Abstract
AIM: To assess the clinicopathological and biological significance of cripto in human colorectal cancer.
METHODS: Real-time reverse-transcription polymerase chain reaction (PCR) was used to examine cripto mRNA levels in primary colon cancer and normal colon tissues as well as normal and metastatic lymph nodes from colon cancers. Human colon cancer LS-174T cells were transfected with cripto small interfering RNA (siRNA), and mRNA and protein levels were evaluated using real-time PCR and western blot analysis, respectively. The growth of cancer cells was evaluated using the MTT assay and colony formation in soft agar. Invasion was examined using a Transwell assay, and the expressions of matrix metalloproteinase (MMP)-7 and MMP-9 were determined using western blot assay.
RESULTS: Cripto was significantly overexpressed in primary colon cancer and metastatic lymph nodes. Silencing cripto gene expression with cripto siRNA resulted in a significant decrease in colony formation in soft agar in the colon cancer cell line LS-174T. Cripto siRNA treatment decreased the migration and invasion capabilities of the colon cancer cell line LS-174T in vitro. Furthermore, cripto siRNA treatment inhibited the expression of matrix MMP-7 and MMP-9.
CONCLUSION: The results provide evidence that cripto siRNA could be an effective approach for the inhibition of cancer cell invasion and migration and thus has potential for use in devising novel preventive and therapeutic strategies for colon cancer metastasis.
Keywords: Colon cancer, Invasion, Metastasis, Cripto, Small interfering RNA, Matrix metalloproteinases
Core tip: To assess the clinicopathologic and biological significance of cripto as a novel target for colon cancer gene therapy, pathological and in vitro studies were carried out. Cripto was significantly overexpressed in primary colon cancer and metastatic lymph nodes. In vitro studies found that cripto siRNA resulted in a significant reduction in colony formation in soft agar and in the migration and invasion abilities of colon cancer cells. Furthermore, cripto siRNA led to an inhibition of MMP-7 and MMP-9. These results suggest that the cripto gene be useful for devising novel preventive and therapeutic strategies for colon cancer.
INTRODUCTION
The cripto gene is a member of the epidermal growth factor-CFC family of signaling proteins first cloned from the human teratocarcinoma cell line NTERA2[1]. Cripto is overexpressed in most malignant solid tumors, including colon, breast, lung, ovarian, and pancreatic cancers[2-7]. In contrast, crypto is generally absent from or found at low levels in normal tissues. In vitro, cripto exhibits many of the properties of an oncogene, including transformation of immortalized cells, induction of cell migration, and stimulation of branching morphogenesis[8]. These findings suggest that cripto serves an important function in carcinogenesis and in the development of some tumors.
Recent reports show that cripto overexpression may be closely related to invasion and metastasis in some human cancers[9,10]. Ertoy et al[9] evaluated cripto expression in matched sets of non-neoplastic cervical epithelium, primary cervical carcinoma, and metastatic tumors in the lymph nodes using immunopositivity staining. Strong cripto immunopositivity was found to be significantly correlated with tumor size and lymphovascular space involvement (P < 0.05). More importantly, the level of cripto expression increased in metastatic lymph nodes compared with their primary tumors. Despite the clear association between cripto overexpression and human breast cancer, the clinicopathological and biological significance of cripto overexpression in human colon cancer remains undiscovered.
Colorectal cancer is the third most common malignant neoplasm worldwide[10] and the second leading cause of death attributed to cancer[11]. Despite recent advances in diagnostic and therapeutic measures, the prognosis of colorectal cancer patients with distant metastasis remains poor. Enhanced understanding of the signaling mechanisms that regulate the metastasis of colon cancer may provide important insights with which to establish improved therapeutic strategies.
In this study, we demonstrate that cripto is highly overexpressed in primary and metastatic colon cancer tissues. In addition, we demonstrate that RNA interference (RNAi) cripto gene expression decreases the proliferation, migration, and invasion capability of colon cancer cell lines in vitro. To define the mechanisms underlying cripto invasion inhibition, we investigate the effect of cripto small interfering RNA (siRNA) transfection on the expression levels of mRNA and proteins of matrix metalloproteinase (MMP)-7 and MMP-9. Based on the results of this study, we conclude that cripto is a potential novel target of gene therapy for colon cancer metastasis.
