SUMMARY
Phenotypic variation is a prerequisite for evolution by natural selection, yet the processes that give rise to the novel morphologies upon which selection acts are poorly understood. We employed a chemical genetic screen to identify developmental changes capable of generating ecologically relevant morphological variation as observed among extant species. Specifically, we assayed for exogenously applied small molecules capable of transforming the ancestral larval foregut of the herbivorous Xenopus laevis to resemble the derived larval foregut of the carnivorous Lepidobatrachus laevis. Appropriately, the small molecules that demonstrate this capacity modulate conserved morphogenetic pathways involved in gut development, including downregulation of retinoic acid (RA) signaling. Identical manipulation of RA signaling in a species that is more closely related to Lepidobatrachus, Ceratophrys cranwelli, yielded even more similar transformations, corroborating the relevance of RA signaling variation in interspecific morphological change. Finally, we were able to recover the ancestral gut phenotype in Lepidobatrachus by performing a reverse chemical manipulation to upregulate RA signaling, providing strong evidence that modifications to this specific pathway promoted the emergence of a lineage-specific phenotypic novelty. Interestingly, our screen also revealed pathways that have not yet been implicated in early gut morphogenesis, such as thyroid hormone signaling. In general, the chemical genetic screen may be a valuable tool for identifying developmental mechanisms that underlie ecologically and evolutionarily relevant phenotypic variation.
Introduction
The emergence of new phenotypic variation is generally assumed to arise from novel mutations (or equivalent environmental perturbations; West-Eberhard 2003; Moczek et al. 2011) or the recombination of standing genetic variation. Yet even with genomic sequence information from hundreds of species, we understand little about the origins of morphological variants that ultimately result in adaptive diversification. To understand the relationship between genetic variants and phenotypic innovation, we must first understand how endogenous and environmental signals are modified in quantity, time or space to produce novel phenotypic variation that can be integrated with preexisting developmental programs (Gilbert 2003). This is currently a formidable task to achieve using non-model systems, and evolutionary innovations are rare among model systems (Kopp 2011).
Several elegant studies have identified developmental underpinnings of novel trait variation by perturbing pathways known to influence the morphogenesis of those traits (Kopp et al. 2000), or of similar traits, in model organisms (Abzhanov et al. 2004; Moczek et al. 2007). These studies disrupted the function of relevant genes through, for instance, RNAi or viral-induced gene silencing, to mimic the form of morphological variation observed in extant species. One limitation of such candidate approaches is that they may preclude the discovery of novel or alternate genetic or epigenetic variants that might also give rise to the same phenotype. Although forward approaches such as quantitative trait locus (QTL) analysis (e.g., Shapiro et al. 2004) or classical mutagenesis screens have the potential to expand a repertoire of candidate pathways underlying novel traits, they are often expensive and time-consuming, and few non-model systems meet the criteria necessary for such analyses. Recently, screening embryos against a library of chemicals has provided an alternative and powerful approach to revealing the role of specific signaling pathways in development (Wheeler and Brandli 2009). Although typically used to produce phenotypes that mimic developmental abnormalities, we propose that chemical genetic screens can also be used to produce phenotypes that mimic naturally occurring interspecific variation.
Chemical genetic screens work by targeting, in vivo, various modulators of the conserved signaling pathways that control metazoan development. This is achieved by the action of cell permeable “small molecule” reagents (i.e., low molecular weight organic compounds) that inhibit or activate specific pathway components. Small molecules thus act as “mutational equivalents,” inducing loss- or gain-of-function phenotypes that often mimic genetic mutants (Wheeler and Brändli 2009; Stockwell 2000). This approach has at least two key advantages when used in an evolutionary context. First, because small molecules are cell permeant and bind to the conserved functional domains of their target proteins, they are largely species-independent and thus can be employed in non-model organisms. Second, embryos can be exposed to small molecules during specific developmental windows, thus avoiding pleiotropic and lethal effects that are commonly encountered in traditional genetic screens. In effect, small molecule exposure might be able to mimic a regulatory change that arose during evolution and resulted in altered gene expression during a particular stage of development; such regulatory changes are seen as an important source of evolutionary novelty (reviewed in Wray 2007; Carroll 2008; Kirschner and Gerhart 2011).
In this study we focus on identifying evolutionary changes in development that may have resulted in adaptive variation in gut phenotypes among anuran larvae. Digestive physiology and morphology is a critical aspect of any organism’s ecological niche (Karasov and Diamond 1988), and evolutionary diversification often goes hand in hand with a change in food resources (Schluter 2000; Grant and Grant 2008). In particular, an individual’s nutritional intake is influenced by the configuration and length of its digestive tract. Generally, carnivores have a capacious, proteolytic stomach and short intestine, while herbivores and omnivores have a long intestine and a stomach that is enzymatically simple (Stevens and Hume 1995). This range of dietary variation and corresponding gut phenotypes can be observed among larvae of the South American anuran clade, Ceratophryinae (e.g., Chacophrys, Ceratophrys and Lepidobatrachus). Like most anuran larvae, Chacophrys larvae are mainly herbivorous (Duellman and Trueb 1986; Rossa-Feres et al. 2004; Wild 1999). In contrast, Ceratophrys larvae are microphagous carnivores that process their tadpole or invertebrate prey orally before swallowing it, whereas Lepidobatrachus larvae are megalophagous carnivores that consume their animal prey whole (Ruibal and Thomas 1988). Both Ceratophrys and Lepidobatrachus have derived stomach and intestine morphologies to accommodate their unique diets (Fig. 1H and I; Fabrezi 2011), although the molecular and developmental origins of these adaptive traits remain elusive.
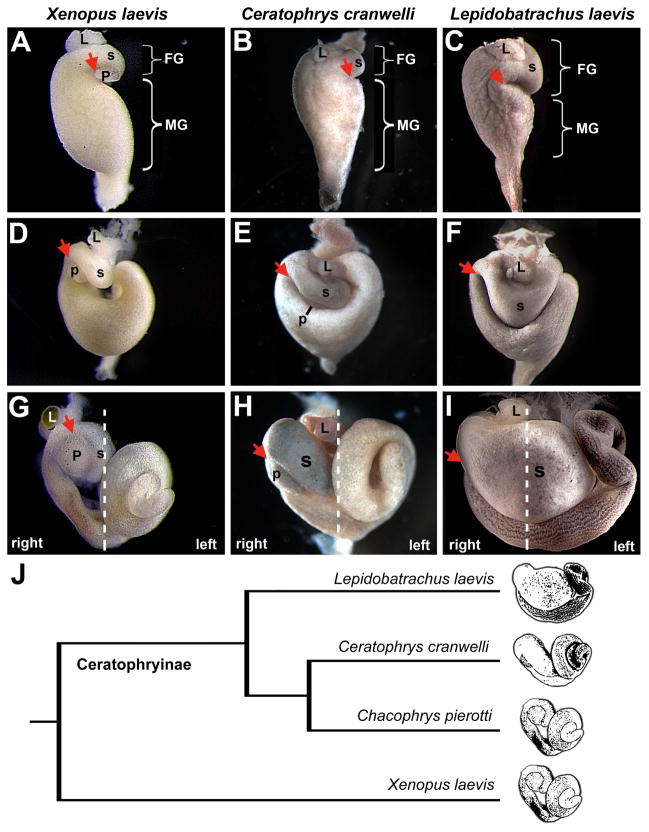
Fig. 1.
