be able to induce pea aphid resistance to parasitic wasps and fungi, respectively127,128. These observations, along with the finding that the reproductive parasites Wolbachia spp. can confer resistance to viruses in flies and mosquitoes129-131, suggest that insects can supplement their innate immune system by exploiting the defence mechanisms of their symbionts.
Integrating mechanisms towards symbiostasis
The examples described above show that, in addition to the classical mechanisms of responding to pathogens, invertebrates are capable of using the innate immune system to interact with symbionts in order to promote the establishment and maintenance of symbiotic associations. The use of invertebrate models gives researchers the opportunity to discover additional regulatory pathways that are likely to be ancient and might reveal novel immunological mechanisms that confer high microbial specificity (outside of the conventional antibody-based adaptive response attributed to vertebrates).
We propose three possible mechanisms by which hosts differentiate between symbionts and pathogens, and these mechanisms would not be mutually exclusive (FIG. 3). First, reciprocal signalling between the partners (mediated by MAMP–PRR interactions) could induce an altered immune response that would lead to host tolerance of the symbiont. Second, symbionts might be sequestered in specialized tissues, organs or cells, where the molecular dialogue leading to stasis could be regulated locally. Last, if the symbiont or other microorganism (such as an invasive pathogen) infiltrates other areas of the host and/or causes tissue damage, signalling events mediated through DAMPS and MAMPs could lead to heightened immune responses and clearance by the host. A lack of such damage signals could also promote maintenance of the symbiont. The outcome of one or all three of these processes would be a healthy and stable association referred to as symbiostasis132.
Figure 3. Model for the establishment of symbiostasis in host–microorganism associations.
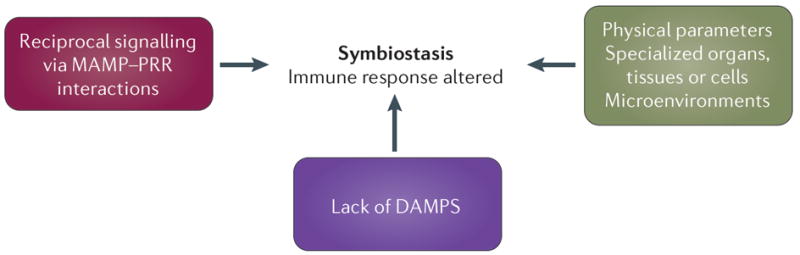
Multiple mechanisms can lead to the successful establishment and maintenance of beneficial symbioses. Here, we propose three possibilities. The first possible mechanism is reciprocal signalling between the partners, often mediated through microorganism-associated molecular pattern (MAMP)–pattern recognition receptor (PRR) interactions, which can lead to an altered immune response and host tolerance to the symbiont. The second possible mechanism is sequestration of the symbionts in specialized tissues, organs or cells, where the molecular dialogue leading to stasis can be regulated locally. The third potential mechanism is the avoidance of combined MAMP–damage-associated molecular pattern (DAMP) signalling, allowing the symbiont to persist in the host at the specialized site; if the symbiont or another microorganism (that is, an invasive pathogen) infiltrate other areas of the host and/or causes tissue damage, then signalling events mediated through DAMPS and/or MAMPs can lead to a heightened immune response and clearance by the host.
The first mechanism requires the sequential recognition of signals from both partners, for example, mediated by the interaction between MAMPs and PRRs. In the E. scolopes–V. fischeri symbiosis and in the classical symbiosis between leguminous plants and rhizobial bacteria, intricate reciprocal signalling is required to establish these associations19,52,133. Multiple signalling events allow colonization by the appropriate microorganisms and modify the immune defence responses accordingly. The pea aphid has apparently taken a different strategy by limiting the number of interactions between MAMPs and PRRs, in addition to having a reduced repertoire of signalling and immune effectors.