MATERIALS AND METHODS
Tissue specimens and treatment
Thirty-nine paired samples of colon cancer and distant normal colon tissue were obtained from 39 inpatients who had undergone surgery from 2009 to 2010 in the Affiliated People’s Hospital of Jiangsu University. Eighteen metastasized lymph nodes were obtained from patients undergoing surgical therapy for the treatment of colon cancer. Tumor histotype and grade of differentiation were defined according to WHO criteria. Clinical and pathological stages were defined according to Dukes Staging’ criteria. Inpatients did not receive any chemotherapy or radiotherapy prior to surgery. Eleven normal lymph nodes without evidence of cancer were obtained from patients undergoing carotid endarterectomy. None of these patients had any history or clinical evidence of cancer.
To facilitate real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis, the specimens were identified and bisected. One portion was processed for real-time RT-PCR, whereas the other portion was sent for routine pathology analysis. All specimens were immediately snap-frozen in liquid nitrogen to prevent RNA degradation. The specimens were then stored at -70 °C until total RNA processing could be performed. This study was conducted with approval from the Medical Ethical Committee, and all patients provided written informed consent to participate in the study (Table 1).
Table 1.
Clinicopathologic characteristics of colon cancer patients n (%)
| Characteristic | No. of patients |
| Sex | |
| Male | 21 (53.8) |
| Female | 18 (46.2) |
| Age (yr), mean (range) | 62 (35–81) |
| ≤ 65 | 20 (51.3) |
| > 65 | 19 (48.7) |
| Dukes staging | |
| A + B | 12 (30.8) |
| C + D | 27 (69.2) |
| Tumor differentiation | |
| Well | 0 |
| Moderate | 29 (74.4) |
| Poor | 10 (25.6) |
Cell lines
The human colon cancer cell lines LS-174T and GEO were obtained from the Institute of Cell Biology, Shanghai, China. Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) at 37 °C under a 5%CO2 atmosphere. For 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, cells were plated in 96-well plates at a density of 2 × 103 cells/well. For real-time RT-PCR, cells were seeded in 6-well plates at a density of 1 × 105 cells/well.
RNA isolation and complementary DNA synthesis
Total cellular RNA was isolated from colon cancer cell lines, normal lymph nodes, and lymph nodes from primary colon cancer patients using Trizol. Final RNA pellets were dissolved in 20 μL of diethyl pyrocarbonate-treated water. RNA yield was determined by spectroscopy. For complementary DNA (cDNA) synthesis, 5 μg of total RNA was transcribed with cDNA transcription reagents using 0.5 μg of oligo(dT)18 primer for subsequent quantitative, RT-PCR.
Real-time RT-PCR
Real-time RT-PCR analyses were performed on an ABI Prism 7700 sequence detection system (Applied Biosystems, CA, United States). Primers and TaqMan probes were designed using Primer ExpressTM 1.0 (Applied Biosystems) software, and probes were labeled at the 5’ end with the reporter dye molecule FAM (6-carboxy-fluorescein) and at the 3’ end with the quencher dye molecule TAMARA (6-carboxytetramethyl-rhodamine). Real-time PCR was conducted in a total volume of 50 μL with 1 × TaqMan Master Mix (Applied Biosystems) and primers. Thermal cycler parameters included one cycle at 95 °C for 3 min, 45 cycles involving denaturation at 95 °C for 30 s, annealing at 52 °C for 45 s, and extension at 72 °C for 45 s, followed by a final extension at 72 °C for 10 min. The relative amount of cDNA in each sample was calculated by dividing the CT value with the corresponding value of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All reactions were performed in triplicate. The data were normalized to the internal control gene, GAPDH, to control for RNA preparation. Real time RT-PCR results were analyzed using Q-Gene software[12], which expresses data as mean normalized expression (MNE). MNE is directly proportional to the amount of RNA of the target gene (cripto and MMPs) relative to the amount of RNA of GAPDH.
Primer design
Primers for cripto, MMP-7, and MMP-9 were designed using Perkin-Elmer Primer Express software. The primer sequences are presented in Table 2.
Table 2.
Primers of cripto, matrix metalloproteinase-7 and matrix metalloproteinase-9
| Genes | Forward(5'-3') | Reverse(5'-3') |
| Cripto | CAATTCGGCCTCGGTCTTC | TTCAGGCAGCAGGTTCTGTTT |
| MMP-7 | AACTCCCGCGTCATAGAAAT | GATACGATCCTGTAGGTGAC |
| MMP-9 | CGGAGTGAGTTGAACCAG | GTCCCAGTGGGGATTTAC |
MMP: Matrix metalloproteinase.
Cripto siRNA sequence
Cripto siRNAs corresponding to cripto mRNA with dTdT on 3’-overhangs were designed and chemically synthesized according to the recommendation of the manufacturer (Dharmacon Research, United States). Six SiRNAs targeting the coding sequence of cripto mRNA (S1-S6) were used in the current experiment. Information on the cripto siRNAs is provided in Table 3. The scrambled siRNA served as a control and its sequences were 5’-UUCUCCGAACGUGUCACGUTdTd-3’ and 5’-ACGUGACACGUUCGGAGAATdTdT-3’.