Gut development in omnivorous and carnivorous anuran larvae. Ventral views of the developing gut of an omnivorous tadpole (Xenopus laevis) at Nieuwkoop and Faber (NF) stages 41 (A), 43 (D) and 46 (G) are compared to the developing guts of carnivorous Ceratophrys cranwellii and Lepidobatrachus laevis tadpoles at comparable Gosner stages (GS) 21 (B and C), 23 (E and F) and 25 (H and I). In Xenopus (A) the GD loop (arrow) is located in a proximal position along the length of the gut tube, the foregut (FG) is small relative to the midgut (MG), and the pancreas is located within the GD concavity. The GD loop is similarly positioned in Ceratophrys (B), although the pancreas is not visible early in development. In Lepidobatrachus the GD loop forms more distally, which leaves the portion of the gut tube proximal to the GD loop of more equal proportion to the prospective midgut (C). The relative positions of the developing stomach (s), liver (L) and pancreas (p) are indicated, where visible. (The pancreas remains dorsal in Lepidobatrachus and is not visible in these ventral views.) Dashed lines in G, H, and I indicate the approximate position of the embryonic midline and the left and right sides of each embryo. Images are not to scale. The cladogram (J) illustrates the relationships among Xenopus and three ceratophryine genera, including Ceratophrys and Lepidobatrachus.
To address the mechanisms underlying the development and evolution of the novel gut phenotype found specifically in Lepidobatrachus, we used a chemical genetic screen to identify developmental changes that are capable of modifying ancestral gut morphogenesis to yield derived gut morphologies. This screen identified modulators of pathways that have been previously implicated in gut development (thereby providing validation for the approach), as well as unexpected pathways. Our results suggest that evolutionary modifications to these developmental pathways might have led to ecologically and evolutionarily important phenotypic variation among anuran larvae. Chemical genetic screening may be a valuable tool for uncovering developmental mechanisms that generate selectable morphological variation.
Materials and Methods
Characterizing Gut Morphogenesis
Xenopus laevis embryos were obtained by in vitro fertilization using established procedures and reared at 16°C in 0.1X MMR (Sive et al 1998). The developmental stages of Xenopus embryos were determined using the morphological criteria of Nieuwkoop and Faber (NF; 1994). Lepidobatrachus laevis embryos were obtained through natural matings of adults that were collected in Salta, Argentina, and maintained in a colony at Harvard University. To induce spawning, male and female adults were injected with luteinizing hormone-releasing hormone (LHRH; Sigma-Aldrich, St. Louis, MO, USA) at a dosage of 0.1 mg per kg body weight. Embryos were reared at 22°C in 10% Holtfreter solution (Holtefreter 1931). Ceratophrys cranwelli embryos were obtained from K. Thomas (The Frog Ranch, Granite Bay, CA), and reared at 28°C in 10% Holtfreter solution. The developmental stages of Lepidobatrachus and Ceratophrys embryos were determined using the morphological criteria of Gosner (GS; 1960). For all species, the morphogenesis of the stomach, gastroduodenal loop (an anatomical feature that occurs between the foregut and midgut; Fig. 1A), intestine, liver and pancreas was characterized at end stage (approximately NF46 in Xenopus, GS23-25 in Ceratophrys and Lepidobatrachus).
Chemical Genetic Screen
To identify molecular signaling changes that might underlie the differential morphogenesis of the carnivorous gut, we screened for small molecules capable of transforming the ancestral tadpole foregut to the derived morphology found in Lepidobatrachus. Xenopus laevis was chosen as the representative model of the ancestral character state because Xenopus larvae possess an ancestral-type digestive tract common to most extant tadpoles (including tadpoles of Chacophrys pierottii, an omnivorous species that is closely related to Ceratophrys and Lepidobatrachus (Maxson and Ruibal 1998; Fabrezi and Quinzio 2008). It is therefore likely that, on the molecular level, Xenopus possess a developmental program similar to that possessed by the ceratophryine ancestor (i.e., of Chacophrys, Ceratophrys and Lepidobatrachus). Approximately 200 small molecules were applied to Xenopus embryos from late neurula stages (NF18-22) through the end of gut morphogenesis (NF46). This panel included 143 custom-synthesized novel compounds (Dush et al., 2011) and 59 commercially available reagents that target pathways known to be involved in morphogenesis (Table S1). The concentrations of each compound were based on published IC50 values in tissue culture or aquatic embryo assays; novel compounds were assayed at 100 μM. At least 4 embryos were assayed per compound and any compounds that yielded carnivore-like gut morphologies were repeated with embryos from different clutches.
Modulation of Retinoic Acid Signaling
All-trans retinoic acid (RA; a diffusible cell signaling molecule) and diethylaminobenzaldehyde (DEAB; an RA signaling antagonist) were prepared in ethanol, and Ro-41-5253 (an RA signaling antagonist) was prepared in DMSO. Stock solutions were diluted to working concentrations in either 0.1X MMR (X. laevis) or 10% Holtfreter (L. laevis and C. cranwelli) media. In each experiment, sibling controls were cultured in an appropriate dilution of ethanol or DMSO to control for any effects of the solvent.
Groups of ten or more Xenopus laevis embryos (stage NF22) were each cultured in 10 ml of DEAB (0.4 mM, 0.5 mM, 0.6 mM or 0.7 mM) or Ro-41-5253 (1 μM or 1.5 μM) solution at 16°C. After 24 h, embryos were washed five times in 0.1X MMR and maintained in 0.1X MMR until stage NF45/46 to evaluate gut phenotypes.
Groups of five to seven C. cranwellii embryos were cultured in 30 ml of DEAB solution (0.4 mM, 0.5 mM, 0.6mM or 0.7mM); Ceratophrys and Lepidobatrachus embryos and tadpoles are significantly larger than Xenopus and require larger culture volumes) at stages GS16, GS17 or GS18. At stage GS19, embryos were washed five times in 10% Holtfreter and maintained in 10% Holtfreter until stage GS23-GS25 (equivalent to NF44-46; McDiarmid and Altig 1999) to evaluate gut phenotypes.
Groups of ten Lepidobatrachus embryos at stages GS16, GS17 or GS18 were each cultured in 100 ml of RA solution (0.05 μM, 0.1 μM, 0.5 μM or 1 μM) at 22°C. At stage GS19, embryos were washed five times in 10% Holtfreter then maintained in 10% Holtfreter until stage GS23-GS25 (equivalent to NF44-46; McDiarmid and Altig 1999) to evaluate gut phenotypes.
In Situ Hybridization
Some embryos of all species were fixed in MEMFA (Sive et al., 1998) at various stages and processed for in situ hybridization. Pitx2 and Nkx2.5 were detected in X. laevis and L. laevis using X. laevis riboprobes as previously described (Lipscomb et al 2006; Muller et al 2003; Smith et al 2000) with the following modifications for large embryos: rehydration washes were carried out for 10 min each, additional washes were performed before and after refixation, and a 1/3000 dilution of alkaline phosphatase-conjugated, anti-digoxygenin anti-FAB fragments was used. The X. laevis Pitx2 riboprobe has the potential to recognize all three Pitx2 isoforms, but only isoform c is expressed asymmetrically in the left lateral plate mesoderm of vertebrate embryos (Schweickert et al. 2000). Pitx2c was detected in Ceratophrys using a Ceratophrys Pitx2c riboprobe. To create the Pitx2c probe, Pitx2c was amplified from complementary DNA synthesized from total RNA collected from stage GS19 Ceratophrys using degenerate primers. The amplified fragment was cloned into the pCR2-TOPO cloning vector (Invitrogen), transfected into chemically competent cells, cultured, purified, and used for riboprobe synthesis.
