Abstract
Prenatal exposure to moderate doses of valproic acid (VPA) produces brainstem abnormalities, while higher doses of this teratogen elicit social deficits in the rat. In this pilot study, we examined effects of prenatal exposure to a moderate-dose of VPA on behavior and on transcriptomic expression in three brain regions that mediate social behavior. Pregnant Long-Evans rats were injected with 350 mg/kg VPA or saline on gestational day 13. A modified social-interaction test was used to assess social behavior and social preference/avoidance during early and late adolescence and in adulthood. VPA-exposed animals demonstrated more social investigation and play fighting than control animals. Social investigation, play fighting, and contact behavior also differed as a function of age; the frequency of these behaviors increased in late adolescence. Social preference and locomotor activity under social circumstances were unaffected by treatment or age. Thus, a moderate prenatal dose of VPA produces behavioral alterations that are substantially different from the outcomes that occur following exposure to a higher dose. At adulthood, VPA-exposed subjects exhibited transcriptomic abnormalities in three brain regions: anterior amygdala, cerebellar vermis, and orbitofrontal cortex. A common feature among the proteins encoded by the dysregulated genes was their ability to be modulated by acetylation. Analysis of the expression of individual exons also revealed that genes involved in post-translational modification and epigenetic regulation had particular isoforms that were ubiquitously dysregulated across brain regions. The vulnerability of these genes to the epigenetic effects of VPA may highlight potential mechanisms by which prenatal VPA exposure alters the development of social behavior.
Keywords: adolescence, autism, gene expression, sex differences, social interaction, teratogen
1. INTRODUCTION
During nervous system development, neurons exhibit periods of vulnerability to teratogens. One period of vulnerability is neuronal birthdate; i.e., the day on which neurons undergo their final mitosis. In the rat, neurons in some of the cranial nerve nuclei are born on gestational day (G) 12 or G13. These include the principal sensory nucleus of the trigeminal nerve and the motor nuclei of the trigeminal, facial, and hypoglossal nerves (Altman & Bayer, 1980a; b; c). Exposure to ethanol on G12 or G13 results in a permanent reduction in the number of neurons in some cranial nerve nuclei (Mooney & Miller, 2007), as well as alterations in social behavior and gene expression (Mooney & Varlinskaya, 2011; Middleton et al., 2012). The pronounced and permanent social deficits seen throughout adolescence and adulthood were most apparent in male offspring (Mooney & Varlinskaya, 2011; Middleton et al., 2012). Specifically, males prenatally exposed to ethanol and tested as early adolescents, late adolescents or young adults exhibited a significant reduction of social investigation, contact behavior, and play fighting regardless of age. Older adolescent and adult males and females demonstrated social anxiety indexed by transformation of social preference into social avoidance.
Administration of another teratogen, valproic acid (VPA) at a dose of 350 mg/kg, during the same critical period also decreases neuronal number in cranial nerve nuclei (Rodier et al., 1996) but the effect on social behavior, anxiety-like responses under social circumstances, or gene expression is unknown. For this reason we decided to use this dose to explore the effects of prenatal VPA. Many studies examining behavioral outcome after exposure to VPA use a high dose of the drug (500 – 800 mg/kg). In these models, animals show altered nociception (Schneider et al., 2001; Schneider & Przewlocki, 2005; Schneider et al., 2008), abnormal fear conditioning and increased anxiety (Markram et al., 2008), repetitive, stereotypic-like behaviors (Schneider & Przewlocki, 2005; Markram et al., 2008; Schneider et al., 2008), decreases in social interactions (Schneider & Przewlocki, 2005; Markram et al., 2008; Schneider et al., 2008; Dufour-Rainfray et al., 2010), and alterations in eye-blink conditioning that are similar to the changes seen in humans with ASD (Stanton et al., 2007). Acute prenatal exposure to a high dose of VPA also has anatomical effects; for example, it reduces the number of Purkinje cells in the cerebellum (Ingram et al., 2000), alters the location of serotonergic cells (Kuwagata et al., 2009), decreases serotonin expression in the hippocampus (Stanton et al., 2007), and alters cortical neuronal connectivity (Rinaldi et al., 2008). Chronic prenatal exposure to a moderate dose (300 or 350 mg/kg) alters hippocampal synaptic plasticity (Zhang et al., 2003), increases complexity of apical dendrite branching in motor cortex (Snow et al., 2008), and decreases complexity of dendrite branching in hippocampal neurons (Raymond et al., 1996). But high doses of this drug can also be toxic to the dam and/or cause fetal death (Vorhees, 1987).
VPA has several mechanisms. Its acute effects may be driven by increases of gamma-aminobutyric acid (GABA) concentrations in the brain (Dufour-Rainfray et al., 2010) via inhibition of GABA transaminase (Rosenberg, 2007b). VPA can also have more sustained effects that are mediated by manipulation of DNA-processing and changes in gene transcription that result from its ability to act as a histone deacetylase inhibitor (HDACi) (Phiel et al., 2001; Rosenberg, 2007a). Histone acetylation is a global mark and facilitator of gene activity (Brownell & Allis, 1996). Acetylation yields a negative charge, acting to neutralize the positive charge on histones and decrease the interaction of the N-termini of histones with the negatively charged phosphate groups of DNA. As a consequence, the condensed chromatin is transformed into a more relaxed structure, which facilitates transcription. An even more prolonged influence proposed for VPA is through the induction of replication-independent DNA demethylation (Detich et al., 2003b; a). VPA can reset stable DNA methylation patterns in established non-dividing cells and, therefore, can have wide-ranging and long-term effects on all cell types found in the brain and at any period of life (Detich et al., 2003b; Rosenberg, 2007a). Equally interesting is the ability of VPA to also affect the level of acetylation of non-histone proteins (Mannaerts et al., 2010).
Brain regions important for social behavior include the orbitofrontal cortex (OFC), the amygdala, and the cerebellar vermis, among others. A lesion made to the OFC or the amygdala can cause deficits in social behavior (Daenen et al., 2002; Diergaarde et al., 2004; Mah et al., 2004; Rudebeck et al., 2007). Cerebellar vermis forms connections with the limbic system (Strick et al., 2009), and abnormalities of the vermis are associated with a number of disorders, including attention-deficit/hyperactivity disorder, schizophrenia, bipolar disorder, depression, anxiety, and autism (DelBello et al., 1999; Ichimiya et al., 2001; Loeber et al., 2001; Kaufmann et al., 2003; Mackie et al., 2007; Picard et al., 2008; Strick et al., 2009).
In the present study, we tested whether acute exposure to a moderate dose of VPA during a critical period of gestation: (1) produces social deficits and increases social anxiety during adolescence and/or adulthood; and (2) alters gene expression in three areas of the brain that are important for normal social behavior. Different forms of social behavior and social preference/avoidance (an index of social anxiety-like behavior) (Varlinskaya & Spear, 2010) were assessed using a modified social interaction test (Varlinskaya et al., 1999; 2001), which allows assessments of different components of social behavior, and of social motivation indexed via a coefficient of social preference/avoidance. Transcriptome expression was also evaluated at two levels: as an aggregate measure of each gene’s overall expression which encompassed all expressed isoforms, and at the level of individual exons to identify particular differentially spliced isoforms of each gene.
2. METHODS
2.1. Animals
Timed-pregnant Long Evans rats (Taconic, Germantown NY) were injected intraperitoneally (i.p.) with 350 mg/kg VPA (Sigma; St Louis MO) or an equivalent volume of saline on gestational day (G) 13. G1 was designated as the first day on which a sperm-positive plug was identified. All procedures were approved by the Committee for Humane Use of Animals at Upstate Medical University and the Institutional Animal Care and Use Committee at the Syracuse Veterans Affairs Medical Center.
Within 24 hours of birth (on postnatal day (P) 0), litters were culled to ten, maintaining the ratio of male and female pups at 1:1 as well as possible. Pups were weaned on P21 and subsequently housed in same-sex groups of four littermates on a reverse light-dark cycle (i.e., lights off from 6am–6pm).
2.2. Social Behavior Study
2.2.1. Modified Social Interaction Test
Animals were tested on P28, P42, or P75 (early and late adolescence and young adulthood, respectively) as described previously (Mooney & Varlinskaya, 2011). One male and one female from each litter were tested at each age, and each animal was only tested once (n=10 per sex/age for saline-exposed animals, n=8 per age for VPA-exposed males, n=9 for VPA-exposed females tested at P28 or P42, n=7 for VPA-exposed females tested at P75).
Testing was conducted under dim light using Plexiglas (Binghamton Plate Glass, Binghamton, NY) test apparatuses. The test apparatus was divided into two compartments of the same size by a clear Plexiglas partition. The partition contained a semi-circular aperture that allowed animals to move between compartments such that only one animal was able to move through the aperture at a time (Varlinskaya et al., 1999; 2001). The box used for adolescent animals was smaller than that used for adult animals (30 × 20 × 20 cm for adolescents, 45 × 30 × 20 cm for adults).
On the day prior to testing, each experimental animal spent 30 min alone in the testing apparatus. Allowing the animal to familiarize itself with the apparatus increases the frequency of social interactions in the later test situation (File & Hyde, 1978; File & Seth, 2003). On the test day, experimental animals were marked with indelible ink for later identification, and placed alone into a holding cage for 30 min, another way to increase social interactions. Animals were then placed into the testing apparatus. Five min later, a non-manipulated same-sex, same-age novel rat was also placed into the testing apparatus (Varlinskaya & Spear, 2002; 2008). Animals were video-taped for 10 min with the investigator outside the room. Between tests, the testing apparatus was cleaned with 3% hydrogen peroxide.