The second mechanism relies on tissue-specific conditions that select for particular microorganisms. Whereas some organs and tissues of metazoans are continuously exposed to microorganisms, others are normally sterile. Spatial or even temporal differences in the host immune response, physiology and/or anatomy can create zones of tolerance where the symbionts thrive. For example, in both the leech crop and the squid light organ, physical conditions exist that promote specificity of the symbionts and sequester them to specific tissues52,134. B. aphidicola and certain other intracellular symbionts are confined to bacteriocytes, which may limit symbiont exposure to the host immune system. Recent evidence from the weevil suggests that host AMPs are crucial for containing endosymbionts within bacteriocytes and that bacteria escape this niche when AMP pressure is removed135. Temporal differences can also occur during development, such that specific stages of the animal are conducive to colonization, whereas other stages are not. For example, the squid light organ briefly allows entry of non-symbiotic bacteria for 30 minutes after hatching52, although these interlopers are promptly removed by mechanisms that might involve the innate immune response. Changes in the composition of the gut micro-biota are also well described in a number of systems136,137. These microbial communities can vary widely depending on a number of parameters, including the age, health and diet of the host, and the presence or absence of antibiotics138. In the event of symbiont translocation from such specialized sites — for example, owing to tissue damage in a diseased host — the symbiont can become pathogenic at the new site and elicit an immune response139,140.
The third mechanism requires the avoidance of stimulating both MAMP- and DAMP-mediated signalling. The requirement for the integration of a MAMP and a DAMP to provoke an immune response was originally proposed by Matzinger141. An excellent example of this can be found in Hydra spp., in which the combination of two signals — bacterial flagellin and tissue damage — is required for the strongest induction of an immune response65. In D. melanogaster, low levels of bacterial PGN negatively regulate NF-κB signalling and subsequent AMP production, as well as ROS pathways (reviewed in REF. 142). During a systemic infection or following tissue damage, such downregulation is inhibited, allowing for the initiation of innate immunity effector mechanisms. In vertebrates, tissue damage is often followed by the activation of several signalling pathways that are induced by the release of cytokines. Although these cytokine-induced pathways are not as well characterized among the invertebrates, cytokine homologues have been described and probably cause similar responses in these hosts143. The requirement for tissue damage in order to trigger an immune response would presumably prevent symbionts (which do not damage the tissue) from stimulating the innate immune system, unless the interaction shifted from beneficial to pathogenic. The generality of this signal integration is currently unknown, and further mechanistic studies on animals from different lineages are required to establish whether it is a common pathway for differentiating between symbionts and non-symbionts.
Conclusions
Since the first ground-breaking phagocytosis experiments by Metchnikoff more than 100 years ago, it has been apparent that the innate immune system is crucial for the response to pathogens and for preventing disease. Here, we present findings from four model symbioses that highlight the importance of the invertebrate innate immune system in establishing and maintaining symbiotic bacteria. Key aspects of an ideal model system for studying symbioses and the immune system include the ability to culture the host and symbiont separately, the availability of genetic tools to manipulate each partner, the availability of genomic information for both partners and experimental tractability of the system144. Each of the systems described here is experimentally tractable and has some genomic information available, although currently only ESTs are available for E. scolopes and H. verbana; furthermore, most of the symbionts discussed, with the notable exception of B. aphidicola, have been cultured. Genetic manipulation of the host is limited to RNAi in Hydra spp., the pea aphid and the leech. Although none of these four models currently fulfils all the criteria of an ideal model system, the study of these models and other invertebrate systems nonetheless provides broad insight into symbiotic relationships and the range of immune interactions that shape such associations.
The combined evidence suggests that the innate immune system can distinguish between and respond differently to symbionts and pathogens, although the mechanisms behind this ability are not fully elucidated. This differential response might be regulated by several mechanisms (FIG. 3), including specific recognition (or lack thereof) of symbionts and MAMPS, and spatial and temporal factors that might prevent a general immune response from being triggered by MAMPs and/or DAMPs. Microbial recognition is at least partially mediated by PRRs that are conserved among metazoans, although the likely crucial role of downstream signal transduction pathways remains to be resolved, but a role for the ancient NF-κB pathway has been implied. Future research should focus on examining an even more diverse array of invertebrate systems for the identification of evolutionarily conserved mechanisms. The exciting explosion of omics techniques has made a plethora of new information available, but these tools often suggest new hypotheses on the basis of only the presence or absence of specific sequences, so these hypotheses will require experimental testing. In addition, bioinformatic analyses are challenging, especially when the identified sequences are not functionally characterized or when the characterized gene comes from a distantly related organism. Thus the use of omics in parallel with the more challenging functional studies is crucial to advance this exciting field. More effort should also be put into understanding how the immune system develops in these diverse invertebrate groups and whether interactions with the microbiota influence its maturation. Each model system has its own benefits and limitations, but only by applying a number of methods to a variety of associations will researchers reveal the breadth of interactions that exist in nature. We predict that these studies will lead to novel discoveries about the role of the innate immune system in fostering symbiotic relationships with microorganisms.