Table 3.
Cripto siRNA sequence
| siRNA | Sense (5' → 3') | Antisense (5' → 3') | MW | Position |
| S1 | UUCGGCCUCGGUCUUCCCATT | UGGGAAGACCGAGGCCGAATT | 13347.2 | 175-195 |
| S2 | CAGAACCUGCUGCCUGAAUTT | AUUCAGGCAGCAGGUUCUGTT | 13317.2 | 236-256 |
| S3 | CUGUGAGCACGAUGUGCGCTT | GCGCACAUCGUGCUCACAGTT | 13347.2 | 314-334 |
| S4 | GAGAACUGUGGGUCUGUGCTT | GCACAGACCCACAGUUCUCTT | 13332.2 | 336-356 |
| S5 | UGCUGGCACGGUCAGCUCCTT | GGAGCUGACCGUGCCAGCATT | 13362.2 | 396-416 |
| S6 | CUACCACCGUCUGCACGUATT | UACGUGCAGACGGUGGUAGTT | 13332.2 | 495-515 |
In vitro transfection
Transfection of siRNA was performed using a commercial reagent, oligofecamine (Invitrogen, United States), in 6-well plates following the manufacturer’s instructions. Briefly, the day before transfection, confluent layers of cells were trypsinized, counted, and resuspended. Thereafter, 1 × 105 of cells were plated into each well of the 6-well plates, such that approximately 70% confluence could be achieved the next day at the time of transfection. Oligofecamine was diluted in serum-free RPMI 1640 and mixed with siRNA at a 1:2 ratio (4 μL of 20 μmol/L of siRNA formulated with 8 μL of oligofecamine). The cells were then incubated for another 48 h. Cell numbers were determined using a hemocytometer before subsequent assays.
MTT assay
Cells plated in 96-well plates were grown in their respective media for 48 h after the addition of siRNA. At each time point, cells were checked visually for growth and proliferation. MTT (Sigma) was then added to the wells, and the cells were incubated at 37 °C for 4 h. MTT solubilization solution (10% Triton X-100 in acidic isopropanol, 0.1 mol/L HCl) was added to the wells, and the cells were incubated overnight. Colorimetric measurements were performed using a microplate reader (Molecular Devices) at 560 nm, and the background at 650 nm was subtracted.
Anchorage-independent growth assay
For the anchorage-independent growth experiments, LS-174T cells (1 × 104 cells/well) were seeded in 0.3% FBS supplemented with complete culture medium. This suspension was layered over 0.5 mL of 0.8% agar-medium base layer in 24-well plates. After 15 d, the colonies were stained with nitroblue tetrazolium, and colonies larger than 50 μm were acquired using a micro-Scopeman camera system (Moritex Europe Ltd.) and analyzed with Image-Pro Plus (Media Cybernetics) software.
In vitro invasion/migration assay
Transwell migration and invasion assays were performed using LS-174T cells cultured in 12-well plates containing either 8 μm pore Biocoat® control inserts (migration assays) or Matrigel-coated inserts (invasion assays) according to the manufacturer’s instructions (Becton Dickinson, Bedford, MA, United States). The membranes were rehydrated with warm serum-free Dulbecco’s modified Eagle’s medium (1.0 mL/chamber) for 2 h. The upper chamber was filled with 1 × 105 cells in L-15 medium containing 5% FBS. The lower chamber was filled with L-15 medium containing 25% FBS as a chemoattractant. After the chambers were incubated for 24 h at 37 °C under a 5% CO2 atmosphere, the non-invading cells were removed from the upper surface of the membrane by scrubbing, and invading cells on the lower surface of the membrane were fixed and stained with hematoxylin and eosin.
The number of cells that penetrated the filter was counted by a technician blinded to the experimental settings in four microscopic fields of each filter under × 20 magnification. The percentage of invasion was expressed as the ratio of the mean cell number from the invasion chamber to the mean cell number from the control chamber according to the manufacturer’s recommendation. The percentage of migration was expressed as the ratio of the mean cell number in control inserts containing S1 siRNA transfected cells to mean cell numbers in control inserts containing untreated cells (untreated cells were given a value of 100%).