Results
Interspecific Variation in Anuran Larval Gut Morphogenesis
In this study, the Xenopus larval digestive system developed as previously described (Fig. 1A, D, G; Chalmers and Slack 1998, 2000). As in other vertebrates, the digestive organs arise from a primitive gut tube that undergoes looping and rotation events to shape the final anatomical configuration. For example, the gastroduodenal loop begins as a left side concavity that rotates rightward as the foregut elongates, ultimately fixing the stomach, liver and pancreas on the embryo’s right side, with the pancreas closely apposed to the concavity of the loop (arrows, Fig. 1A, D, G; Muller at al. 2003). As in most anuras, larval X. laevis form a rudimentary stomach (Griffiths 1961) that does not develop a pyloric sphincter or produce pepsinogen, a proteolytic enzyme (Ishizuya-Oka et al. 1998). Simultaneously with foregut morphogenesis, the midgut elongates extensively (up to nine times the snout-to-vent length of the larva), forming several concentric intestinal coils that occupy the left side of the visceral cavity (Fig. 1G; Chalmers and Slack 1998). This long intestine facilitates nutrient absorption and bacterial fermentation (Pryor and Bjorndal 2005).
Gut morphogenesis proceeds differently in the carnivorous larvae of Ceratophrys and Lepidobatrachus. As observed in X. laevis, the Ceratophrys embryonic foregut exhibits a left side concavity, that eventually loops rightward (Fig. 1B, E, H). However, in contrast to the anteroposteriorly oriented foregut of Xenopus, Ceratophrys’ foregut remains oriented transversely before moving rightward and becomes substantially larger than Xenopus. Further, the appearance of the dorsal pancreatic rudiment in the concavity of the loop is delayed in Ceratophrys relative to Xenopus (cf. Fig. 1D and E), although the organ ultimately forms and resides in the same position (Fig. 1G and H).
The Lepidobatrachus embryonic foregut exhibits an even more extreme phenotype. Most notably, the left side concavity forms more posteriorly along the gut tube (Fig. 1C). Consequently, a larger segment of the gut tube is situated anterior to the loop, the stomach becomes fixed in a transverse orientation and the liver remains closer to the midline (Fig 1I). As in Ceratophrys, the stomach is enlarged; however, the expansion is dramatic in Lepidobatrachus, filling most of the anterior visceral cavity (Fig 1I). Although the dorsal pancreas eventually becomes visible in the concavity of the Ceratophrys GD loop, the pancreas is greatly reduced in Lepidobatrachus (a small group of insulin-positive cells was detected dorsal to the stomach; data not shown) and invisible in ventral view as it is displaced from the concavity of the GD loop. Finally, midgut elongation in Lepidobatrachus is uniquely attenuated, and the intestine remains practically uncoiled (Fig. 1I; Fabrezi 2011).
Modulating Specific Signaling Pathways Produces Interspecific Variation in X. laevis
In a chemical genetic screen of approximately 200 small molecules applied to Xenopus embryos, five structurally distinct compounds resulted in the formation of a more carnivore-like foregut that resembles that of Lepidobatrachus: cyclopamine (Fig S1), diethylaminobenzaldehyde (DEAB; Fig. 2), latrunculin (Fig S1), Ro-41-5253 (Fig. 2), and triiodothyronine (T3; Fig S1). Cyclopamine is an alkaloid that inhibits signaling via hedgehogs, a highly conserved family of secreted morphogens that regulates key features of development and patterning (Ryan and Chiang 2012). DEAB inhibits retinaldehyde dehydrogenase, which catalyzes the conversion of retinal, a Vitamin A derivative, to retinoic acid (RA), a diffusible signaling molecule that plays essential roles in cell growth, differentiation and patterning during development (Blomhoff and Blomhoff, 2006). Latrunculin is a compound that inhibits the polymerization of actin, a process essential for cytoskeletal dynamics during cell migration (Ridley 2011). Ro-41-5253 is a soluble antagonist of a retinoic acid receptor, RARα (Keidel et al. 1994), which binds RA and transduces the RA signal to the nucleus to control gene expression. Finally, T3 is the metabolically active form of thyroid hormone, which controls numerous physiological processes, including anuran metamorphosis (Laudet 2011).
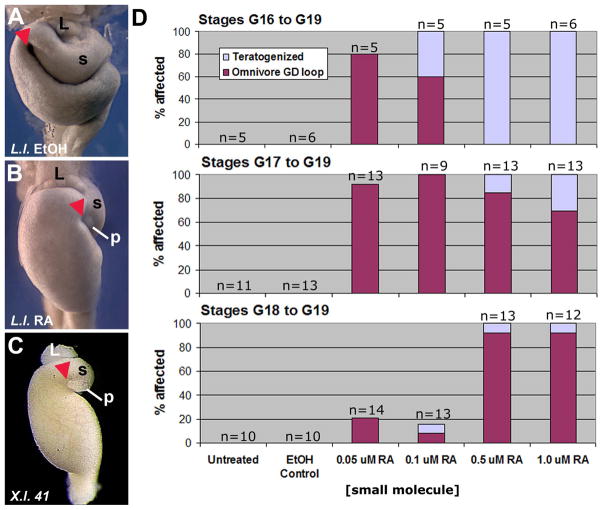
Fig. 2.
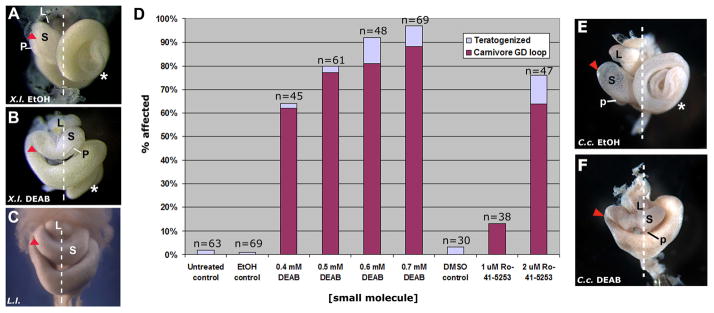
Treatment of anuran embryos with a retinoic acid synthesis inhibitor results in the formation of a more derived/carnivore-like GD loop morphology. Xenopus laevis embryos were subjected to an acute chemical treatment with solvent control (ethanol, EtOH; A) or an RA synthesis inhibitor, DEAB (0.4 mM; B). The Xenopus GD loop (arrowhead; NF46) shifts posteriorly upon exposure to DEAB (B) and the final foregut anatomy appears similar to the normal morphology of Lepidobatrachus laevis (C; G 23; Although Xenopus NF46 is most equivalent to GS25, the relative anatomical topology of the foregut organs is already established by GS23 and is more easily visualized at this stage, i.e., before stomach expansion.). D: Effects on gut morphogenesis after treatment with small molecule inhibitors of retinoic acid synthesis (DEAB) or signaling (Ro-41-5253) are concentration dependent. The percentage of embryos with the derived/carnivore-like GD loop and organ placement (NF46) is indicated for different concentrations of each molecule. Embryos that exhibit severely disrupted development (e.g., massive edema, tail curvature) or abnormal, uninterpretable phenotypes not resembling either species are classified as “teratogenized.” Results are pooled from 5 different experiments. DMSO was used as the solvent control for Ro-41-5253. Ceratophrys cranwellii embryos were subjected to an acute chemical treatment with solvent control (EtOH; E) or an RA synthesis inhibitor, DEAB (0.5 mM; F). As observed in Xenopus, the Ceratophrys GD loop (arrowhead; GS25) shifts posteriorly upon exposure to DEAB (F) and the intestine (*) does not elongate, phenotypes remarkably similar to the morphological features found in Lepidobatrachus (C; GS23). The relative positions of the developing stomach (s), liver (L) and pancreas (p) are indicated, where visible.