Acknowledgments
The authors thank the anonymous reviewers for helpful suggestions. Research in the S.V.N. laboratory was supported by the US National Science Foundation (NSF) (grant IOS-0958006) and the University of Connecticut Research Foundation. Research in the J.G. laboratory was supported by the NSF (Career Award MCB 0448052), the US National Institutes of Health (grant RO1 GM095390) and the University of Connecticut Research Foundation.
Glossary
- Symbiont
As used in this Review: a microorganism that forms a specific, stable and beneficial association with a particular host. Although this is now a commonly accepted definition, the original definition of symbionts, by Anton de Bary, included pathogenic, commensal and mutualistic symbionts
- Pattern recognition receptors
Eukaryotic proteins that bind to microorganism-associated molecular patterns, activating downstream signalling and, ultimately, innate immunity effector responses. Examples include Toll-like receptors, peptidoglycan recognition proteins and Gram-negative bacteria-binding proteins
- Peptidoglycan
The polymer that makes up the bacterial cell wall, and consists of β-(1-4)-linked N-acetylglucosamine and N-acetylmuramic acid linked to small peptide chains
- Lipopolysaccharide
A major component of the outer membrane of Gram-negative bacteria. It is composed of an O antigen, a core oligosaccharide and lipid A
- Outer-membrane proteins
Proteins that are common to the outer membrane of Gram-negative bacteria
- Flagellins
Bacterial major flagellar proteins
- Type III secretion system
A needle-like secretion system that allows Gram-negative bacteria to inject toxins directly into eukaryotic cells
- Microorganism-associated molecular patterns
Molecular motifs that are unique to microorganisms (both pathogens and non-pathogens) and are often recognized by components of the innate immune system
- Toll-like receptors (TLRs)
A diverse group of evolutionarily conserved pattern recognition receptors that are characterized by extracellular leucine-rich repeats and a cytoplasmic Toll–interleukin-1 receptor motif. Binding of microorganism-associated molecular patterns to TLRs often leads to downstream signalling via the nuclear factor-κB pathway or related pathways and results in the regulation of immune effectors
- Peptidoglycan recognition proteins
Eukaryotic proteins that can bind and sometimes degrade bacterial peptidoglycan, and are expressed in various locations within a cell
- Endoymbioses
As used in this Review: associations between hosts and intracellular symbionts (endosymbionts). These symbionts fall into two groups: obligate or primary symbionts, which are always found associated with the host, and secondary symbionts, which are not always present
- Complement system
A humoral component of the innate immune system; the complement pathway leads to the opsonization of microorganisms. There are three types of complement pathway: classical, alternative and lectin
- Nuclear factor-κB pathway
An evolutionarily conserved signalling cascade involving multiple protein complexes that control the transcription of immunity genes
- Damage-associated molecular pattern
A host-derived non-microbial factor that is released following tissue damage or necrosis and can activate the immune system in a similar way to microorganism-associated molecular patterns
- Toll–interleukin-1 receptor domain
A cytosolic domain that is common to a diverse group of receptors, including Toll and Toll-like receptors, and is found in proteins of the basal Hydra spp. discussed in this Review
- Expressed sequence tag (EST)
A cDNA sequence that has been obtained from a reverse-transcribed mRNA
- Immune deficiency pathway
An immune pathway that is found in most insects and responds to bacteria via peptidoglycan recognition proteins binding to diaminopimelic acid-containing peptidoglycan. Subsequent signalling pathways can lead to AMP production
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Contributor Information
Spencer V. Nyholm, Email: spencer.nyholm@uconn.edu.
Joerg Graf, Email: joerg.graf@uconn.edu.