2.2.2. Behavioral Measures
Four separate behavioral measures were scored and analyzed: frequency of social investigation (sniffing of the partner), contact behavior (crawling under or over the partner, social grooming), and play fighting (following, chasing, playful nape attacks and pinning), and also social preference/avoidance (Mooney & Varlinskaya, 2011). Social preference/avoidance was assessed by scoring the number of crossovers (movements between compartments) demonstrated by the experimental animal toward the partner and away from the partner (for details see 5). In addition, the total number of crossovers from one compartment to another was used as an index of locomotor activity in the social context. The frequency of each behavior for each test subject within the 10 min period was noted (Meaney & Stewart, 1981; Thor & Holloway, 1984; Vanderschuren et al., 1997; Varlinskaya et al., 1999; 2001; Mooney & Varlinskaya, 2011). Behavioral data were scored by a trained observer without knowledge of experimental condition of any animal.
2.2.3. Statistical Analysis
Statistical analysis of each behavioral measure was performed using separate 2 (prenatal exposure) × 3 (test age) × 2 (sex) between-group analyses of variance (ANOVAs). Changes in social behavior induced by prenatal exposure to VPA were assessed by post-hoc comparisons (Fisher’s planned least significant difference tests) between VPA-exposed animals and their age-matched saline-exposed controls. Statistical analyses were performed using Statistica software.
2.3. Microarray Study
2.3.1. Tissue Samples
Sixty to 90 minutes after behavioral testing, 75-day-old rats were anesthetized (100 mg/kg ketamine and 10 mg/kg xylazine, i.p.) and decapitated. Brains were removed and the forebrain was separated from the brainstem and cerebellum by a coronal cut between the colliculi. Forebrains were separated by a cut in the mid-sagittal plane. The cerebellum was removed from the brainstem by cutting through the peduncles. Brain pieces were frozen rapidly on dry ice, then stored at −80°C. Three male and three female rats, each from a different litter, were taken from each of the two treatment groups.
2.3.2. RNA Extraction
Segments enriched in three brain regions were obtained from each of the six saline-exposed animals and six VPA-exposed animals. Target brain regions included those that had been previously associated with social behavior: orbitofrontal cortex, amygdala, and cerebellar vermis. All tissue was taken from the left hemisphere, and dissections were performed using a rat atlas as a guide (Paxinos G., 2009). To isolate orbitofrontal cortex, the anterior 1 mm was sectioned from the hemisphere, the olfactory tubercle was removed, then a triangle of tissue was cut from the central part of the ventral half of the remaining tissue. The amygdala tissue was dissected from a 2 mm-thick slab taken from the middle of the forebrain. The slab was laid flat and a cut was made parallel and slightly lateral to the internal capsule. A second cut was made approximately 2 mm lateral to the first to separate much of the piriform cortex. A final cut was made in the horizontal plane 1 mm below the rhinal fissure. This resulted in a block of tissue enriched in amygdala. The cerebellar vermis was defined as the middle one third of the cerebellum; i.e., the tissue separating the cerebellar hemispheres. The cerebellum and brainstem were isolated from the rest of the brain by a coronal cut made between the superior and inferior colliculi, then the cerebellum was removed by severing the peduncles. The hemispheres were cut away to reveal the vermis.
Samples were prepared for RNA extraction using a mortar and pestle to disrupt the tissue, then a QIAshredder (Qiagen, Valencia, CA) to homogenize it. RNA was extracted from each sample using an AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA). The purity, yield, and quality of RNA were evaluated via ultraviolet spectrophotometry and comparison of 28S:18S ratios using a Bioanalyzer RNA Nanochip (Agilent, Palo Alto CA). All RNA samples were of high quality and showed no signs of DNA contamination. RNA amplification was performed using the WT Expression Kit (Ambion, Austin, TX) in preparation for fragmentation and labeling.
2.3.3. Gene Expression Quantification
Genome-wide quantification of mRNA expression was accomplished using Affymetrix GeneChip Rat Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA). This chip contained approximately 40 probes per gene and four probes per exon, thus providing information sufficient for determining differential gene expression as well as alternative splicing between the control- and VPA-exposed rat groups. Hybridization and scanning of microarrays were performed as previously described (Glatt et al., 2009).
2.3.4. Statistical Analysis
CEL files containing microarray probe intensities were imported into Partek Genomics Suite Software (Partek Inc., St. Louis, MO) for all subsequent analyses. Data for 80,908 probes on the microarray were summarized in two steps: 1) probes for the same exon were summarized by Tukey's biweight to get one measurement per exon; and 2) exons of the same gene were also summarized by Tukey's biweight to get one measurement per gene. Chips were quantile-normalized by robust multi-array average and probe intensities were log-2 transformed to yield data that more closely approximated normality. Probes with a signal:noise ratio less than 3.0 were not used in the analysis.
For analyses of gene (i.e., full-length transcript) expression, data in each brain region (within-subjects) were analyzed jointly in a mixed analysis of variance (ANOVA) to isolate regionally specific and ubiquitous effects of treatment group (between-subjects). In addition to the main effects of prenatal exposure and brain region, we also modeled the interaction of these two terms, and compared the results of post-hoc analyses (one-way ANOVAs with prenatal treatment as the lone factor) in each brain region via intersection/union tests (IUTs). Due to the large number of tests performed and the small number of subjects, we would not anticipate observing results that surpassed a Bonferroni-corrected threshold for statistical significance, nor would we ascribe much weight to nominally significant results for individual genes found to be dysregulated in only one brain region; therefore, the main outcome of these analyses involved the identification of gene ontologies, pathways, and protein domains that were over-represented among sets of genes dysregulated in each brain region and across brain regions. These pathway/ontology/domain analyses were conducted using DAVID (Dennis et al., 2003).
For analyses of alternatively spliced transcript isoforms and their individual exons, separate models were employed. Prenatal VPA- and saline-exposed groups were compared on mean expression levels of all exons in each gene on a gene-by-gene basis through ANCOVAs and inspection of interaction terms, as described previously (Partek Incorporated, 2008; Glatt et al., 2009). Briefly, treatment was added as a between-subjects factor, and since not all exons in a gene express at the same level, exon identity (ID) was added to the model to account for exon-to-exon differences. Also, since multiple measurements (on the multiple exons) come from the same subject, subject ID was added to the model to accommodate the assumption of independence that is fundamental to ANCOVA. The final term included was the interaction of exon ID with treatment group, which allowed for the detection of differences in the expression of one or more (but not all) exons in a gene between the two treatment groups (Partek Incorporated, 2008). The significance of these interaction terms (one per gene) was judged against a stringent Bonferroni-corrected threshold, and post-hoc F-tests were used to identify the specific dysregulated exon(s) in the genes showing significant interactions. The analysis was performed separately in each of the three brain regions and ubiquitously dysregulated isoforms were identified by IUTs of the results from each region. Ontological analysis of genes that show dysregulated alternative splicing was also conducted using DAVID for each brain region in order to discover enriched biological themes.
3. RESULTS
3.1. Litter outcomes
One-way ANOVA identified a treatment-induced difference in average pup weight (untreated animals 7.85 g, saline-exposed animals 8.07 g, VPA-exposed animals 7.20 g; F2,25=3.627; p=0.043). A post-hoc Tukey B-test found that the difference in body weight of animals exposed to VPA compared with saline-exposed animals was just shy of significance (p=0.051). The number of pups and the proportion of males was unaffected by treatment. For all treatment groups, there was an average of 11.3 pups per litter (F2,25=0.0402; p=0.961), and 50% of untreated or saline-exposed pups and 55% of VPA-exposed pups were male (F2,25=0.339; p=0.716). We observed similar maternal culling between the different treatments, and dams seemed to normally retrieve pups into nest regardless of maternal exposure. This is in agreement with others who report that exposure to VPA during pregnancy does not alter maternal behavior (Mychasiuk et al., 2012)
3.2. Social behavior study
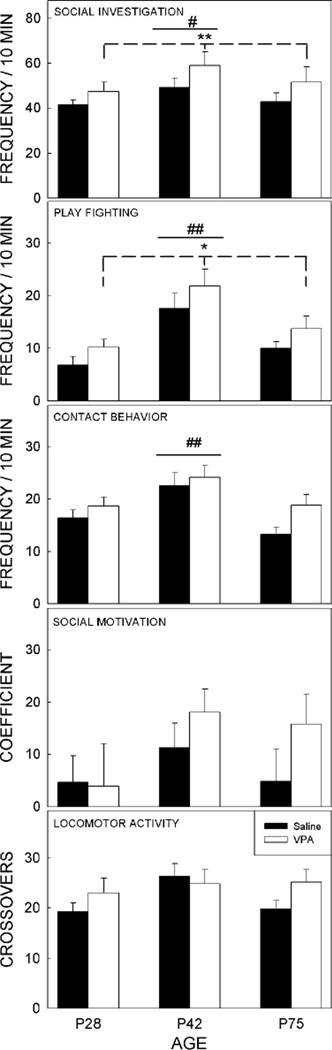
A modified social interaction test was performed and four indices of social behavior as well as locomotor activity were scored and analyzed (Fig. 1). Play fighting differed as a function of age (F2,97=13.54, p<0.001), with 42-day-old animals showing more play than their older and younger counterparts. Prenatal exposure to VPA increased this form of social interaction regardless of age (main effect of treatment, F1,97=4.09, p<0.05).
Figure 1.
Effects of Exposure to Valproic Acid on Social Behavior.
Four features of social behavior as well as locomotor activity were analyzed from a modified social interaction test. Both social investigation and play fighting, were significantly increased in animals exposed to valproic acid when the data were collapsed across age. Three features, social investigation, play fighting, and contact behavior, were significantly increased in 42-day-old animals compared with those tested on P28 or P75 (data collapsed across treatment). Neither social motivation nor locomotor activity showed a significant effect of age or prenatal treatment.