In this study, we chose to investigate more closely the effects of DEAB and Ro-41-4253 since they both target different components of RA signaling, thus corroborating the potential evolutionary relevance of this pathway for the novel trait of interest. In embryos exposed to DEAB and Ro-41-5253, the gastroduodenal loop is displaced posteriorly with respect to the stomach, liver and pancreas, a phenotype that is strikingly reminiscent of what is found in the Lepidobatrachus embryo (Fig. 2A–C). Moreover, the stomach is oriented transversely and the liver and pancreas become fixed more medially, rather than adopting their normal right-sided orientations (cf. Fig. 2A, B). Finally, the pancreas is often reduced and positioned more dorsally and medially; i.e., it is displaced from the concavity of the GD loop (cf. Fig. 2A, B). Among the treated X. laevis individuals there was some variation in the degree of this displacement, but any displacement always co-occurred with a transverse orientation of the stomach, a condition not observed in untreated X. laevis. This carnivore-like phenotypic profile is induced by small molecules in a concentration-dependent manner (Fig. 2D).
We reasoned that the same signaling modulations would not only elicit similar morphological changes in Ceratophrys, but that they could actually produce a more accurate form of the carnivore gut morphology in these animals, since Ceratophrys might possess any requisite changes in developmental programming that may have occurred, evolutionarily, after Xenopus and ceratophryine ancestors diverged. Indeed, we observe the same profile of foregut changes in Ceratophrys embryos exposed to DEAB, including the transversely oriented stomach, medial accessory organ orientation, and reduced pancreas (Fig. 2E, F), but, additionally, the attenuated midgut elongation of Lepidobatrachus is also induced by DEAB in Ceratophrys (cf Fig. 2C, F).
Increased Retinoic Acid Signaling Produces Interspecific Variation in L. laevis
The above results show that multiple components of the RA pathway produce similar phenotypic variation when perturbed, implicating decreased RA availability or RARα-mediated activity in the development of the altered gastroduodenal loop found in Lepidobatrachus laevis. If decreased RA signaling underlies the development of a carnivore foregut, then increasing RA signaling in Lepidobatrachus embryos might restore the foregut to a more herbivorous, i.e., ancestral, state. To test this hypothesis, we exposed Lepidobatrachus embryos to exogenous all-trans RA, an agonist for RARα. As predicted, RA-treated Lepidobatrachus tadpoles exhibit several anatomical features that are reminiscent of an herbivorous larval foregut, including a more anterior gastroduodenal loop, with the liver oriented slightly posterior to the loop (Fig. 3A, B). In addition, the pancreas, which normally is hidden dorsally, became visible in ventral view, closely apposed to the concavity of the gastroduodenal loop, as in most herbivores (cf. Fig. 3B, C). These effects were induced at high frequency over multiple doses and stages (Fig. 3D).
Fig. 3.
Treatment of Lepidobatrachus laevis with ectopic RA results in a more ancestral/omnivore-like GD loop morphology. Lepidobatrachus embryos were exposed to solvent control (ethanol, EtOH; A) or RA (B). RA-treated Lepidobatrachus embryos (B; GS23) exhibit a lack of midgut elongation, a known teratogenic effect of RA exposure in vertebrates. In this context, the GD loop (arrowhead) shifts anteriorly upon exposure to ectopic RA (B) and foregut morphology appears remarkably similar to that of Xenopus laevis at a comparable degree of midgut elongation (C; NF41, reproduced from Fig. 1A). D: Effects on gut morphogenesis after treatment with ectopic RA at successively later developmental stages are both concentration- and stage-dependent. Results are pooled from two different breedings. “Teratogenized” classification is as in Fig. 2. The relative positions of the developing stomach (s), liver (L) and pancreas (p) are indicated, where visible.
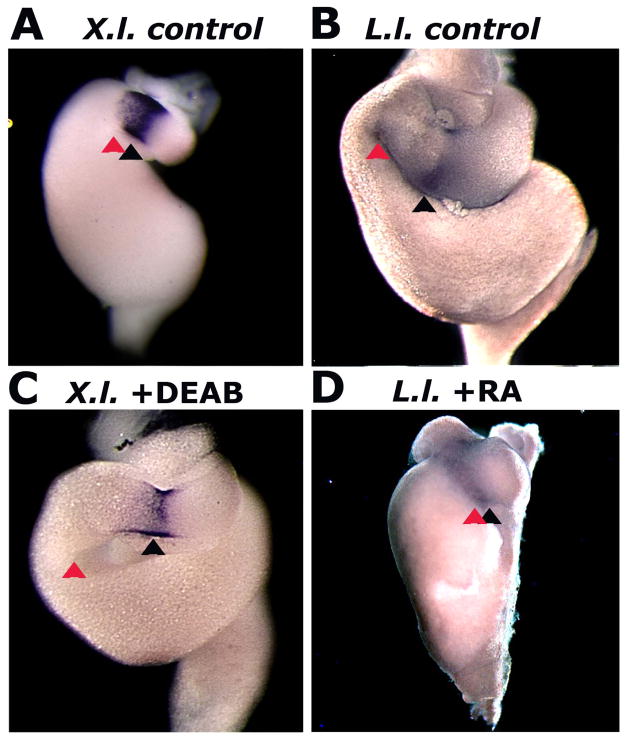
Molecular markers further indicate the authenticity of these small-molecule induced phenocopies. In Xenopus, as in other vertebrates, gastroduodenal looping occurs adjacent to the expression domain of Nkx2.5, a conserved marker for the boundary between the stomach and duodenum (Fig. 4A; Smith et al. 2000). In Lepidobatrachus embryos and DEAB-treated Xenopus embryos, Nkx-2.5 is still expressed at the stomach-duodenal boundary, but the loop forms more posteriorly relative to the Nxk-2.5 expression domain (cf. Fig. 4A–C). In contrast, in RA-treated Lepidobatrachus embryos, as in Xenopus, the loop forms adjacent to the expression of Nkx2.5 (Fig. 4D). Variation in RA signaling levels can generate evolutionarily relevant variation in the topography of the foregut loop.
Fig. 4.
Chemically modulating RA signaling in anuran embryos shifts the location of the GD loop along the anterioposterior axis of the gut tube. The stomach-duodenal boundary in each species is indicated by expression of the homeobox transcription factor Nkx2.5. The GD loop (red arrowhead) is located adjacent to the Nkx2.5 expression domain (black arrowhead) in Xenopus controls (A; NF42), but it is shifted posteriorly following DEAB treatment (C; NF45). Conversely, the GD loop is located posterior to Nkx2.5 expression in Lepidobatrachus controls (B; GS23), but it is shifted anteriorly following RA treatment (D, GS23).