Western blot assay for MMP-7 and MMP-9
Seventy-two hours after transfection, cells were washed twice in PBS, and total protein was extracted in 150 mmol/L NaCl, 50 mmol/L Tris·HCl (pH 7.5), 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 5 mmol/L EDTA, 10 mg/mL leupeptin, 1% aprotinin, and 2 mmol/L PMSF. Ten micrograms of protein sample was loaded onto a 10% SDS-PAGE and electroblotted onto a PVDF nylon membrane (Millipore, Bedford). Membranes were blocked in 0.05% Tween 20 (v/v) PBS containing 5% skimmed milk and then incubated with MMP-7, MMP-9, and β-actin antibodies (Santa Cruz Biotechnology). Membranes were then incubated with a HRP-linked goat anti-rabbit IgG secondary antibody (Santa Cruz Biotechnology). Finally, the membrane was reacted with DAB reagent and washed with PBS once protein bands had appeared.
Statistical analysis
Statistical analyses included the independent t-test and analysis of variance. Statistical analyses were performed using SPSS 11.5 software (SPSS Inc., Chicago, IL, United States.
RESULTS
Cripto is highly expressed in primary colon cancer
To determine whether or not cripto was expressed in human colon cancer, real-time RT-PCR was conducted on 39 paired samples to determine cripto mRNA expression levels in clinical tissues. The results showed that cripto expression in primary colon cancer samples was significantly higher (mean expression in cancer tissue was more than 16-fold higher; P < 0.001) than that in normal tissues.
Cripto is significantly overexpressed in lymph nodes containing metastatic colon cancer
Real-time RT-PCR analysis was performed on 11 normal lymph nodes and 18 lymph nodes containing metastatic colon cancer. The results indicated that cripto expression in metastatic lymph nodes was significantly higher (mean expression in cancer tissue was approximately 150-fold higher; P < 0.001) than that in normal lymph nodes.
Screening of cripto siRNA
The capability of siRNA to inhibit cripto expression was quantified by real-time RT-PCR analysis 48 h following siRNA exposure. S1-S6 (targeting the coding sequence of cripto mRNA) showed various suppressant effects on cripto mRNA expression in LS-174T cells which express high levels of cripto mRNA[13,14]. Among them, S1, which targets nucleotides 175-195, exhibited the strongest effect. At a concentration of 100 nmol/L, S1 reduced the cripto mRNA level by 89% 48 h after the start of transfection (Figure 1). In contrast, the control-scrambled siRNA treatment showed no effect on cripto mRNA levels, thus supporting the specificity of cripto siRNA. To characterize the potency of S1 further, the dose and time dependency of its effects on cripto mRNA in LS-174T cells were examined, and results indicated that S1 downregulated the cripto mRNA level in a dose-dependent manner (Figure 2).
Figure 1.
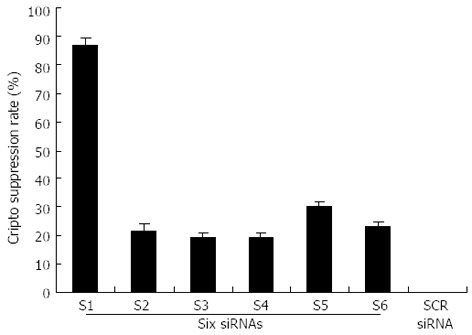
Suppression of cripto mRNA by six siRNA transfection in LS-174T cells. The colon cancer cell line LS-174T was transfected with 100 nmol/L of six siRNA after 48-h incubation. Real-time polymerase chain reaction quantification of the relative amount of cripto transcript was performed using the cripto primers and probe set as described in “Materials and Methods,” using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal standard. All data are presented as the mean ± SD of three independent experiments; the level of cripto mRNA relative to GAPDH in untreated cells maintained under identical experimental conditions was taken as 100%.
Figure 2.
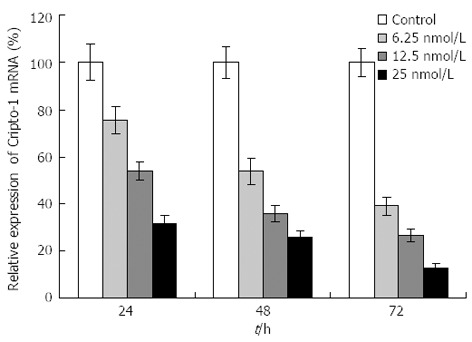
Effects of S1 siRNA on cripto mRNA of colon cancer LS-174T cells. The LS-174T colon cancer cell line was transfected with different dose of S1 siRNA for 24, 48, 72 h. Cripto mRNA was evaluated by real-time reverse transcription-polymerase chain reaction as described in “Materials and Methods,” using GAPDH as internal standard. All data are presented as the mean ± SD of three independent experiments; the level of cripto mRNA relative to GAPDH in untreated cells maintained under identical experimental conditions was taken as 100%.