Significant differences between saline- and valproic acid-exposed animals: * p < 0.05, ** p<0.01. Significant differences between age groups: # p < 0.01, ## p<0.001. Bars show the mean for each group, t-bars depict the standard error of the mean. No differences between the sexes were identified for any of the measures, thus data were collapsed across sex.
For social investigation, there was also a significant main effect of test age, (F2,97=5.55, p<0.01) with 42-day-old animals demonstrating more social investigation than rats tested on P28 or P75. Social investigation also differed as a function of prenatal treatment (F1,97=7.05, p<0.01), with animals exposed to VPA demonstrating more social investigation than controls.
Contact behavior was not affected by prenatal treatment and differed as a function of age only (F2,97=7.52, p<0.001), with 42-day-old adolescents demonstrating more contact behavior than younger and older animals.
The coefficient of social preference did not differ as a function of age (F2,97=1.82, p=0.1673) or treatment (F1,97=1.40, p=0.2398), with all animals demonstrating equivalent levels of social preference. Locomotor activity within the social context, as determined by the number of crossings from one part of the test box to the other, was not altered by treatment (F1,97=1.51, p=0.2215) or by age (F2,97=1.79, p=0.1716).
3.3. Microarray Study
Gene-expression analysis was done in the 75-day-old animals as this age targets those genes that most likely are permanently altered. No sex differences were seen in the behavioral analyses; thus, gene expression data from male and female rats were combined to maximize statistical power to detect the main effect of interest (VPA exposure).
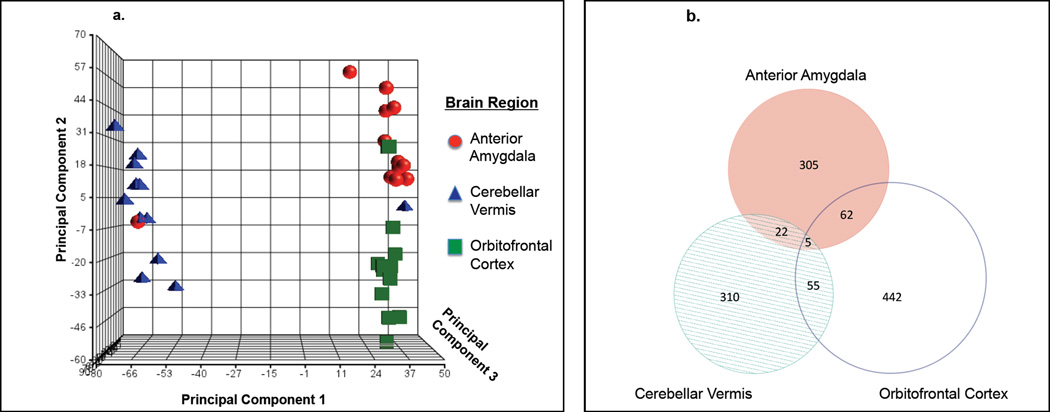
Quality-control analyses on normalized transformed data involved inspection of sample histograms of log2 expression intensities of each probe and box-plots of the mean and variance of log2 expression intensities, neither of which identified any outliers. The high degree of overlap in these parameters across all samples demonstrated that, after transformation and normalization, all samples had highly comparable distributions, thus allowing for their comparison via parametric inferential statistics. The first three principal components of transcriptome-wide gene expression were also visualized, identifying three distinct clusters of data segregated by brain region (Fig. 2a). Two samples (from two brain regions of the same subject) did not map to the appropriate cluster, which most likely reflected an experimental error of labeling or processing, therefore these two data points (as well as the third brain-region data point from this same subject) were removed from all further analyses. Data from each of the three brain regions clustered tightly together (Fig. 2a), but there was no clustering by prenatal treatment, suggesting that the exposure to VPA in pregnancy does not alter the fundamental and biologically critical correlations between expression levels of most genes. In other words, when certain genes are highly expressed, we would expect, biologically, that others would be highly expressed while some others would be expressed at very low levels. VPA exposure during pregnancy does not alter this fundamental biological co-expression and joint regulation; however, clearly the co-expression and joint regulation of sets of genes is different in discrete brain regions.
Figure 2.
a. Principal Components Analysis of Rat Brain Transcriptome.
Individual data points (three per subject, one per brain region) were mapped to the first three principal components (P. C.) of gene expression variance derived from the entire rat transcriptome. Data points from each brain region showed very strong correspondence with each other but no relation to prenatal treatment, while data from each individual subject was clearly distributed across the three clusters defined by brain region, with the exception of a single subject that was removed from all subsequent analyses. This demonstrates the brain-regional specificity of the co-expression of genes in the rat transcriptome and the relative integrity of its global characteristics and gene-wise correlations despite prenatal VPA exposure.
b. Venn Diagram of Whole-Gene Clusters Significantly Dysregulated by VPA Treatment in Each Brain Region
Most genes dysregulated by prenatal VPA treatment were only affected in one of the three evaluated brain regions. In contrast to these regionally specific effects of prenatal VPA exposure on gene expression, there were also 139 instances where prenatal VPA exposure had long-lasting effects on gene expression in two of the three regions, with the anterior amygdala and orbitofrontal cortex highest in similarity. There were also 5 instances where the effects of VPA were persistent and ubiquitous across all three brain regions
702 (8.0%) of the 8,762 genes represented by the nearly 81,000 probes on the utilized microarray showed a nominally significant main effect of prenatal treatment (p<0.05). Although none surpassed a strict Bonferroni-corrected significance level, this is not surprising given the small sample size of this pilot study. Critically, however, the top 163 results had a false-discovery rate q-value of just 0.40, meaning that while 40% of these results could be false-positives, 60% could be true discoveries. Yet, beyond simply identifying individual genes that were dysregulated by prenatal VPA exposure, we primarily hypothesized that VPA exposure would affect biologically related groups of genes, which was assessed using annotation-enrichment analyses in DAVID. Among the 702 nominally significantly dysregulated genes, there was a 1.5-fold enrichment of genes (121) that encode acetylation-sensitive proteins, relative to the ratio of acetylation-sensitive genes expected based on their number in the rat genome. This enrichment was highly significant (p=2.3e−6), and remained significant even after applying the Bonferroni correction for the number of pathways, ontologies, and protein domains evaluated in DAVID (corrected p=1.0e−3). This finding lends support to our principal hypothesis that prenatal VPA exposure affects gene expression via its actions as a deacetylation inhibitor. Two other annotation categories survived Bonferroni correction, including “FAD-dependent pyridine nucleotide-disulphide oxidoreductase” (corrected p=5.6e−3) which showed a 9.9-fold enrichment, and “negative regulation of catalytic activity” (corrected p=2.8e−2) which showed a 2.6-fold enrichment.
Beyond the main effect of prenatal VPA treatment across brain regions, we also found that 247 (2.8%) of the 8,762 genes assayed showed a nominally significant interaction between prenatal treatment and brain region (p<0.05). Again, because of the small sample size and the relatively lower power to detect interactions than main effects in general, none of the detected interactions surpassed a Bonferroni-corrected significance level. Closer inspection of the regional distribution of those genes that did evince a significant interaction of brain region with prenatal treatment revealed some quite strong and consistent regional differences between the two prenatal treatment groups. As shown in figure 2b, 394 genes were significantly dysregulated in AA, 392 in CV, and 564 in OFC (Supplementary Tables 1, 2, and 3 respectively). There are also instances (k=149) where prenatal VPA exposure had effects on gene expression in two of the three regions (with greatest similarity between anterior amygdala and orbitofrontal cortex), as well as some instances (k=5) where the effects of VPA were persistent and ubiquitous across all three brain regions examined (Fig. 2b). Each of the five ubiquitously dysregulated genes exhibited a consistent effect (i.e., changed in the same direction) across all brain regions (Table 1).
Table 1.
Genes Significantly Dysregulated in All Three Evaluated Brain Regions after Prenatal Valproic Acid Treatment
| Anterior Amygdala | Cerebellar Vermis | Orbitofrontal Cortex | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcript Cluster ID |
Reference Sequence |
Gene Symbol |
Gene Product |
p |
Log2 Fold- Change |
p |
Log2 Fold- Change |
p |
Log2 Fold- Change |
| 7043913 | NM_013073 | Pcmt1 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase | 0.0243 | −1.19 | 0.0127 | −1.25 | 0.0419 | −1.19 |
| 7059188 | NM_021868 | Cttn | Src substrate cortactin | 0.0059 | −1.17 | 0.0324 | −1.09 | 0.0379 | −1.16 |
| 7066168 | NM_019273 | Kcnmb1 | Calcium-activated potassium channel subunit beta-1 | 0.0288 | 1.25 | 0.0448 | 1.31 | 0.0423 | 1.44 |
| 7216980 | NM_212529 | Hsd17b8 | Estradiol 17-beta-dehydrogenase 8 | 0.0297 | −1.37 | 0.0093 | −1.33 | 0.0263 | −1.34 |
| 7288291 | NM_001007144 | Plin2 | Perilipin-2 | 0.0002 | −1.36 | 0.0222 | −1.47 | 0.0207 | −1.33 |
Brain region-specific ontological analysis of all nominally significant whole genes did not uncover enrichments that surpassed Bonferroni-corrected significance at any brain region. However, independent analysis of genes that displayed increased expression and genes that displayed decreased expression did identify enrichments that surpassed this correction (Table 3).
Table 3.