Chemically Modulating RA Signaling Shifts Pitx2 Expression
To investigate the possibility that left-right asymmetric development has diverged between species possessing herbivore and carnivore gut phenotypes, we compared the embryonic expression of the gene Pitx2. Pitx2 is a homeobox transcription factor that is expressed exclusively on the left side in vertebrate embryos, where it serves as a key effector in the formation of left-right asymmetries in internal organs (Ryan et al. 1998; Campione et al. 1999). RA is known to influence Pitx2 expression patterns during development (Matt et al. 2008; Chazaud et al. 1999; Matt et al. 2005). Moreover, in Xenopus embryos, asymmetric Pitx2 expression correlates with the segment of the prospective gut tube that will form the concavity of the gastroduodenal loop (Fig. 5A; Muller et al. 2003).
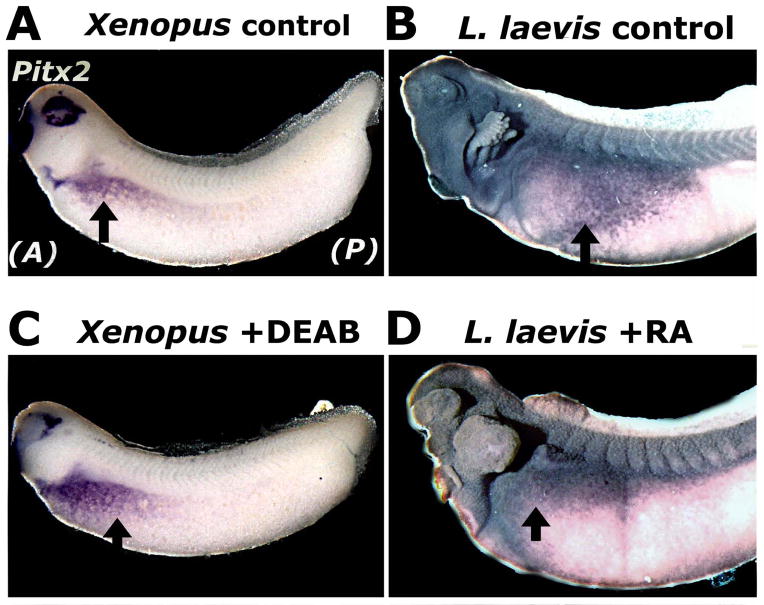
Fig. 5.
Chemically modulating RA synthesis and signaling shifts the expression of Pitx2, a left-side determinant of asymmetric gut looping. Compared to the domain of Pitx2 expression (purple) revealed by in situ hybridization in Xenopus embryos (A), the Pitx2 domain is shifted posteriorly and ventrally in the Lepidobatrachus embryo (B). C–D: Pitx2 expression is shifted posteriorly and ventrally in Xenopus embryos exposed to DEAB (C), and anteriorly and dorsally in Lepidobatrachus embryos exposed to RA (D). Arrows in A and C indicate the posterior limit of Pitx2 expression; those in B and D indicate the anterior limit.
As in Xenopus and other vertebrates, we found that Pitx2 is expressed exclusively on the left side of Ceratophrys and Lepidobatrachus embryos (Fig. S2) at a stage immediately preceding gut morphogenesis (equivalent stages NF32 and GS20 in Xenopus and Ceratophryines, respectively; McDiarmid and Altig 1999); however, in Lepidobatrachus the expression domain is positioned considerably more posteriorly and extends farther ventrally (cf Fig. S2C with A, B and Fig. 5B with A). According to published fate maps of amphibian embryos, this region is fated to give rise to a more posterior region of the gut tube, consistent with the more posterior origin of the carnivorous gut loop in Lepidobatrachus (Muller et al. 2003; Tahara and Nakamura 1961; Chalmers and Slack 2000).
To determine whether a shift in Pitx2 expression also accompanies the shifted gut looping observed in Xenopus treated with small molecule RA inhibitors, Pitx2 expression was evaluated in the context of DEAB. Exposure of Xenopus embryos to DEAB resulted in a posterior and ventral expansion of the Pitx2 expression domain (Fig. 5C). This new domain correlates with a more posterior position along the gut tube and is consistent with the more posterior gastroduodenal loop induced by inhibition of RA signaling. Conversely, treatment of Lepidobatrachus embryos with RA resulted in an anterior and dorsal shift in the expression domain of Pitx2 (Fig. 5D). This new domain is fated to give rise to a more anterior position along the gut tube, consistent with the more anterior gastroduodenal loop observed in RA-treated Lepidobatrachus tadpoles.
Discussion
Selection on phenotypic variation may yield novel adaptive traits, but little is known about the proximate developmental mechanisms by which the requisite phenotypic diversity arises. In this study, we performed a small molecule screen to interrogate these mechanisms, reasoning that small molecule inhibitors or activators could alter development in an extant species with the ancestral character state and “reproduce” the morphogenetic context that generated a novel phenotype in a derived lineage. Our results implicate specific and possibly interdependent developmental pathways in the generation of novel gut morphologies.
The molecules identified by our screen have the potential to modify developmental processes as diverse as actin polymerization (via latrunculin; Yarmola et al. 2000), cell differentiation (via T3) and the many morphogenetic processes controlled by hedgehog signaling (via cyclopamine). In particular, our screen identified two structurally distinct compounds, DEAB and Ro-41-5253, that inhibit different steps in retinoic acid (RA) signaling, thus strongly implicating this pathway in the generation of the novel larval gut morphologies possessed by Lepidobatrachus. Treating Xenopus embryos with these molecules promotes the development of a foregut phenotype that is remarkably similar to that of Lepidobatrachus, including the exaggerated gastroduodenal loop, transversely oriented foregut, medially positioned liver and reduced, dorsally situated pancreas. Importantly, this transformation is even more accurate when the embryos of Ceratophrys – a lineage more closely related to Lepidobatrachus – are treated with DEAB. Along with the expected conversions of foregut morphology obtained with Xenopus, the same treatment in Ceratophrys prevents the midgut from extending and coiling, a phenotypic profile that is, overall, remarkably similar to that possessed by Lepidobatrachus.
Molecular evidence from this study corroborates our hypothesis that altered RA signaling may have promoted the morphological evolution of the larval foregut that accompanied the transition to larval carnivory in Lepidobatrachus. An RA-inhibitory small molecule treatment (DEAB) that produces a carnivore-like gut in Xenopus also produces a heterotopic shift in Pitx2 expression which mimics the normal pattern of Pitx2 found in Lepidobatrachus. The more posterior domain of Pitx2 found in Lepidobatrachus and DEAB-treated Xenopus is correlated with a more posterior position of the gastroduodenal loop. Conversely, increasing RA signaling in Lepidobatrachus embryos results in an anteriorly restricted Pitx2 domain, which is similar to that observed in unmanipulated Xenopus. Since RA receptor complexes have been shown to bind to cis-regulatory regions of the Pitx2 gene (Kumar and Duester, 2010), these heterotopic shifts, together with evidence that Pitx2 regulates the looping topography of the vertebrate gut tube (e.g., Muller et al. 2003; Logan et al. 1998; Latacha et al. 2005), suggest that simple RA-mediated shifts in asymmetric gene expression might have facilitated the emergence of a novel gut phenotype without compromising basic digestive function.