References
- 1.Relman DA. ‘Til death do us part’: coming to terms with symbiotic relationships. Forward. Nature Rev Microbiol. 2008;6:721–724. doi: 10.1038/nrmicro1990. [DOI] [PubMed] [Google Scholar]
- 2.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 5.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silver AC, et al. Complex evolutionary history of the Aeromonas veronii group revealed by host interaction and DNA sequence data. PLoS ONE. 2011;6:e16751. doi: 10.1371/journal.pone.0016751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KH, Ruby EG. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol. 1994;176:1985–1991. doi: 10.1128/jb.176.7.1985-1991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regmi PR, Metzler-Zebeli BU, Ganzle MG, van Kempen TA, Zijlstra RT. Starch with high amylose content and low in vitro digestibility increases intestinal nutrient flow and microbial fermentation and selectively promotes bifidobacteria in pigs. J Nutr. 2011;141:1273–1280. doi: 10.3945/jn.111.140509. [DOI] [PubMed] [Google Scholar]
- 13.Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 15.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 16.Silver AC, et al. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci USA. 2007;104:9481–9486. doi: 10.1073/pnas.0700286104. This study shows that a bacterial T3SS is required for both symbiotic competence and virulence in different animal hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 18.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. This work finds that colonization of the squid light organ might contribute to immune tolerance of the symbiont V. fischeri. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes—Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraune S, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells JM, Loonen LM, Karczewski JM. The role of innate signaling in the homeostasis of tolerance and immunity in the intestine. Int J Med Microbiol. 2010;300:41–48. doi: 10.1016/j.ijmm.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nature Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 26.Watson FL, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uematsu S, Fujimoto K. The innate immune system in the intestine. Microbiol Immunol. 2010;54:645–657. doi: 10.1111/j.1348-0421.2010.00267.x. [DOI] [PubMed] [Google Scholar]
- 29.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nature Rev Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 32.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. This paper highlights the concept of PAMPs and the importance of pattern recognition in innate immunity. [DOI] [PubMed] [Google Scholar]
- 33.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 35.Dale C, Young SA, Haydon DT, Welburn SC. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci USA. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004M;306:1186–1188. doi: 10.1126/science.1102218. This article coins the term MAMP and shows that a bacterial toxin can be used as a signalling molecule to induce normal host development. [DOI] [PubMed] [Google Scholar]
- 37.Leulier F, Lemaitre B. Toll-like receptors -— taking an evolutionary approach. Nature Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 38.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 39.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 40.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2010;708:105–121. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 41.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss BL, Wang J, Aksoy S. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 2011;9:e1000619. doi: 10.1371/journal.pbio.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans. 2011;39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowling DK, Simmons LW. Reactive oxygen species as universal constraints in life-history evolution. Proc Biol Sci. 2009;276:1737–1745. doi: 10.1098/rspb.2008.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 47.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway – its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 48.Augustin R, Fraune S, Franzenburg S, Bosch TC. Where simplicity meets complexity: hydra, a model for host–microbe interactions. Adv Exp Med Biol. 2012;710:71–81. doi: 10.1007/978-1-4419-5638-5_8. [DOI] [PubMed] [Google Scholar]
- 49.Bosch TC. What hydra has to say about the role and origin of symbiotic interactions. Biol Bull. 2012;223:78–84. doi: 10.1086/BBLv223n1p78. [DOI] [PubMed] [Google Scholar]
- 50.Galliot B. Hydra, a fruitful model system for 270 years. Int J Dev Biol. 2012;56:411–423. doi: 10.1387/ijdb.120086bg. [DOI] [PubMed] [Google Scholar]
- 51.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 52.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid–Vibrio symbiosis. Nature Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 53.Rader BA, Nyholm SV. Host/microbe interactions revealed through “omics” in the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri. Biol Bull. 2012;223:103–111. doi: 10.1086/BBLv223n1p103. [DOI] [PubMed] [Google Scholar]