Significant Annotation Clustering Enrichments In Response to VPA by Brain-Region-Specific Ontological Analysis
| Anterior Amygdala | Cerebellar Vermis | Orbitofrontal Cortex | ||||
|---|---|---|---|---|---|---|
| Alternative Splicing Dysregulated | Neuron projection | Neuron projection | Cell projection | |||
| Cell-cell signaling | Cellular homeostasis (ion/channel) | Homeostatic process | ||||
| Phosphoprotein | Phosphoprotein | Phosphoprotein | ||||
| Protein domain specific binding | Calcium ion binding and transport | Purine nucleotide binding | ||||
| Synaptic transmission | Plasma membrane | Plasma membrane | ||||
| Vesicle (membrane-bound/cytoplasmic) | Vesicle (membrane-bound/cytoplasmic) | |||||
| Whole Gene Transcript Dysregulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated |
| Glycoprotein | Acetylation | Glycoprotein | Acetylation (p=0.09) | Neurological system | Nucleotide binding (p=0.06) | |
| Nucleoside binding | Sensory perception | |||||
| Galactose metabolism | Cognition (p=0.09) | |||||
Gene-annotation enrichments that surpassed Bonferroni-corrected level of significance (p-value indicated for marginally significant enrichments)
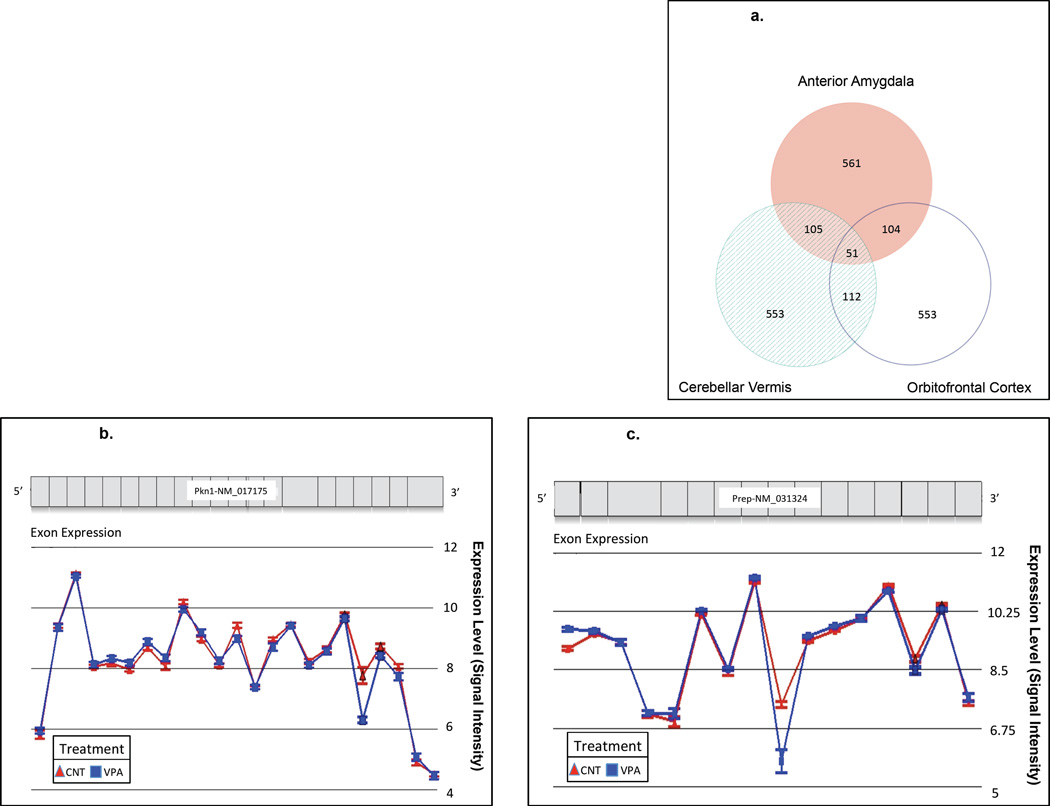
Alternative splicing was also evaluated by identifying genes that have differential expression of one or more exons in response to prenatal VPA exposure (Fig. 3). Unlike the analysis of full-length gene expression aggregating all isoforms, the exon-level analysis was able to detect several effects that surpassed Bonferroni-corrected significance thresholds, perhaps due in part to the fact that exon expression is better controlled via comparison to other exons in the same gene, and perhaps because exons (which index particular alternatively spliced isoforms) are more biologically meaningful elements than genes. Nominally significant interaction for alternative splicing and treatment was seen in 821, 821, and 820 genes in AA, CV, and OFC; of these 22, 11, and 11, respectively, survived Bonferroni correction (Supplementary Tables 4, 5, and 6 respectively). The differential exon expression of prolyl endopeptidase (Prep) in VPA-exposed subjects surpassed a Bonferroni-corrected level of significance in all three brain regions (AA, CV, and OFC). Further analysis of ubiquitous alternative-splicing anomalies revealed 51 genes that were nominally significant in all three brain regions (Table 2), many of which surpassed Bonferroni-corrected significance levels in one or two of the three regions.
Figure 3.
a. Venn Diagram of Genes with Significantly Dysregulated Alternative Splicing by VPA Treatment in Each Brain Region
Most genes with dysregulated alternative splicing by prenatal VPA treatment were only affected in one of the three evaluated brain regions. In contrast to these regionally specific effects of prenatal VPA exposure on gene expression, there were also 321 instances where prenatal VPA exposure had long-lasting effects on gene expression in two of the three regions, with the cerebellar vermis and orbitofrontal cortex having highest in similarity. There were also 51 instances where the effects of VPA were persistent and ubiquitous across all three brain regions.
b. Exonic expression of Serine/threonine-protein kinase N1 (Pkn1).
Exonic expression of Pkn1 across all three brain regions: anterior amygdala (AA), cerebellar vermis (CV), and oribitofrontal cortex (OFC) from 5 prenatally valproic acid (VPA) exposed rats and 6 saline control (CNT) rats. Differential expression of exon 19 is seen between exposure groups.
c. Exonic expression of Prolyl endopeptidase (Prep)
Exonic expression of Prep across all three brain regions: anterior amygdala (AA), cerebellar vermis (CV), and oribitofrontal cortex (OFC) from 5 prenatally valproic acid (VPA) exposed rats and 6 saline control (CNT) rats. Differential expressions of exons 1 and 8 are seen between exposure groups for all three brain regions.
Table 2.
Alternative Splicing Significantly Dysregulated in All Three Evaluated Brain Regions after Prenatal Valproic Acid Treatment
| Anterior Amygdala | Cerebellar Vermis | Orbitofrontal Cortex | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcript Cluster ID |
Reference Sequence |
Gene Symbol |
Gene Product | F | p | F | p | F | p |
| 7213582 | NM_012804 | Abcd3 | ATP-binding cassette sub-family D member 3 | 3.481 | 6.93E-08* | 1.532 | 4.73E-02 | 2.574 | 5.54E-05 |
| 7082684 | NM_016987 | Acly | ATP-citrate synthase | 1.914 | 4.95E-03 | 1.721 | 1.61E-02 | 1.657 | 2.35E-02 |
| 7123967 | NM_012493 | Afp | alpha-fetoprotein | 1.973 | 1.61E-02 | 3.672 | 7.32E-06 | 1.983 | 1.54E-02 |
| 7337058 | NM_012903 | Anp32a | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | 4.110 | 7.62E-03 | 5.783 | 1.06E-03 | 4.233 | 6.55E-03 |
| 7377029 | NM_001024367 | Armcx1 | Armadillo repeat-containing X-linked protein 1 | 3.937 | 2.46E-03 | 2.45 | 3.62E-02 | 4.330 | 1.23E-03 |
| 7349069 | NM_012913 | Atp1b3 | ATPase, Na+ | 5.745 | 4.01E-02 | 10.564 | 9.99E-03 | 5.188 | 4.88E-02 |
| 7268482 | NM_012508 | Atp2b2 | Plasma membrane calcium-transporting ATPase 2 | 4.219 | 1.75E-08* | 2.605 | 2.42E-04 | 2.217 | 2.14E-03 |
| 7378504 | NM_133288 | Atp2b3 | Plasma membrane calcium-transporting ATPase 3 | 2.751 | 4.02E-05 | 2.628 | 8.95E-05 | 3.634 | 1.12E-07* |
| 7385140 | NM_031785 | Atp6ap1 | ATPase, H+ transporting, lysosomal accessory protein 1 | 2.437 | 9.86E-03 | 3.778 | 1.56E-04 | 3.458 | 4.21E-04 |
| 7384924 | NM_001004224 | Bcap31 | B-cell receptor-associated protein 31 | 2.272 | 3.15E-02 | 2.597 | 1.49E-02 | 2.138 | 4.29E-02 |
| 7137201 | NM_012827 | Bmp4 | bone morphogenetic protein 4 | 2.235 | 4.28E-02 | 2.307 | 3.69E-02 | 2.722 | 1.57E-02 |
| 7335781 | NM_001012201 | Cadm1 | cell adhesion molecule 1 | 3.122 | 7.81E-04 | 3.65 | 1.32E-04 | 5.360 | 4.69E-07* |
| 7092876 | NM_019195 | Cd47 | Cd47 molecule | 2.314 | 2.27E-02 | 3.021 | 3.69E-03 | 2.480 | 1.49E-02 |
| 7215277 | NM_201419 | Clca4 | chloride channel accessory 4 | 8.108 | 9.65E-16* | 2.093 | 6.89E-03 | 2.371 | 1.82E-03 |
| 7065253 | NM_133381 | Crebbp | CREB binding protein | 2.778 | 5.13E-07* | 1.909 | 1.48E-03 | 1.991 | 7.44E-04 |
| 7083616 | NM_001007613 | Ddx5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 2.503 | 3.28E-02 | 2.000 | 3.09E-02 | 1.892 | 4.30E-02 |
| 7089864 | NM_001012472 | Dgcr14 | DiGeorge syndrome critical region protein 14 | 2.849 | 4.00E-03 | 2.047 | 3.73E-02 | 2.067 | 3.53E-02 |
| 7198615 | NM_013057 | F3 | Tissue factor Precursor | 5.375 | 3.39E-06* | 3.932 | 1.84E-04 | 3.595 | 4.78E-04 |
| 7340407 | NM_021660 | Ip6k2 | Inositol hexakisphosphate kinase 2 | 9.178 | 9.05E-08* | 3.964 | 1.20E-03 | 4.646 | 3.05E-04 |
| 7231634 | NM_031045 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | 2.185 | 2.56E-02 | 2.495 | 1.09E-02 | 2.672 | 6.61E-03 |
| 7326239 | NM_212523 | Kif5a | kinesin family member 5A | 1.850 | 6.62E-03 | 1.634 | 2.49E-02 | 1.639 | 2.42E-02 |
| 7070380 | NM_001012011 | Lig3 | DNA ligase | 2.548 | 4.40E-04 | 1.891 | 1.35E-02 | 2.154 | 3.59E-03 |