Effects of cripto siRNA on proliferation in LS-174T cells in vitro
We then evaluated the biological effects of cripto suppression on colon cancer LS-174T cells using different types of assays. The MTT assay showed that cellular proliferation in the monolayer culture was unaffected by either siRNA (Figure 3A). However, colony formation in soft agar was strongly inhibited by treatment with cripto siRNA but not by control siRNA (Figure 3B). Figure 3B shows that treatment with cripto siRNA induced significant anchorage-independent growth inhibition in a dose-dependent manner (Figure 3).
Figure 3.
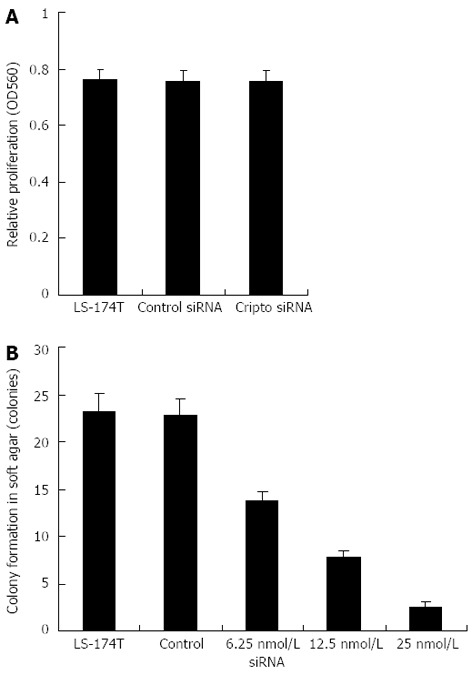
Effects of S1 siRNA on cell proliferation and anchorage-independent growth of colon cancer LS-174T cells. A: S1 siRNA treatment decreases cell proliferation. Cells plated in 96-well plates were grown in their respective media for 48 h after the addition of siRNA. Cell proliferation was examined by MTT assay; B: S1 siRNA treatment reduces anchorage-independent growth of LS-174T cells. The anchorage-independent growth was evaluated by colony formation in soft agar.
Downregulation of cripto decreased cancer cell invasion and migration capabilities of colon cancer cell lines in vitro
Colony formation in soft agar is a property closely associated with malignancy. Given the known role of cripto siRNA in the downregulation of anchorage-independent growth of LS-174T cells, we attempted to determine whether or not the cripto gene contributes to cell invasion and migration in colon cancer. Cell migration and invasion studies were performed using Matrigel matrix assays. Tumor cells require both migration and invasion properties to invade through the Matrigel matrix. Two independent experiments were performed. The results showed that S1 treatment, but not scrambled siRNA treatment, resulted in a significant low level of migration and invasion potential of LS-174T cells (Figure 4).
Figure 4.
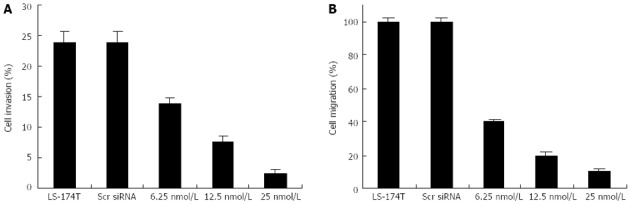
Effects of S1 siRNA on migration and invasion of colon cancer LS-174T cells. The LS-174T colon cancer cell line was used in this experiment. Cell migration was assessed in BioCoat control cell culture chambers. Transwell migration and invasion assays were performed using LS-174T cells cultured in 12-well plates containing either Matrigel-coated inserts (invasion assays) or 8 μm pore Biocoat® control inserts (migration assays), according to the manufacturer’s instructions. Control, scrambled siRNA-, and S1 siRNA-treated cells were added to control and Matrigel chambers. A: The percentage of invasion was expressed as the ratio of mean cell number from invasion chamber to mean cell number from control chamber according to the manufacturer’s recommendation; B: The percentage of migration was expressed as the ratio of mean cell number in control inserts containing siRNA-treated cells to mean cell number in control inserts containing untreated control cells. Control cells were given a value of 100%.
Effects of cripto siRNA on MMP-7 and MMP-9 expression in colon cancer cells
To explore whether or not the invasiveness of transfected cells was associated with MMP induction, real-time PCR and western blot assays were conducted to detect alterations in the expression level of MMP-7 and MMP-9. As illustrated in Figure 5, cripto suppression resulted in decreases in both mRNA and MMP-7 and MMP-9 protein levels compared with those in control cells. Another experiment on transfection with cripto siRNA in the colon cancer cell line GEO also exhibited invasion inhibition and downregulation of MMP-7 and MMP-9 expression (data not shown). These results indicate that cripto suppression by RNAi could inhibit invasion and migration capabilities by reducing MMP-7 and MMP-9 expression in human colon cancer cells.