In addition to RA, our chemical genetic screen also implicated increased thyroid hormone (TH) signaling in the development of the carnivore gut morphology. TH mediates the formation of adult morphology during amphibian metamorphosis (Ishizuya-Oka et al. 1998; Schreiber et al. 2005), and many craniofacial and digestive features that represent metamorphic changes in most frogs develop precociously in larval Lepidobatrachus (Hanken 1992; Fabrezi and Quinzio 2008). For example, early Lepidobatrachus tadpoles have adult-like, pepsinogen-producing stomach glands (Carroll et al. 1991) and pancreas hypoplasia (a prerequisite to metamorphic remodeling; Mukhi et al. 2008), suggesting that embryos or larvae have higher TH levels, increased TH receptor (TR) availability or increased TR sensitivity.
It is plausible that the pathways identified by our screen may interact to produce the derived carnivore phenotype. Both TRs and RARs (Retinoic Acid Receptors) require RXR (Retinoid X Receptor) as a heterodimerizing partner in order to promote transcription of target genes (Rowe 1997), and elevated TR expression decreases RA responsiveness in anuran embryos (Banker and Eisenman 1993). Increased TH activity in Lepidobatrachus could diminish the available pool of RXR heterodimerizing partners and thereby reduce RA signaling. Furthermore, since hedgehog signaling is controlled by TH in the metamorphosing anuran digestive tract (Ishizuya-Oka et al. 2001; Hasebe et al. 2008), increased TH activity may upregulate hedgehog signaling, which antagonizes RA activity during gut morphogenesis (Tehrani and Lin 2011). Future studies are needed to determine whether any of these pathways were targeted by natural selection during the evolution of the carnivore phenotype, and how they might interact.
The pathways identified in this study exemplify the benefits of using a chemical genetic screen as a tool for discovering developmental routes to evolutionary change. The fact that RA signaling was repeatedly the target of small molecules capable of transforming the ancestral, herbivorous gut phenotype into the derived, carnivorous gut phenotype provides confirmation that the chemical genetic approach works. RA signaling has been implicated in both gut and pancreas development (Lipscomb et al. 2006; Pearl et al. 2009), and we might expect molecules affecting this pathway to be identified by our screen. Likewise, cyclopamine disrupts hedgehog signaling, which plays numerous roles in foregut and pancreas morphogenesis (Tsukui et al. 1999); and actin polymerization, which is inhibited by latrunculin, has been implicated in organ looping (Itasaki et al. 1989; Itasaki et al. 1991; Ramasubramanian et al. 2006). Yet, the fact that thyroid hormone also produced the carnivore phenotype reveals the ability of a chemical genetic screen to identify potential novel pathways. While thyroid hormone is known to be involved with gut remodeling later during metamorphosis (Hasebe et al. 2008; Ishizuya-Oka et al. 2001; Stolow and Shi 1995), it has not yet been implicated in early gut development (Wheeler and Liu 2012). Thus, a chemical genetic screen has the ability to both corroborate existing candidate pathways and mechanisms and suggest new ones.
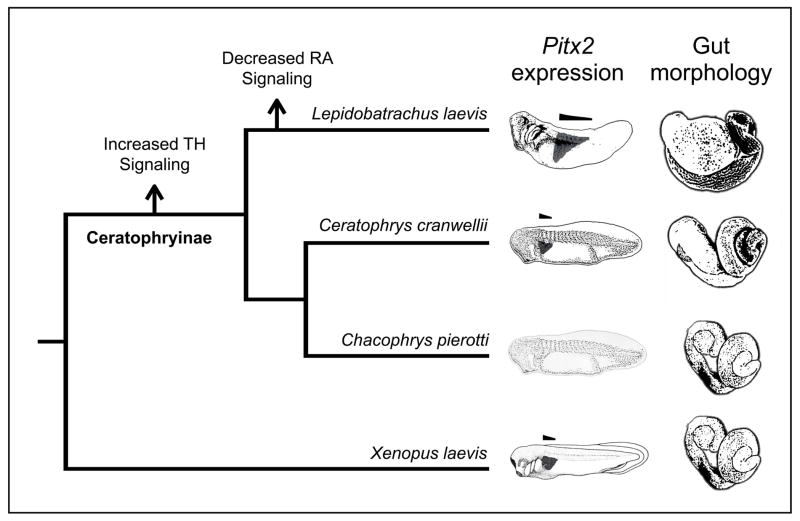
Traditional or chemical genetic screens are a springboard for asking evolutionary questions, but fully addressing a hypothesis regarding the origins of a novel trait requires ancillary lines of evidence. First, a screen may identify equivalent mechanisms for producing a particular phenotype, but one or more might actually underlie phenotypic variation in natural populations (True and Haag 2001). Thus, while forward approaches are capable of phenocopying novel traits, demonstrating that specific developmental pathways are relevant to the derived lineage in question (e.g., by performing a reciprocal manipulation in the derived species as in Fig. 3) will considerably strengthen an evolutionary hypothesis. Second, if several pathways are implicated in the emergence of a novel trait, one must consider whether evolutionary changes in these pathways occurred simultaneously or sequentially. Future investigations will further illuminate the evolutionary sequence in which increased TH or decreased RA signaling arose in ceratophryine lineages. The fact that all ceratophryine lineages including Chacophrys (the herbivorous outgroup) exhibit accelerated larval development (a process that is typically TH dependent; Fabrezi 2011), and that Ceratophrys respond to inhibited RA synthesis (via DEAB) by developing the more derived, carnivore phenotype, suggests that increased TH signaling arose in the common ancestor of all three ceratophryine lineages but decreased RA signaling arose only in Lepidobatrachus (Fig. 6).
Fig. 6.
An evolutionary hypothesis regarding the sequence in which increased thyroid hormone (TH) or decreased retinoic acid (RA) signaling arose in ceratophryine lineages. The domain of Pitx2 expression (depicted with dark shading on the embryo diagrams) is anteriorly restricted in Xenopus and Ceratophrys, but begins and extends more posteriorly in Lepidobatrachus (as highlighted by the horizontal bars over each diagram). These domains are correlated with the position of the gastroduodenal loop, which is positioned more dorsally in Lepidobatrachus. Lepidobatrachus possesses the most extreme larval carnivore morphology, with an enlarged and transversely oriented stomach and a severely reduced, dorsally positioned pancreas. Lepidobatrachus and Ceratophrys share several thyroid hormone dependent traits, such as rapid development and precocious pepsinogen production (see Discussion; data not shown), which suggests that increased TH signaling occurred in the ancestors of all ceratophryine lineages. In contrast, only Lepidobatrachus larvae possess a posteriorally shifted Pitx2 domain (a pattern that can be reproduced in Xenopus using an RA synthesis inhibitor), and Ceratophrys respond to inhibited RA synthesis by developing the more derived, carnivore phenotype, which suggests that decreased RA signaling occurred only in Lepidobatrachus. The domain of Pitx2 expression is unknown for Chacophrys.
In our study, a panel of small molecules generated phenotypes that mimic interspecific morphological variation. Applying chemical genetic screens to embryos of other amenable model and non-model species with limited genetic tools or molecular resources may reveal the role of specific regulatory networks and their target genes in the development of new adaptive forms, providing a novel route to uncovering general principles of morphological evolution.