| 7333434 | NM_001025054 | LOC500956 | hypothetical protein LOC500956 | 2.512 | 1.04E-02 | 4.395 | 5.00E-05 | 5.167 | 5.96E-06* |
| 7322009 | NM_021859 | Matk | Megakaryocyte-associated tyrosine-protein kinase | 4.746 | 1.47E-06* | 2.455 | 5.55E-03 | 2.407 | 6.57E-03 |
| 7059820 | NM_001015013 | Mtvr2 | mammary tumor virus receptor 2 | 3.766 | 4.30E-02 | 5.004 | 1.87E-02 | 4.918 | 1.98E-02 |
| 7322533 | NM_020087 | Notch3 | Notch homolog 3 (Drosophila) | 1.647 | 1.02E-02 | 1.474 | 3.65E-02 | 1.433 | 4.83E-02 |
| 7233312 | NM_012746 | Pcsk2 | Neuroendocrine convertase 2 Precursor | 2.616 | 4.23E-03 | 2.02 | 2.90E-02 | 2.379 | 9.22E-03 |
| 7289582 | NM_199253 | Pcsk9 | proprotein convertase subtilisin | 1.742 | 4.50E-02 | 2.085 | 1.19E-02 | 1.756 | 4.28E-02 |
| 7320471 | NM_031715 | Pfkm | 6-phosphofructokinase | 2.652 | 1.85E-04 | 2.008 | 6.53E-03 | 2.029 | 5.85E-03 |
| 7179966 | NM_017175 | Pkn1 | Serine/threonine-protein kinase N1 | 4.128 | 3.01E-08* | 3.43 | 1.92E-06* | 3.129 | 1.14E-05 |
| 7329136 | NM_001164298 | Plec | plectin | 1.454 | 1.61E-02 | 1.504 | 9.55E-03 | 1.565 | 4.99E-03 |
| 7339147 | NM_057194 | Plscr1 | phospholipid scramblase 1 | 2.510 | 1.82E-02 | 2.148 | 4.19E-02 | 2.956 | 6.46E-03 |
| 7139367 | NM_053999 | Ppp2r2a | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha | 2.092 | 9.72E-03 | 3.057 | 1.28E-04 | 2.888 | 2.79E-04 |
| 7219948 | NM_031324 | Prep | Prolyl endopeptidase | 5.697 | 6.12E-09* | 4.335 | 1.46E-06* | 5.013 | 9.27E-08* |
| 7300035 | NM_019163 | Psen1 | Presenilin-1 | 5.727 | 4.40E-07* | 7.000 | 1.20E-08* | 4.297 | 3.15E-05 |
| 7302329 | NM_031600 | Ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | 1.845 | 9.58E-03 | 1.782 | 1.38E-02 | 2.220 | 9.66E-04 |
| 7042063 | NM_001004261 | Pyroxd2 | Pyridine nucleotide-disulfide oxidoreductase domain 2 | 3.345 | 5.41E-05 | 2.558 | 1.66E-03 | 4.071 | 2.22E-06* |
| 7119529 | NM_022390 | Qdpr | quinoid dihydropteridine reductase | 3.468 | 5.68E-03 | 4.999 | 3.89E-04 | 5.954 | 7.97E-05 |
| 7174465 | NM_017315 | Slc23a1 | solute carrier family 23 (nucleobase transporters), member 1 | 2.071 | 1.49E-02 | 2.049 | 1.61E-02 | 2.348 | 5.07E-03 |
| 7138066 | NM_053442 | Slc7a8 | solute carrier family 7 (cationic amino acid transporter, y+ sys | 2.471 | 6.82E-03 | 1.927 | 3.87E-02 | 2.626 | 4.09E-03 |
| 7365728 | NM_173338 | Slco6c1 | solute carrier organic anion transporter family, member 6c1 | 2.108 | 1.84E-02 | 1.828 | 4.64E-02 | 2.984 | 8.41E-04 |
| 7348411 | NM_031728 | Snap91 | Clathrin coat assembly protein AP180 | 1.983 | 3.18E-03 | 2.041 | 2.18E-03 | 2.249 | 5.42E-04 |
| 7045017 | NM_175583 | Taar4 | Trace amine-associated receptor 4 | 3.576 | 4.68E-03 | 3.161 | 9.91E-03 | 5.199 | 2.77E-04 |
| 7320452 | NM_001008358 | Tmem106c | transmembrane protein 106C | 2.468 | 2.65E-02 | 2.719 | 1.57E-02 | 2.952 | 9.69E-03 |
| 7303979 | NM_001014195 | Tmem214 | transmembrane protein 214 | 2.277 | 8.30E-03 | 2.045 | 1.93E-02 | 2.778 | 1.26E-03 |
| 7219299 | NM_001013033 | Tspyl1 | TSPY-like 1 | 21.453 | 1.23E-03 | 5.538 | 4.31E-02 | 16.525 | 2.82E-03 |
| 7378941 | NM_001013933 | Ube2a | ubiquitin-conjugating enzyme E2A | 3.788 | 3.21E-03 | 3.404 | 6.38E-03 | 5.201 | 2.76E-04 |
| 7084520 | NM_138844 | Unc13d | Protein unc-13 homolog D | 1.841 | 4.03E-03 | 1.69 | 1.18E-02 | 1.884 | 2.91E-03 |
| 7154203 | NM_001001516 | Usp19 | ubiquitin specific peptidase 19 | 1.737 | 1.34E-02 | 1.985 | 2.73E-03 | 1.858 | 6.25E-03 |
| 7331440 | NM_017058 | Vdr | Vitamin D3 receptor | 3.091 | 4.71E-03 | 3.426 | 2.16E-03 | 3.630 | 1.34E-03 |
| 7295338 | NM_017154 | Xdh | xanthine dehydrogenase | 1.495 | 4.03E-02 | 2.690 | 2.96E-06* | 2.267 | 1.17E-04 |
Genes that surpassed Bonferroni corrected significance for that brain region.
Genes that may be relevant to social interaction behavior are indicated in bold.
Brain region-specific ontological analysis of genes that showed dysregulated alternative splicing revealed Bonferroni-corrected significant enrichments that were unique to the specialized brain region; however, most seem to span at least two, if not all three, brain regions (Table 3). While, “calcium ion binding” and “calcium ion transport” enrichments were found exclusively in the CV, “cell projection” and “vesicle” enrichments were found in three and two brain regions, respectively.
4. DISCUSSION
4.1 Behavior
In our study, acute prenatal exposure to a moderate dose of VPA appeared to enhance social behavior. Social investigation and play fighting occurred at a significantly (p<0.05) higher frequency in animals exposed to VPA than in saline-exposed animals at all ages of testing. In addition, social investigation, play fighting, and contact behavior differed as a function of age, with higher frequencies of these behaviors apparent in 42-day-old animals. All animals, regardless of prenatal treatment, showed social preference suggesting that prenatal exposure to VPA had no anxiogenic effects. Locomotor activity under social circumstances was also not affected by treatment or age.
The fact that exposure to a moderate dose of VPA (350 mg/kg) enhances social behavior is intriguing as it is in direct contrast to reports that social behavior is negatively affected by exposure to higher doses of VPA given at the same gestational age (Schneider et al., 2001; Dennis et al., 2003; Schneider & Przewlocki, 2005; Stanton et al., 2007; Markram et al., 2008; Schneider et al., 2008; Dufour-Rainfray et al., 2010). The different outcomes may be partly attributable to differences in the social behavior tests, but are also likely to be a function of the different doses used. It should also be noted that increased social behavior is as aberrant as decreased and thus, should not be considered an auspicious outcome. Combining the current data with that reported previously by others, we show that the dose of this teratogen defines the outcome.
4.2.1 Gene Expression: Whole gene
The microarray analysis identified a nominally significant main effect of prenatal treatment in 8.0% (702) of the 8,762 genes probed, and 2.8% (247) of genes showed a nominally significant interaction between prenatal treatment and brain region. Among the 702 genes that show main effect of treatment, there was a 1.5-fold enrichment of genes encoding acetylation-sensitive proteins, consistent with the role of VPA as a histone and non-histone deacetylase inhibitor (Phiel et al., 2001; Mannaerts et al., 2010). Our experimental outcomes represent the long-term effects of prenatal manipulations, thus, it is reasonable to consider these as epigenetic effects of VPA; however, manipulation of protein activity during critical periods can also lead to lifelong changes in gene expression (Detich et al., 2003b; a). Brains for mRNA analysis were taken from animals after completion of a social behavior test; thus, the described gene effects may be an interaction of prenatal exposure to VPA and the sum of all postnatal experiences, including the social interaction test.