Figure 5.
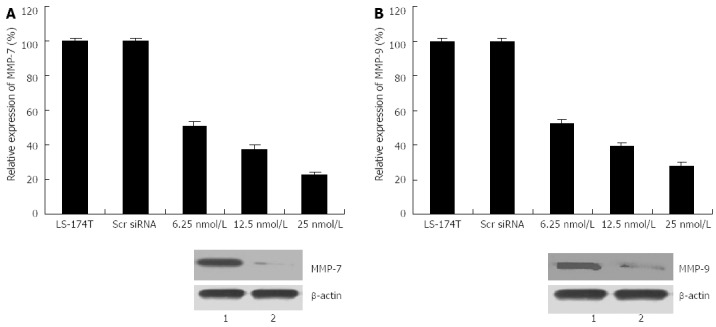
Effects of S1 treatment on matrix metalloproteinase-7 and -9 expression of colon cancer line LS-174T. After cancer cells were treated with S1 siRNA for different times, cells were harvested, and total RNA and proteins were extracted. Real-time reverse transcription-polymerase chain reaction and western blotting were performed to detect mRNA and protein levels, respectively, of matrix metalloproteinase (MMP)-7 and -9. A: Expression level of mRNA and protein of MMP-7 (2 h); B: Expression level of mRNA and protein of MMP-9 (72 h). 1: Scr siRNA; 2: S1(25 nmol/L).
DISCUSSION
The primary modalities for colon cancer therapy are surgery, radiotherapy, and chemotherapy. One of the main limitations of current treatment modalities is that systemic therapies for metastatic disease are not curative. To improve the choice of therapeutic strategy, the mechanism of invasion and metastasis of colon cancer must be clarified.
The cripto gene is known to be overexpressed in numerous solid cancers, and its overexpression appears to be associated with enhanced proliferation and malignant potential[1,2,5]. We found that cripto was significantly overexpressed in primary and metastatic colon cancer tissues through real-time RT-PCR. To determine whether or not cripto was a potential target for colon cancer gene therapy, the colon cancer cell line LS-174T was treated with cripto siRNA, and anchorage-independent growth and the capacity for invasion and migration were determined using different assays. The results showed that cancer cells transfected with cripto siRNA inhibited the anchorage-independent growth, invasive capacity, and migration capability of colon cancer cells. Finally, our results provide mechanistic insight into the function of cripto in the regulation of invasion and migration through the suppression of MMP-7 and MMP-9 expression, which suggests that cripto may serve as a novel tumor marker for colon cancer metastasis.
The mechanisms by which cripto regulates invasive potential and migration capability remain unclear. Normanno et al[15] recently showed that crypto overexpression enhanced invasion capability by inhibiting anoikis in breast cancer. The process of metastasis is complex, occurring in a series of steps, including cell invasion and degradation of the basement membranes and stromal extracellular matrix, ultimately leading to tumor cell invasion and metastasis. MMPs are critically involved in the processes of tumor cell invasion and metastasis[16-19]. The MMPs comprise a family of related enzymes that degrade the extracellular matrix and are considered to be important factors in facilitating tumor invasion. Among the MMPs, MMP-7 and MMP-9 have been considered important factors in facilitating invasion and metastases in human colon cancer[20-24]. We further investigated whether or not cripto-induced invasion of LS-174T cells is mediated through MMP-7 and MMP-9. Real-time RT-PCR and Western blot analysis were performed to detect MMP expression in the colon cancer cell line LS-174T. Cripto siRNA-transfected cells showed significantly low levels of mRNA and MMP-7 and MMP-9 proteins. Data from these results show that the downregulation of cripto expression after siRNA treatment decreases MMP expression. To the best of our knowledge, this study is the first to report on the primary mechanism responsible for the decrease in invasion potential observed after cripto siRNA treatment.
Based on our studies, we speculate that transfection with cripto siRNA decreases invasion and metastasis in human colon cancer through MMP downregulation. Thus, targeting cripto for molecular intervention may be an attractive therapeutic strategy for colon cancer. Many studies have proven that RNAi technology siRNA can be used successfully for gene silencing in vivo[25,26]. Thus, the application of RNAi mediated by siRNA to knock down cripto expression in colon cancer may prove to be a valuable strategy for patients with advanced colon cancer.