- 54.Nelson M, Graf J. Bacterial symbioses of the medicinal leech Hirudo verbana. Gut Microbes. 2012;3:322–331. doi: 10.4161/gmic.20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graf J. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect Immun. 1999;67:1–7. doi: 10.1128/iai.67.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worthen PL, Gode CJ, Graf J. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol. 2006;72:4775–4781. doi: 10.1128/AEM.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong CN, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol. 2011;13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 59.Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nature Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- 60.Unterman BM, Baumann P, McLean DL. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol. 1989;171:2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brinza L, et al. Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. C R Biol. 2009;332:1034–1049. doi: 10.1016/j.crvi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Bosch TC. Understanding complex host-microbe interactions in hydra. Gut Microbes. 2012;3:345–351. doi: 10.4161/gmic.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller DJ, et al. The innate immune repertoire in Cnidaria – ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. The results from this study suggest that Hydra spp. can actively determine the composition of their microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 66.Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 67.Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 68.Ford LA. Host defense mechanisms of cephalopods. Annu Rev Fish Dis. 1992;2:25–41. [Google Scholar]
- 69.Beuerlein K, Lohr S, Westermann B, Ruth P, Schipp R. Components of the cellular defense and detoxification system of the common cuttlefish Sepia officinalis (Mollusca, Cephalopoda) Tissue Cell. 2002;34:390–396. doi: 10.1016/s0040816602000708. [DOI] [PubMed] [Google Scholar]
- 70.Collins AJ, Schleicher TR, Rader BA, Nyholm SV. Understanding the role of host hemocytes in a squid/Vibrio symbiosis using transcriptomics and proteomics. Front Immunol. 2012;3:91. doi: 10.3389/fimmu.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 72.McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 73.Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid Euprymna scolopes Berry (Mollusca: Cephalopoda) Marine Biol. 2004;144:1151–1155. [Google Scholar]
- 74.Ruby EG, Lee KH. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee KH, Ruby EG. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl Environ Microbiol. 1992;58:942–947. doi: 10.1128/aem.58.3.942-947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Troll JV, et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodson MS, et al. Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Troll JV, et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal–bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 83.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2009;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goldman WE, Klapper DG, Baseman JB. Detection, isolation, and analysis of a released Bordetella pertussis product toxic to cultured tracheal cells. Infect Immun. 1982;36:782–794. doi: 10.1128/iai.36.2.782-794.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melly MA, McGee ZA, Rosenthal RS. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J Infect Dis. 1984;149:378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- 86.Chun CK, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci USA. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heath-Heckman EA, McFall-Ngai MJ. The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 2011;114:191–198. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 89.Mathis D, Benoist C. Microbiota and autoimmune disease: the hosted self. Cell Host Microbe. 2011;10:297–301. doi: 10.1016/j.chom.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 90.Wlodarska M, Finlay BB. Host immune response to antibiotic perturbation of the microbiota. Mucosal Immunol. 2010;3:100–103. doi: 10.1038/mi.2009.135. [DOI] [PubMed] [Google Scholar]
- 91.Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna–Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 93.Schleicher TR, Nyholm SV. Characterizing the host and symbiont proteomes in the association between the bobtail squid Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS ONE. 2011;6:e25649. doi: 10.1371/journal.pone.0025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- 95.Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Ruby EG. The roles of NO in microbial symbioses. Cell Microbiol. 2011;13:518–526. doi: 10.1111/j.1462-5822.2011.01576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. Attenuation of host NO production by MAMPs potentiates development of the host in the squid–vibrio symbiosis. Cell Microbiol. 2011;13:527–537. doi: 10.1111/j.1462-5822.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Castillo MG, Goodson MS, McFall-Ngai M. Identification and molecular characterization of a complement C3 molecule in a lophotrochozoan, the Hawaiian bobtail squid Euprymna scolopes. Dev Comp Immunol. 2009;33:69–76. doi: 10.1016/j.dci.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fujito NT, Sugimoto S, Nonaka M. Evolution of thioester-containing proteins revealed by cloning and characterization of their genes from a cnidarian sea anemone, Haliplanella lineate. Dev Comp Immunol. 2010;34:775–784. doi: 10.1016/j.dci.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 100.Zhang H, et al. Molecular cloning and characterization of a thioester-containing protein from Zhikong scallop Chlamys farreri. Mol Immunol. 2007;44:3492–3500. doi: 10.1016/j.molimm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 101.Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40:903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 102.Kikuchi Y, Graf J. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl Environ Microbiol. 2007;73:1984–1991. doi: 10.1128/AEM.01833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Graf J. The effect of symbionts on the physiology of Hirudo medicinalis, the medicinal leech. Invertebr Reprod Dev. 2002;41:269–275. [Google Scholar]
- 104.Silver AC, Graf J. Innate and procured immunity inside the digestive tract of the medicinal leech. Invertebrate Surv J. 2011;8:173–178. [PMC free article] [PubMed] [Google Scholar]
- 105.Sawyer RT. Leech Biology and Behavior. Clarendon Press; 1986. [Google Scholar]
- 106.Tasiemski A. Antimicrobial peptides in annelids. Invertebrate Surv J. 2008;5:75–82. [Google Scholar]