Brain region-specific whole-gene dysregulations are particularly interesting when considering the specialized behavioral-related function of each of the brain regions analyzed, however, none of our transcripts surpassed a Bonferroni-corrected level of significance when both up-regulated and down-regulated genes were used. VPA acts as a histone deacetylase inhibitor, thereby promoting gene expression, thus, we divided each brain-region analysis into separate groups of up- and down-regulated genes. Interestingly, acetylation-sensitive proteins showed enrichment in the down-regulated genes. This enrichment was significant in the AA and marginally significant in the CV. The AA and CV also showed similarities in enrichments from the up-regulated genes, perhaps signifying similar pathologies (Table 3). The enrichments found from whole-genes dysregulated in the OFC seemed to be the most localized, and were more obviously relevant to social interaction behaviors. This is likely due to the specialized cognitive function of the OFC. A closer look at the individual genes that make up these enrichments and their interaction with each other will be an interesting avenue for future work.
Five genes were ubiquitously dysregulated by prenatal exposure to VPA ; that is, they changed in the same direction across all brain regions examined. Of these, one was up-regulated and four were down-regulated in all three regions (Table 1). The gene that was up-regulated, Kcnmb1 (Calcium-activated potassium channel subunit beta-1), is a regulatory subunit of the calcium activated potassium KCNMA1 channel, controlling both the sensitivity and gating kinetics of this channel (Meera et al., 1996). Genes that were down-regulated did not fall into any defined categories: Cttn (Src substrate cortactin) plays a role in cell migration and adhesion (von Holleben et al., 2011),, Pcmt1 (Protein-L-isoaspartate (D-aspartate) O-methyltransferase) is involved in protein repair and degradation, Plin2 (Perlipin-2) is involved with maintenance of adipose tissue, and Hsp17b8 (Estradiol 17-beta-dehydrogenase 8) is important for biosysthesis of fatty acids (Chen et al., 2009; McIntosh et al., 2012). In mice, Hsp17b8 has been found to regulate estradiol and testosterone activity (Ohno et al., 2008).
4.2.2 Gene Expression: Exon-level analysis
Results from the individual exon-level analysis showed that each brain region had a distinct pattern of significantly dysregulated exons. Most gene-annotation enrichments that surpassed a Bonferroni-corrected level of significance, including “neuronal projection”, “vesicle”, and “plasma membrane”, were not localized in one particular brain region (Table 3). However, calcium-related enrichments were found exclusively in the CV and could suggest a localized disturbance in that pathway.
Genes that showed differential exon expression across all brain regions should perhaps take priority for follow-up studies since they signify a more global, and thus, a more traceable effect of exposure (Table 2). CREB binding protein, encoded by the Crebbp gene, showed an interaction of treatment and exon ID that surpassed Bonferroni-corrected significance in AA and had nominal significance in the CV and OFC. This gene plays a key role in embryonic development. Deletions of chromosome region 16p13.3 which harbors this gene causes Rubinstein-Taybi syndrome (RTS) in humans (Hennekam et al., 1993), and duplication 16p13.3 syndrome is thought to be directly caused by over-activity of the CREBBP gene (Demeer et al., 2013). This highlights phenotypes that are highly dosage dependent. CREBBP binds to phosphorylated cAMP response element binding protein (CREB) further synergizing its activity and increasing the expression of cAMP-responsive genes. CREB is a central player in dendritic spine organization of pyramidal neurons in the hippocampus and, consequently, of memory formation (Middei et al., 2012). CREBBP also plays an important role in controlling transcription by acetylation of histones and non-histone proteins (Chen et al., 1999; van der Horst et al., 2004). Synergy between the acetylating role of CREBBP and the deacetylation-inhibiting role of VPA remains to be explored.
The only gene with dysregulated alternative splicing that surpassed Bonferroni-corrected significance in all three brain regions was prolyl endopeptidase (Prep). Prep is a serine protease that cleaves prolyl bonds of small (less than 3 kDa) hormones and neuropeptides, including oxytocin, arginine vasopressin, neurotensin, and substance P (Welches et al., 1993). These peptides are key regulators of social behavior and therefore, impaired PREP activity has been linked to several psychiatric disorders (Peltonen & Mannisto, 2011). While activity of PREP has been negatively correlated with the severity of depression, an increased activity is seen in serum collected from individuals with schizophrenia and mania (Maes et al., 1995; Peltonen & Mannisto, 2011). The discovery that VPA inhibits PREP activity has allowed some speculation about the therapeutic mechanisms of VPA (Maes et al., 1995; Cheng et al., 2005). However, how this activity is regulated in vivo, and the long-term effects of VPA exposure during developmental stages are still unclear.
Exon-level analysis of Prep revealed several differentially expressed exons for this gene (Fig. 3c). The two exons that showed differential expression across all three brain regions were exon 1 and 8. In the VPA group exon 1 had a 1.5-fold increase in expression, while exon 8 had a 3-fold decrease. Although these don’t seem to affect any of the amino acids in the catalytic triad (Ser554, Asp641, and His680), other properties such as ligand specificity may be affected (Fulop et al., 1998). Dysregulation of an exon that could disturb the catalytic triad (exon 13) was only seen in the anterior amygdala. Exon 13 harbors Ser554 and therefore is crucial for Prep’s protease activity.
4.2.3 Gene Expression: Comparison of whole-gene and exon-level changes
An interesting interaction is seen when comparing genes that showed differential exonic expression with genes that showed differential whole-gene expression. We observed ubiquitous dysregulation of an exon in the Pkn1 gene coding for the PKC-related serine/threonine-protein kinase and ubiquitous dysregulation of the aggregated transcripts of cortactin (Cttn). In humans, PKN1 is involved in a wide range of cellular processes, including actin polymerization, cell migration, and dendritic spine morphogenesis. PKN1 also has epigenetic influence and can directly phosphorylate histones and co-activate androgen receptor-dependent transcription (Metzger et al., 2003); it also phosphorylates HDACs, which restricts their import to the nucleus (Harrison et al., 2010). Meanwhile, CTTN is involved in actin polymerization and activation of the Arp2/3 complex, and thus is also involved in cell migration (Ammer & Weed, 2008). Combining our data with evidence that PKN1 directly phosphorylates CTTN (Grassart et al., 2010), further strengthens the association of these two genes and suggests that their expression patterns may be interconnected.
Our results specify an exclusion of exon 19 of Pkn1, which in the human, corresponds to amino acids (aas) 766–812 (Fig. 3b). Dysregulation of this exon surpassed a Bonferroni-corrected threshold for statistical significance in the AA and the CV and had nominal significance in the OFC. This region does not contain the whole catalytic domain (aas 615–874); however, it does contain two known phosphorylation sites (phosphothreonine at aa 774 and aa 778). Threonine 774 is phosphorylated by phosphoinositide-dependent protein kinase-1 (PDK1) in response to insulin, and facilitates cytoskeleton reorganization (Dong et al., 2000). Other studies report that complete inhibition of the catalytic activity of Pkn1 ameliorates the cellular and behavioral deficits commonly seen in fragile × mental retardation 1 (Fmr1) knockout mice, an animal model of fragile × syndrome. (Hayashi et al., 2007). Thus, the dendritic spine abnormalities and behavioral deficits in this animal model are thought to be, at least partly, due to the catalytic activity of Pkn1. Indeed, Fmr1 knockout mice show reduced interaction of Cttn with actin (Seese et al., 2012), consistent with increased phosophorylation of this protein (Webb et al., 2006). This may suggest that VPA alters developmental alternative splicing patterns or perhaps causes a compensatory mechanism that involves exclusion of exons with catalytic domains.
4.2.4 VPA-associated Differential Expression Considerations
It is important to remember that changes in whole-gene or exon expression may be a compensatory response to VPA and not a direct effect. Alternative splicing can manipulate transcripts in response to environmental and other cellular signals in order to allow an organism to adapt. Therefore, exons that show consistent dysregulation in response to exposure may be important for cellular “coping” mechanisms and should be considered for replication and follow up studies.
Certain cellular factors, such as differential microRNA expression and protein degradation, could cause mRNA expression that is inconsistent with protein expression. However, a dysregulated exon indicates that the amino acids that are coded in that exon are missing in the expressed protein. Alternative splicing regulators are activated and directed to exclude or include a particular exon or exons from the mature mRNA transcript in order to control the type of isoform that is translated. Therefore, this cellular behavior is a functional effort, regardless of the amount of active protein found in the cell. Furthermore, it is important to note that genes that showed dysregulated exon expression (alternative splicing) did not show whole-gene expression differences. That is, the whole gene cluster averages were not different between VPA- and Saline-exposed animals. The only difference in expression that is expected is for the amino acids that are coded by the particular exon that is dysregulated. These results underscore the importance of exon-level analysis, as without it biologically important changes in expression will be missed in the analysis of whole-gene expression data.
5. Conclusion
In contrast to prior studies with higher doses of prenatal VPA exposure, none of the behavioral changes observed in this study appeared homologous to autistic behavior in humans; however, this is perhaps not unexpected due to the lower dose and acute exposure we employed. It is quite remarkable, however, that this relatively mild exposure to VPA had long-lasting behavioral and neuromolecular consequences. The opposite behavioral outcomes seen between high- and moderate-dose prenatal VPA exposures clearly illustrate the need for a more comprehensive dose-response curve. Aside from what we can learn about the teratogenicity of VPA, region-specific and ubiquitous changes in gene- and exon-expression may give insight into the mechanism(s) of VPA’s influence on social behavior.