COMMENTS
Background
The cripto gene is always overexpressed in most malignant solid tumors, including colon, breast, lung, ovarian, and pancreatic cancers. Recent reports showed that cripto overexpression may very well be closely related to invasion and metastasis in a few of human cancers, including cervical carcinoma and breast cancer, but the clinicopathologic and biological significance of cripto overexpression in human colon cancer remains unclear.
Research frontiers
To explore the clinicopathologic and biological significance of cripto as a novel target for colon cancer gene therapy, pathological and in vitro studies were carried out using RNA interference (RNAi). The results suggest that the cripto gene may be useful for devising novel preventive and therapeutic strategies for colon cancer.
Innovations and breakthroughs
The results showed that RNAi cripto can decrease cell migration and invasion by inhibiting matrix metalloproteinases.
Applications
The results here suggested that the detection of cripto is helpful in understanding the development of colorectal carcinoma, and that cripto siRNA could be an effective approach for the inhibition of invasion and migration of human colon cancer.
Terminology
RNAi’s main function is to adjust and shut down gene expression and regulate all kinds of activities of cells.Small interfering RNA (siRNA) is generated within the cell in the process of RNAi, which consists of fragments of about 21-25 nuclear acids of the double-stranded RNA molecule.
Peer review
The biological significance of cripto in the occurrence and development of colorectal carcinoma in vivo, in vitro were investigated. The results showed that cripto was significantly overexpressed in primary colon cancer and metastatic lymph nodes. Furthermore, the molecular mechanism of invasion and the regulation of cripto gene expression in colon cancer cells were explored by RNA interference technology. The study found, MMP-7 and -9 gene is involved in the regulation of cripto invasion of colorectal cancer cells. There is important innovation and scientific significance of the study.
Footnotes
Supported by The National Natural Science Fund, No. 81250035; The Jiangsu Province of 333 Engineering talent fund, No. RA2012103, and Zhenjiang City Social Development Fund No. SH2011031 and SH2011034
P- Reviewers: Meyers BM, Meropol NJ, Van Cutsem E S- Editor: Wen LL L- Editor: Cant MR E- Editor: Zhang DN
References
- 1.Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, Persico MG. Molecular characterization of a gene of the ‘EGF family’ expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989;8:1987–1991. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciardiello F, Kim N, Saeki T, Dono R, Persico MG, Plowman GD, Garrigues J, Radke S, Todaro GJ, Salomon DS. Differential expression of epidermal growth factor-related proteins in human colorectal tumors. Proc Natl Acad Sci USA. 1991;88:7792–7796. doi: 10.1073/pnas.88.17.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi CF, Liscia DS, Normanno N, Merlo G, Johnson GR, Gullick WJ, Ciardiello F, Saeki T, Brandt R, Kim N. Expression of transforming growth factor alpha, amphiregulin and cripto-1 in human breast carcinomas. Br J Cancer. 1994;69:903–910. doi: 10.1038/bjc.1994.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friess H, Yamanaka Y, Büchler M, Kobrin MS, Tahara E, Korc M. Cripto, a member of the epidermal growth factor family, is over-expressed in human pancreatic cancer and chronic pancreatitis. Int J Cancer. 1994;56:668–674. doi: 10.1002/ijc.2910560511. [DOI] [PubMed] [Google Scholar]
- 5.Panico L, D’Antonio A, Salvatore G, Mezza E, Tortora G, De Laurentiis M, De Placido S, Giordano T, Merino M, Salomon DS, et al. Differential immunohistochemical detection of transforming growth factor alpha, amphiregulin and CRIPTO in human normal and malignant breast tissues. Int J Cancer. 1996;65:51–56. doi: 10.1002/(SICI)1097-0215(19960103)65:1<51::AID-IJC9>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Fontanini G, De Laurentiis M, Vignati S, Chinè S, Lucchi M, Silvestri V, Mussi A, De Placido S, Tortora G, Bianco AR, et al. Evaluation of epidermal growth factor-related growth factors and receptors and of neoangiogenesis in completely resected stage I-IIIA non-small-cell lung cancer: amphiregulin and microvessel count are independent prognostic indicators of survival. Clin Cancer Res. 1998;4:241–249. [PubMed] [Google Scholar]