- 107.Macagno ER, et al. Construction of a medicinal leech transcriptome database and its application to the identification of leech homologs of neural and innate immune genes. BMC Genomics. 2010;11:407. doi: 10.1186/1471-2164-11-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Indergand S, Graf J. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl Environ Microbiol. 2000;66:4735–4741. doi: 10.1128/aem.66.11.4735-4741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Silver AC, Rabinowitz N, Küffer S, Graf J. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J Bacteriol. 2007;189:6763–6722. doi: 10.1128/JB.00685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sha J, et al. Further characterization of a type III secretion system (T3SS) and of a new effector protein 3 from a clinical isolate of Aeromonas hydrophila — Part I. Microbiol Pathog. 2007;43:127–146. doi: 10.1016/j.micpath.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 111.Silver AC, Graf J. Prevalence of genes encoding the type three secretion system and the effectors AexT and AexU in the Aeromonas veronii group. DNA Cell Biol. 2009;28:383–388. doi: 10.1089/dna.2009.0867. [DOI] [PubMed] [Google Scholar]
- 112.Burr SE, Stuber K, Frey J. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J Bacteriol. 2003;185:6583–6591. doi: 10.1128/JB.185.22.6583-6591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burr SE, Wahli T, Segner H, Pugovkin D, Frey J. Association of Type III secretion genes with virulence of Aeromonas salmonicida subsp. salmonicida. Dis Aquat Organ. 2003;57:167–171. doi: 10.3354/dao057167. [DOI] [PubMed] [Google Scholar]
- 114.Bomar LGJ. Investigation into the physiologies of Aeromonas veronii in vitro and inside the digestive tract of the medicinal leech using RNA-seq. Biol Bull. 2012;223:155–166. doi: 10.1086/BBLv223n1p155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schikorski D, et al. Microbial challenge promotes the regenerative process of the injured central nervous system of the medicinal leech by inducing the synthesis of antimicrobial peptides in neurons and microglia. J Immunol. 2008;181:1083–1095. doi: 10.4049/jimmunol.181.2.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tasiemski A, et al. Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J Biol Chem. 2004;279:30973–30982. doi: 10.1074/jbc.M312156200. [DOI] [PubMed] [Google Scholar]
- 117.Tasiemski A, et al. Proenkephalin A-derived peptides in invertebrate innate immune processes. Brain Res Mol Brain Res. 2000;76:237–252. doi: 10.1016/s0169-328x(00)00005-x. [DOI] [PubMed] [Google Scholar]
- 118.Baumann P, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 119.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:RESEARCH0054. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Braendle C, et al. Developmental origin and evolution of bacteriocytes in the aphi–Buchnera symbiosis. PLoS Biol. 2003;1:e21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Douglas AE, Bouvaine S, Russell RR. How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci. 2011;278:333–338. doi: 10.1098/rspb.2010.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gerardo NM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. This investigation shows that the pea aphid has a reduced innate immunity repertoire compared to other insects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmitz A, et al. The cellular immune response of the pea aphid to foreign intrusion and symbiotic challenge. PLoS ONE. 2012;7:e42114. doi: 10.1371/journal.pone.0042114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Laughton AM, Garcia JR, Altincicek B, Strand MR, Gerardo NM. Characterisation of immune responses in the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 2011;57:830–839. doi: 10.1016/j.jinsphys.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 126.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci USA. 2005;102:16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 129.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6:e1000833. doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reid RGB. Biological Emergences: Evolution by Natural Experiment. MIT Press; 2007. [Google Scholar]
- 133.Murray JD. Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- 134.Graf J, Kikuchi Y, Rio RV. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 2006;14:365–371. doi: 10.1016/j.tim.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 135.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 136.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 137.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 139.Mason KL, et al. From commensal to pathogen: translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. MBio. 2011;2:e00065–e00011. doi: 10.1128/mBio.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Daskin JH, Alford RA. Context-dependent symbioses and their potential roles in wildlife diseases. Proc Biol Sci. 2012;279:1457–1465. doi: 10.1098/rspb.2011.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. This paper highlights the danger model and the importance of DAMPs in the immune response. [DOI] [PubMed] [Google Scholar]
- 142.Ryu JH, Ha EM, Lee WJ. Innate immunity and gut–microbe mutualism in Drosophila. Dev Comp Immunol. 2010;34:369–376. doi: 10.1016/j.dci.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 143.Ottaviani E, Malagoli D, Franceschi C. Common evolutionary origin of the immune and neuroendocrine systems: from morphological and functional evidence to in silico approaches. Trends Immunol. 2007;28:497–502. doi: 10.1016/j.it.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 144.Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nature Rev Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Tomaso AW, et al. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]