Supplementary Material
A moderate dose of the VPA has long-term effects on social interaction behaviors.
Behavioral effects are opposite of those described after a high dose of VPA.
Transcriptomic analysis identified some intriguing differences.
Differential whole-genes and individual exons expression were identified
Acknowledgments
We thank Karen Gentile, Renee Mezza, Wendi Burnette, and Terri Novak for their technical assistance. This research was supported by Autism Speaks (SMM), the National Institute of Alcohol Abuse and Alcoholism (AA018693 and AA178231 to SMM; AA012453 and AA178231 to EIV), the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, and the National Institute of Child Health and Human Development (1P50MH 081755-020003 to SJG), and the Gerber Foundation (SJG).
ABBREVIATIONS
- aa
amino acid
- AA
anterior amygdala
- ANCOVA
analysis of covariance
- ANOVA
analysis of variance
- CREB
cAMP response element binding protein
- CREBBP
CREB binding protein
- CV
cerebellar vermis
- DNA
deoxyribose nucleic acid
- G
gestational day
- GABA
gamma-aminobutyric acid
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- ID
identity
- i.p.
intraperitoneal
- IUTs
intersection/union tests
- mg/kg
milligrams per kilogram body weight
- mRNA
messenger RNA
- OFC
orbitofrontal cortex
- P
postnatal day
- P. C.
principal component
- RNA
ribonucleic acid
- RTS
Rubinstein-Taybi syndrome
- VPA
valproic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altman J, Bayer SA. Development of the brain stem in the rat. I. Thymidine-radiographic study of the time of origin of neurons of the lower medulla. J Comp Neurol. 1980a;194:1–35. doi: 10.1002/cne.901940102. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. II. Thymidine-radiographic study of the time of origin of neurons of the upper medulla, excluding the vestibular and auditory nuclei. J Comp Neurol. 1980b;194:37–56. doi: 10.1002/cne.901940103. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. IV. Thymidine-radiographic study of the time of origin of neurons in the pontine region. J Comp Neurol. 1980c;194:905–929. doi: 10.1002/cne.901940411. [DOI] [PubMed] [Google Scholar]
- Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran O, Nergiz Y, Tuncer MC. The effects of valproic acid, vitamin E and folic acid on ribs of rat fetuses in the prenatal period. Ann Anat. 2006;188:117–125. doi: 10.1016/j.aanat.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen Z, Kastaniotis AJ, Miinalainen IJ, Rajaram V, Wierenga RK, Hiltunen JK. 17beta-hydroxysteroid dehydrogenase type 8 and carbonyl reductase type 4 assemble as a ketoacyl reductase of human mitochondrial FAS. Faseb J. 2009;23:3682–3691. doi: 10.1096/fj.09-133587. [DOI] [PubMed] [Google Scholar]
- Cheng L, Lumb M, Polgar L, Mudge AW. How can the mood stabilizer VPA limit both mania and depression? Mol Cell Neurosci. 2005;29:155–161. doi: 10.1016/j.mcn.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Daenen EW, Wolterink G, Gerrits MA, Van Ree JM. The effects of neonatal lesions in the amygdala or ventral hippocampus on social behaviour later in life. Behav Brain Res. 2002;136:571–582. doi: 10.1016/s0166-4328(02)00223-1. [DOI] [PubMed] [Google Scholar]
- DelBello MP, Strakowski SM, Zimmerman ME, Hawkins JM, Sax KW. MRI analysis of the cerebellum in bipolar disorder: a pilot study. Neuropsychopharmacology. 1999;21:63–68. doi: 10.1016/S0893-133X(99)00026-3. [DOI] [PubMed] [Google Scholar]
- Demeer B, Andrieux J, Receveur A, Morin G, Petit F, Julia S, Plessis G, Martin-Coignard D, Delobel B, Firth HV, Thuresson AC, Lanco Dosen S, Sjors K, Le Caignec C, Devriendt K, Mathieu-Dramard M. Duplication 16p13.3 and the CREBBP gene: confirmation of the phenotype. Eur J Med Genet. 2013;56:26–31. doi: 10.1016/j.ejmg.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003a;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003b;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Diergaarde L, Gerrits MA, Stuy A, Spruijt BM, van Ree JM. Neonatal amygdala lesions and juvenile isolation in the rat: differential effects on locomotor and social behavior later in life. Behav Neurosci. 2004;118:298–305. doi: 10.1037/0735-7044.118.2.298. [DOI] [PubMed] [Google Scholar]
- Dong LQ, Landa LR, Wick MJ, Zhu L, Mukai H, Ono Y, Liu F. Phosphorylation of protein kinase N by phosphoinositide-dependent protein kinase-1 mediates insulin signals to the actin cytoskeleton. Proc Natl Acad Sci U S A. 2000;97:5089–5094. doi: 10.1073/pnas.090491897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour-Rainfray D, Vourc'h P, Le Guisquet AM, Garreau L, Ternant D, Bodard S, Jaumain E, Gulhan Z, Belzung C, Andres CR, Chalon S, Guilloteau D. Behavior and serotonergic disorders in rats exposed prenatally to valproate: a model for autism. Neurosci Lett. 2010;470:55–59. doi: 10.1016/j.neulet.2009.12.054. [DOI] [PubMed] [Google Scholar]
- File SE, Hyde JR. Can social interaction be used to measure anxiety? Br J Pharmacol. 1978;62:19–24. doi: 10.1111/j.1476-5381.1978.tb07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File SE, Seth P. A review of 25 years of the social interaction test. Eur J Pharmacol. 2003;463:35–53. doi: 10.1016/s0014-2999(03)01273-1. [DOI] [PubMed] [Google Scholar]
- Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Chandler SD, Bousman CA, Chana G, Lucero GR, Tatro E, May T, Lohr JB, Kremen WS, Everall IP, Tsuang MT. Alternatively Spliced Genes as Biomarkers for Schizophrenia, Bipolar Disorder and Psychosis: A Blood-Based Spliceome-Profiling Exploratory Study. Curr Pharmacogenomics Person Med. 2009;7:164–188. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassart A, Meas-Yedid V, Dufour A, Olivo-Marin JC, Dautry-Varsat A, Sauvonnet N. Pak1 phosphorylation enhances cortactin-N-WASP interaction in clathrin-caveolin-independent endocytosis. Traffic. 2010;11:1079–1091. doi: 10.1111/j.1600-0854.2010.01075.x. [DOI] [PubMed] [Google Scholar]
- Harrison BC, Huynh K, Lundgaard GL, Helmke SM, Perryman MB, McKinsey TA. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 2010;584:1103–1110. doi: 10.1016/j.febslet.2010.02.057. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Rao BS, Seo JS, Choi HS, Dolan BM, Choi SY, Chattarji S, Tonegawa S. Inhibition of p21-activated kinase rescues symptoms of fragile × syndrome in mice. Proc Natl Acad Sci U S A. 2007;104:11489–11494. doi: 10.1073/pnas.0705003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekam RC, Tilanus M, Hamel BC, Voshart-van Heeren H, Mariman EC, van Beersum SE, van den Boogaard MJ, Breuning MH. Deletion at chromosome 16p13.3 as a cause of Rubinstein-Taybi syndrome: clinical aspects. Am J Hum Genet. 1993;52:255–262. [PMC free article] [PubMed] [Google Scholar]
- Ichimiya T, Okubo Y, Suhara T, Sudo Y. Reduced volume of the cerebellar vermis in neuroleptic-naive schizophrenia. Biol Psychiatry. 2001;49:20–27. doi: 10.1016/s0006-3223(00)01081-7. [DOI] [PubMed] [Google Scholar]
- Ingram JL, Peckham SM, Tisdale B, Rodier PM. Prenatal exposure of rats to valproic acid reproduces the cerebellar anomalies associated with autism. Neurotoxicol Teratol. 2000;22:319–324. doi: 10.1016/s0892-0362(99)00083-5. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Cooper KL, Mostofsky SH, Capone GT, Kates WR, Newschaffer CJ, Bukelis I, Stump MH, Jann AE, Lanham DC. Specificity of cerebellar vermian abnormalities in autism: a quantitative magnetic resonance imaging study. J Child Neurol. 2003;18:463–470. doi: 10.1177/08830738030180070501. [DOI] [PubMed] [Google Scholar]
- Kolozsi E, Mackenzie RN, Roullet FI, deCatanzaro D, Foster JA. Prenatal exposure to valproic acid leads to reduced expression of synaptic adhesion molecule neuroligin 3 in mice. Neuroscience. 2009;163:1201–1210. doi: 10.1016/j.neuroscience.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Kuwagata M, Ogawa T, Shioda S, Nagata T. Observation of fetal brain in a rat valproate-induced autism model: a developmental neurotoxicity study. Int J Dev Neurosci. 2009;27:399–405. doi: 10.1016/j.ijdevneu.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Loeber RT, Cintron CM, Yurgelun-Todd DA. Morphometry of individual cerebellar lobules in schizophrenia. Am J Psychiatry. 2001;158:952–954. doi: 10.1176/appi.ajp.158.6.952. [DOI] [PubMed] [Google Scholar]
- Mackie S, Shaw P, Lenroot R, Pierson R, Greenstein DK, Nugent TF, 3rd, Sharp WS, Giedd JN, Rapoport JL. Cerebellar development and clinical outcome in attention deficit hyperactivity disorder. Am J Psychiatry. 2007;164:647–655. doi: 10.1176/ajp.2007.164.4.647. [DOI] [PubMed] [Google Scholar]
- Maes M, Goossens F, Scharpe S, Calabrese J, Desnyder R, Meltzer HY. Alterations in plasma prolyl endopeptidase activity in depression, mania, and schizophrenia: effects of antidepressants, mood stabilizers, and antipsychotic drugs. Psychiatry Res. 1995;58:217–225. doi: 10.1016/0165-1781(95)02698-v. [DOI] [PubMed] [Google Scholar]
- Mah L, Arnold MC, Grafman J. Impairment of social perception associated with lesions of the prefrontal cortex. Am J Psychiatry. 2004;161:1247–1255. doi: 10.1176/appi.ajp.161.7.1247. [DOI] [PubMed] [Google Scholar]