Supplementary Material
Xenopus embryos form a more derived/carnivore-like GD loop morphology upon exposure to small molecules. Xenopus laevis embryos were subjected to an acute chemical treatment with cyclopamine (A; a hedgehog signaling inhibitor; 10–40 μM; p<0.001), latrunculin (B; an actin polymerization inhibitor; 60–100 μM; p<0.01) or triiodothyronine (C; thyroid hormone; 0.25–1 mM; p<0.05). The percentage of embryos with the derived/carnivore-like GD loop and organ placement (NF46) is indicated for each molecule. Embryos that exhibit severely disrupted development (e.g., massive edema, tail curvature) or abnormal, uninterpretable phenotypes not resembling either species are classified as “teratogenized.” Controls for cyclopamine and latrunculin included DMSO (solvent).
The expression domain of Pitx2 in anuran embryos. In situ hybridization patterns (purple stain) indicate the expression of Pitx2 mRNA in the left lateral plate mesoderm of A) Xenopus laevis (NF 34), B) Ceratophrys cranwellii (GS 21), and C) Lepidobatrachus laevis (GS 21) embryos. Arrows delimit expression domains, which reveal the heterotopic shift in Lepidobatrachus. Expression was not detected in the right side (not shown).
Acknowledgments
This work was supported by the U.S. National Science Foundation (IOB0642012 to NN-Y, and EF-0334846—AmphibiaTree—to JH), a Putnam Expeditionary Grant from the Harvard University Museum of Comparative Zoology (to CI), a North Carolina State University NIH Molecular Biotechnology Training Grant Fellowship (to SB), and an NSF Postdoctoral Fellowship (1003035; to CL-R).
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin C. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;3:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Banker DE, Eisenman RN. Thyroid hormone receptor can modulate retinoic acid-mediated axis formation in frog embryogenesis. Mol Cell Biol. 1993;13:7540–7552. doi: 10.1128/mcb.13.12.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Carroll EJ, Seneviratne AM, Ruibal R. Gatric pepsin in an anuran larva. Dev Growth Differ. 1991;33:499–507. doi: 10.1111/j.1440-169X.1991.00499.x. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Slack JM. Development of the gut in Xenopus laevis. Dev Dyn. 1998;212:509–521. doi: 10.1002/(SICI)1097-0177(199808)212:4<509::AID-AJA4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Slack JM. The Xenopus tadpole gut: fate maps and morphogenetic movements. Development. 2000;127:381–392. doi: 10.1242/dev.127.2.381. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Chambon P, Dolle P. Retinoic acid is required in the mouse embryo for left-right asymmetry determination and heart morphogenesis. Development. 1999;126:2589–2596. doi: 10.1242/dev.126.12.2589. [DOI] [PubMed] [Google Scholar]
- Duellman E, Trueb L. Biology of amphibians. John Hopkins University Press; Baltimore: 1986. [Google Scholar]
- Dush MK, McIver AL, Parr MA, Young DD, Fisher J, Newman DR, Sannes PL, Hauck ML, Deiters A, Nascone-Yoder N. Heterotaxin: a TGF-β signaling inhibitor identified in a multi-phenotype profiling screen in Xenopus embryos. Chem Biol. 2011;18:252–63. doi: 10.1016/j.chembiol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrezi M, Quinzio S. Morphological evolution in Ceratophryinae frogs (Anura, Neobatrachia): the effects of heterochronic changes during larval development and metamorphosis. Zool J Linn Soc. 2008;154:752–780. [Google Scholar]
- Fabrezi M. Heterochrony in growth and development in anurans from the Chaco of South America. Evol Biol. 2011;38:390–411. [Google Scholar]
- Faivovich J, Carrizo GR. Descripción de la larva de Chacophrys pierottii (Vellard 1948) (Leptodactylidae, Ceratophryinae) Alytes. 1992;10:81–89. [Google Scholar]
- Gilbert SF. The morphogenesis of evolutionary developmental biology. Int J Dev Biol. 2003;46:467–477. [PubMed] [Google Scholar]
- Gosner KL. A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica. 1960;16:183–190. [Google Scholar]
- Grant PR, Grant BR. How and why species multiply: The radiation of Darwin’s finches. Princeton University Press; Princeton: 2008. [Google Scholar]
- Griffiths I. The form and function of the fore-gut in anuran larvae (Amphibia, Salientia) with particular reference to the manicotto glandulare. Proc Zool Soc Lond. 1961;137:249–283. [Google Scholar]
- Hanken J. Life history and morphological evolution. J Evol Biol. 1992;5:549–557. [Google Scholar]
- Hasebe T, Kajita M, Shi YB, Ishizuya-Oka A. Thyroid hormone-up-regulated hedgehog interacting protein is involved in larval-to-adult intestinal remodeling by regulating sonic hedgehog signaling pathway in Xenopus laevis. Dev Dyn. 2008;237:3006–3015. doi: 10.1002/dvdy.21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtfreter J. Über die Aufsucht isolierter Teile des Amphibienkeimes. II. Züchten von Keimen und Keimteilen in Salzlösung. Roux’ Arch Bd. 1929;124:404–466. doi: 10.1007/BF00652482. [DOI] [PubMed] [Google Scholar]
- Ishizuya-Oka A, Ueda S, Inokuchi T, Amano T, Damjanovski S, Stolow M, Shi YB. Thyroid hormone-induced expression of sonic hedgehog correlates with adult epithelial development during remodeling of the Xenopus stomach and intestine. Differentiation. 2001;69:27–37. doi: 10.1046/j.1432-0436.2001.690103.x. [DOI] [PubMed] [Google Scholar]
- Ishizuya-Oka A, Inokuchi T, Ueda S. Thyroid hormone-induced apotosis of larval cells and differentiation of pepsinogen-producing cells in the stomach of Xenopus laevis in vitro. Differentiation. 1998;63:59–68. doi: 10.1046/j.1432-0436.1998.6320059.x. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Nakamura H, Sumida H, Yasuda M. Actin bundles on the right side in the caudal part of the heart tube play a role in dextro-looping in the embryonic chick heart. Anat Embryol. 1991;183:29–39. doi: 10.1007/BF00185832. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Nakamura H, Yasuda M. Changes in the arrangement of actin bundles during heart looping in the chick embryo. Anat Embryol. 1989;180:413–420. doi: 10.1007/BF00305115. [DOI] [PubMed] [Google Scholar]
- Keidel S, LeMotte P, Apfel C. Different agonist- and antagonist-induced conformational changes in retinoic acid receptors analyzed by protease mapping. Mol Cell Biol. 1994;14:287–298. doi: 10.1128/mcb.14.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov WH, Diamond JM. Interplay between physiology and ecology in digestion. BioScience. 1988;38:602–611. [Google Scholar]
- Kirschner MW, Gerhart JC. Facilitated variation. In: Pigliucci M, Müller GB, editors. Evolution: the extended synthesis. MIT Press; Cambrige, MA: 2010. pp. 253–80. [Google Scholar]
- Kopp A, Duncan I, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kopp A. Drosophila sex combs as a model of evolutionary innovations. Evol Dev. 2011;13:504–522. doi: 10.1111/j.1525-142X.2011.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latacha KS, Rémond MC, Ramasubramanian A, Chen AY, Elson EL, Taber AL. Role of actin polymerization in bending of the early heart tube. Dev Dyn. 2005;233:1272–1286. doi: 10.1002/dvdy.20488. [DOI] [PubMed] [Google Scholar]