- 7.D’Antonio A, Losito S, Pignata S, Grassi M, Perrone F, De Luca A, Tambaro R, Bianco C, Gullick WJ, Johnson GR, et al. Transforming growth factor alpha, amphiregulin and cripto-1 are frequently expressed in advanced human ovarian carcinomas. Int J Oncol. 2002;21:941–948. [PubMed] [Google Scholar]
- 8.Adamson ED, Minchiotti G, Salomon DS. Cripto: a tumor growth factor and more. J Cell Physiol. 2002;190:267–278. doi: 10.1002/jcp.10072. [DOI] [PubMed] [Google Scholar]
- 9.Ertoy D, Ayhan A, Saraç E, Karaağaoğlu E, Yasui W, Tahara E, Ayhan A. Clinicopathological implication of cripto expression in early stage invasive cervical carcinomas. Eur J Cancer. 2000;36:1002–1007. doi: 10.1016/s0959-8049(00)00033-2. [DOI] [PubMed] [Google Scholar]
- 10.Shike M, Winawer SJ, Greenwald PH, Bloch A, Hill MJ, Swaroop SV. Primary prevention of colorectal cancer. The WHO Collaborating Centre for the Prevention of Colorectal Cancer. Bull World Health Organ. 1990;68:377–385. [PMC free article] [PubMed] [Google Scholar]
- 11.Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH, et al. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112:594–642. doi: 10.1053/gast.1997.v112.agast970594. [DOI] [PubMed] [Google Scholar]
- 12.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374, 1376. [PubMed] [Google Scholar]
- 13.Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL, et al. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J Clin Invest. 2003;112:575–587. doi: 10.1172/JCI17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Y, Zheng S, Ding JY. Inhibition of telomerase activity of colon cancer LS-174T cell with liposome-mediated cripto antisense oligodeoxynucleotide. Zhongguo Binglishengli Zazhi. 2006;22:761–765. [Google Scholar]
- 15.Normanno N, De Luca A, Bianco C, Maiello MR, Carriero MV, Rehman A, Wechselberger C, Arra C, Strizzi L, Sanicola M, et al. Cripto-1 overexpression leads to enhanced invasiveness and resistance to anoikis in human MCF-7 breast cancer cells. J Cell Physiol. 2004;198:31–39. doi: 10.1002/jcp.10375. [DOI] [PubMed] [Google Scholar]
- 16.Curran S, Murray GI. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur J Cancer. 2000;36:1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 17.John A, Tuszynski G. The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol Oncol Res. 2001;7:14–23. doi: 10.1007/BF03032599. [DOI] [PubMed] [Google Scholar]
- 18.Heslin MJ, Yan J, Johnson MR, Weiss H, Diasio RB, Urist MM. Role of matrix metalloproteinases in colorectal carcinogenesis. Ann Surg. 2001;233:786–792. doi: 10.1097/00000658-200106000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubbe WJ, Zhou ZY, Fu W, Zuzga D, Schulz S, Fridman R, Muschel RJ, Waldman SA, Pitari GM. Tumor epithelial cell matrix metalloproteinase 9 is a target for antimetastatic therapy in colorectal cancer. Clin Cancer Res. 2006;12:1876–1882. doi: 10.1158/1078-0432.CCR-05-2686. [DOI] [PubMed] [Google Scholar]
- 20.Mori M, Barnard GF, Mimori K, Ueo H, Akiyoshi T, Sugimachi K. Overexpression of matrix metalloproteinase-7 mRNA in human colon carcinomas. Cancer. 1995;75:1516–1519. doi: 10.1002/1097-0142(19950315)75:6+<1516::aid-cncr2820751522>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 21.Adachi Y, Yamamoto H, Itoh F, Hinoda Y, Okada Y, Imai K. Contribution of matrilysin (MMP-7) to the metastatic pathway of human colorectal cancers. Gut. 1999;45:252–258. doi: 10.1136/gut.45.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masaki T, Matsuoka H, Sugiyama M, Abe N, Goto A, Sakamoto A, Atomi Y. Matrilysin (MMP-7) as a significant determinant of malignant potential of early invasive colorectal carcinomas. Br J Cancer. 2001;84:1317–1321. doi: 10.1054/bjoc.2001.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida H, Murata N, Tada M, Okada N, Hashimoto D, Kubota S, Shirakawa K, Wakasugi H. Determining the levels of matrix metalloproteinase-9 in portal and peripheral blood is useful for predicting liver metastasis of colorectal cancer. Jpn J Clin Oncol. 2003;33:186–191. doi: 10.1093/jjco/hyg035. [DOI] [PubMed] [Google Scholar]
- 24.Illemann M, Bird N, Majeed A, Sehested M, Laerum OD, Lund LR, Danø K, Nielsen BS. MMP-9 is differentially expressed in primary human colorectal adenocarcinomas and their metastases. Mol Cancer Res. 2006;4:293–302. doi: 10.1158/1541-7786.MCR-06-0003. [DOI] [PubMed] [Google Scholar]
- 25.Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 26.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]