- Laudet V. The origins and evolution of vertebrate metamorphosis. Curr Biol. 2011;21:R726–37. doi: 10.1016/j.cub.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Lipscomb K, Sablyak AR, Schmitt CE, Yoder JA, Nascone-Yoder N. A role for retinoid signaling in left-right asymmetric digestive organ morphogenesis. Dev Dyn. 2006;235:2266–2275. doi: 10.1002/dvdy.20879. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagán-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Matt N, Dupé V, Garnier JM, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- Matt N, Ghyselinck NB, Pellerin I, Dupé V. Impairing retinoic acid signalling in the neural crest cells is sufficient to alter entire eye morphogenesis. Dev Biol. 2008;320:140–148. doi: 10.1016/j.ydbio.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Maxson RL, Ruibal R. Relationships of frogs in the leptodactylid subfamily Ceratophryinae. J of Herpetol. 1998;22:228–231. [Google Scholar]
- Moczek AP. Emerging model systems in evo-devo: horned beetles and the origins of diversity. Evol Dev. 2007;9:323–328. doi: 10.1111/j.1525-142X.2007.00168.x. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Sultan S, Foster S, Ledón-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig D. The role of developmental plasticity in evolutionary innovation. Proc Roy Soc B. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhi S, Mao J, Brown DD. Remodeling the exocrine pancreas at metamorphosis in Xenopus laevis. PNAS. 2008;105:8962–8967. doi: 10.1073/pnas.0803569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Prather D, Nascone-Yoder NM. Left-right asymmetric morphogenesis in the Xenopus digestive system. Dev Dyn. 2003;228:672–682. doi: 10.1002/dvdy.10415. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) Garland Publishing Inc; New York: 1994. [Google Scholar]
- Pearl EJ, Bilogan CK, Mukhi S, Brown DD, Horb ME. Xenopus pancreas development. Dev Dyn. 2009;238:1271–1286. doi: 10.1002/dvdy.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor GS, Bjorndal KA. Symbiotic fermentation, digesta passage, and gastrointestinal morphology in bullfrog tadpoles (Rana catesibiana) Physiol Biochem Zool. 2005;78:201–215. doi: 10.1086/427050. [DOI] [PubMed] [Google Scholar]
- Ramasubramanian A, Latacha KS, Benjamin JM, Voronov DA, Ravi A, Taber LA. Computational model for early cardiac looping. Ann Biomed Eng. 2006;34:1655–1669. doi: 10.1007/s10439-005-9021-4. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–22. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Rossa-Feres DC, Jim J, Fonseca MG. Diets of tadpoles from a temporary pond in southeastern Brazil (Amphibia, Anura) Revista Brasileira de Zoologia. 2004;21:745–754. [Google Scholar]
- Rowe A. Retinoid X receptors. Int J Biochem Cell Biol. 1997;29:275–278. doi: 10.1016/s1357-2725(96)00101-x. [DOI] [PubMed] [Google Scholar]
- Ruibal R, Thomas E. The obligate carnivorous larvae of the frog Lepidobatrachus laevis (Leptodactylidae) Copeia. 1988;3:591–604. [Google Scholar]
- Ryan AK, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. J Biol Chem. 2012;287:17905–13. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford University Press; Oxford: 2000. [Google Scholar]
- Schreiber AM, Cai L, Brown DD. Remodeling of the intestine during metamorphosis of Xenopus laevis. PNAS. 2005;102:3720–3725. doi: 10.1073/pnas.0409868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A, Campione M, Steinbeisser H, Blum M. Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mech Dev. 2000;90:41–51. doi: 10.1016/s0925-4773(99)00227-0. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks M, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis. Cold Spring Harbor Laboratory Press; New York: 1998. [Google Scholar]
- Smith DM, Grasty RC, Theodosiou NA, Tabin CJ, Nascone-Yoder NM. Evolutionary relationships between the amphibian, avian, and mammalian stomachs. Evol Dev. 2000;2:348–359. doi: 10.1046/j.1525-142x.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- Stevens CE, Hume ID. Comparative Physiology of the Vertebrate Digestive System. Cambridge University Press; Cambridge: 1995. [Google Scholar]
- Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet. 2000;1:116–25. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolow MA, Shi YB. Xenopus sonic hedgehog as a potential morphogen during embryogenesis and thyroid hormone-dependent metamorphosis. Nucleic Acids Res. 1995;23:2555–2562. doi: 10.1093/nar/23.13.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Nakamura O. Topography of the presumptive rudiments in the endoderm of the anuran neurula. J Embryol Exp Morphol. 1961;9:138–158. [PubMed] [Google Scholar]
- Tehrani Z, Lin S. Endocrine pancreas development in zebrafish. Cell Cycle. 2011;10:3466–3472. doi: 10.4161/cc.10.20.17764. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Tsukui T, et al. Multiple left-right asymetry defects in Shh(−/−) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. PNAS. 1999;96:11376–11381. doi: 10.1073/pnas.96.20.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford University Press; New York: 2003. [Google Scholar]
- Wheeler GN, Brandli AW. Simple vertebrate models for chemical genetics and drug discovery screens: lessons from zebrafish and Xenopus. Dev Dyn. 2009;238:1287–1308. doi: 10.1002/dvdy.21967. [DOI] [PubMed] [Google Scholar]
- Wheeler GN, Liu KJ. Xenopus: An ideal system for chemical genetics. Genesis. 2012;50:207–218. doi: 10.1002/dvg.22009. [DOI] [PubMed] [Google Scholar]
- Wilder ER. Description of the chondrocranium and osteogenesis of the Chacoan burrowing frog, Chacophrys pierotti (Anura: Leptodactylidae) J Morphol. 1999;242:229–46. doi: 10.1002/(SICI)1097-4687(199912)242:3<229::AID-JMOR3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yarmola EG, Somasundaram T, Boring TA, Spector I, Bubb MR. Actin-latrunculin A Structure and Function. J Biol Chem. 2000;275:28120–28127. doi: 10.1074/jbc.M004253200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Xenopus embryos form a more derived/carnivore-like GD loop morphology upon exposure to small molecules. Xenopus laevis embryos were subjected to an acute chemical treatment with cyclopamine (A; a hedgehog signaling inhibitor; 10–40 μM; p<0.001), latrunculin (B; an actin polymerization inhibitor; 60–100 μM; p<0.01) or triiodothyronine (C; thyroid hormone; 0.25–1 mM; p<0.05). The percentage of embryos with the derived/carnivore-like GD loop and organ placement (NF46) is indicated for each molecule. Embryos that exhibit severely disrupted development (e.g., massive edema, tail curvature) or abnormal, uninterpretable phenotypes not resembling either species are classified as “teratogenized.” Controls for cyclopamine and latrunculin included DMSO (solvent).
The expression domain of Pitx2 in anuran embryos. In situ hybridization patterns (purple stain) indicate the expression of Pitx2 mRNA in the left lateral plate mesoderm of A) Xenopus laevis (NF 34), B) Ceratophrys cranwellii (GS 21), and C) Lepidobatrachus laevis (GS 21) embryos. Arrows delimit expression domains, which reveal the heterotopic shift in Lepidobatrachus. Expression was not detected in the right side (not shown).