- Mannaerts I, Nuytten NR, Rogiers V, Vanderkerken K, van Grunsven LA, Geerts A. Chronic administration of valproic acid inhibits activation of mouse hepatic stellate cells in vitro and in vivo. Hepatology. 2010;51:603–614. doi: 10.1002/hep.23334. [DOI] [PubMed] [Google Scholar]
- Markram K, Rinaldi T, La Mendola D, Sandi C, Markram H. Abnormal fear conditioning and amygdala processing in an animal model of autism. Neuropsychopharmacology. 2008;33:901–912. doi: 10.1038/sj.npp.1301453. [DOI] [PubMed] [Google Scholar]
- McIntosh AL, Senthivinayagam S, Moon KC, Gupta S, Lwande JS, Murphy CC, Storey SM, Atshaves BP. Direct interaction of Plin2 with lipids on the surface of lipid droplets: a live cell FRET analysis. Am J Physiol Cell Physiol. 2012;303:C728–C742. doi: 10.1152/ajpcell.00448.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Stewart J. Neonatal-androgens influence the social play of prepubescent rats. Horm Behav. 1981;15:197–213. doi: 10.1016/0018-506x(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Meera P, Wallner M, Jiang Z, Toro L. A calcium switch for the functional coupling between alpha (hslo) and beta subunits (Kv,cabeta) of maxi K channels. FEBS Lett. 1996;385:127–128. doi: 10.1016/0014-5793(96)83884-1. [DOI] [PubMed] [Google Scholar]
- Metzger E, Muller JM, Ferrari S, Buettner R, Schule R. A novel inducible transactivation domain in the androgen receptor: implications for PRK in prostate cancer. Embo J. 2003;22:270–280. doi: 10.1093/emboj/cdg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middei S, Spalloni A, Longone P, Pittenger C, O'Mara SM, Marie H, Ammassari-Teule M. CREB selectively controls learning-induced structural remodeling of neurons. Learn Mem. 2012;19:330–336. doi: 10.1101/lm.025817.112. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Varlinskaya EI, Mooney SM. Molecular substrates of social avoidance seen following prenatal ethanol exposure and its reversal by social enrichment. Dev Neurosci. 2012;34:115–128. doi: 10.1159/000337858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Miller MW. Time-specific effects of ethanol exposure on cranial nerve nuclei: gastrulation and neuronogenesis. Exp Neurol. 2007;205:56–63. doi: 10.1016/j.expneurol.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Varlinskaya EI. Acute prenatal exposure to ethanol and social behavior: effects of age, sex, and timing of exposure. Behav Brain Res. 2011;216:358–364. doi: 10.1016/j.bbr.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mychasiuk R, Richards S, Nakahashi A, Kolb B, Gibb R. Effects of Rat Prenatal Exposure to valproic acid on bBehavior and neuro-anatomy. Dev Neurosci. 2012;34:268–276. doi: 10.1159/000341786. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kuwagata M, Hori Y, Shioda S. Valproate-induced developmental neurotoxicity is affected by maternal conditions including shipping stress and environmental change during early pregnancy. Toxicol Lett. 2007;174:18–24. doi: 10.1016/j.toxlet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Ohno S, Nishikawa K, Honda Y, Nakajin S. Expression in E. coli and tissue distribution of the human homologue of the mouse Ke 6 gene, 17beta-hydroxysteroid dehydrogenase type 8. Mol Cell Biochem. 2008;309:209–215. doi: 10.1007/s11010-007-9637-9. [DOI] [PubMed] [Google Scholar]
- Partek Incorporated. Partek Documentation. Partek Incorporated; 2008. [Google Scholar]
- Paxinos GWC. The Rat Brain In Stereotaxic Coordinates. Burlington MA: 2009. [Google Scholar]
- Peltonen I, Mannisto PT. Effects of diverse psychopharmacological substances on the activity of brain prolyl oligopeptidase. Basic Clin Pharmacol Toxicol. 2011;108:46–54. doi: 10.1111/j.1742-7843.2010.00626.x. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Picard H, Amado I, Mouchet-Mages S, Olie JP, Krebs MO. The role of the cerebellum in schizophrenia: an update of clinical, cognitive, and functional evidences. Schizophr Bull. 2008;34:155–172. doi: 10.1093/schbul/sbm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol. 1996;91:117–119. doi: 10.1007/s004010050401. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Silberberg G, Markram H. Hyperconnectivity of local neocortical microcircuitry induced by prenatal exposure to valproic acid. Cereb Cortex. 2008;18:763–770. doi: 10.1093/cercor/bhm117. [DOI] [PubMed] [Google Scholar]
- Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. J Comp Neurol. 1996;370:247–261. doi: 10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg G. The mechanisms of action of valproate in neuropsychiatric disorders: can we see the forest for the trees? Cell Mol Life Sci. 2007a;64:2090–2103. doi: 10.1007/s00018-007-7079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg G. The mechanisms of action of valproate in neuropsychiatric disorders: can we see the forest for the trees? Cell Mol Life Sci. 2007b;64:2090–2103. doi: 10.1007/s00018-007-7079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roullet FI, Wollaston L, Decatanzaro D, Foster JA. Behavioral and molecular changes in the mouse in response to prenatal exposure to the anti-epileptic drug valproic acid. Neuroscience. 2010;170:514–522. doi: 10.1016/j.neuroscience.2010.06.069. [DOI] [PubMed] [Google Scholar]
- Rudebeck PH, Walton ME, Millette BH, Shirley E, Rushworth MF, Bannerman DM. Distinct contributions of frontal areas to emotion and social behaviour in the rat. Eur J Neurosci. 2007;26:2315–2326. doi: 10.1111/j.1460-9568.2007.05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Labuz D, Przewlocki R. Nociceptive changes in rats after prenatal exposure to valproic acid. Pol J Pharmacol. 2001;53:531–534. [PubMed] [Google Scholar]
- Schneider T, Przewlocki R. Behavioral alterations in rats prenatally exposed to valproic acid: animal model of autism. Neuropsychopharmacology. 2005;30:80–89. doi: 10.1038/sj.npp.1300518. [DOI] [PubMed] [Google Scholar]
- Schneider T, Roman A, Basta-Kaim A, Kubera M, Budziszewska B, Schneider K, Przewlocki R. Gender-specific behavioral and immunological alterations in an animal model of autism induced by prenatal exposure to valproic acid. Psychoneuroendocrinology. 2008;33:728–740. doi: 10.1016/j.psyneuen.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Seese RR, Babayan AH, Katz AM, Cox CD, Lauterborn JC, Lynch G, Gall CM. LTP induction translocates cortactin at distant synapses in wild-type but not Fmr1 knock-out mice. J Neurosci. 2012;32:7403–7413. doi: 10.1523/JNEUROSCI.0968-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow WM, Hartle K, Ivanco TL. Altered morphology of motor cortex neurons in the VPA rat model of autism. Dev Psychobiol. 2008;50:633–639. doi: 10.1002/dev.20337. [DOI] [PubMed] [Google Scholar]
- Stanton ME, Peloso E, Brown KL, Rodier P. Discrimination learning and reversal of the conditioned eyeblink reflex in a rodent model of autism. Behav Brain Res. 2007;176:133–140. doi: 10.1016/j.bbr.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- Thor DH, Holloway WR., Jr Social play in juvenile rats: a decade of methodological and experimental research. Neurosci Biobehav Rev. 1984;8:455–464. doi: 10.1016/0149-7634(84)90004-6. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Niesink RJ, Van Ree JM. The neurobiology of social play behavior in rats. Neurosci Biobehav Rev. 1997;21:309–326. doi: 10.1016/s0149-7634(96)00020-6. [DOI] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP. Acute effects of ethanol on social behavior of adolescent and adult rats: role of familiarity of the test situation. Alcohol Clin Exp Res. 2002;26:1502–1511. doi: 10.1097/01.ALC.0000034033.95701.E3. [DOI] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP. Social interactions in adolescent and adult Sprague-Dawley rats: impact of social deprivation and test context familiarity. Behav Brain Res. 2008;188:398–405. doi: 10.1016/j.bbr.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP. Sensitization to social anxiolytic effects of ethanol in adolescent and adult Sprague-Dawley rats after repeated ethanol exposure. Alcohol. 2010;44:99–110. doi: 10.1016/j.alcohol.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP, Spear NE. Social behavior and social motivation in adolescent rats: role of housing conditions and partner's activity. Physiol Behav. 1999;67:475–482. doi: 10.1016/s0031-9384(98)00285-6. [DOI] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP, Spear NE. Acute effects of ethanol on behavior of adolescent rats: role of social context. Alcohol Clin Exp Res. 2001;25:377–385. [PubMed] [Google Scholar]
- von Holleben M, Gohla A, Janssen KP, Iritani BM, Beer-Hammer S. Immunoinhibitory adapter protein Src homology domain 3 lymphocyte protein 2 (SLy2) regulates actin dynamics and B cell spreading. J Biol Chem. 2011;286:13489–13501. doi: 10.1074/jbc.M110.155184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorhees CV. Teratogenicity and developmental toxicity of valproic acid in rats. Teratology. 1987;135:195–202. doi: 10.1002/tera.1420350205. [DOI] [PubMed] [Google Scholar]
- Webb BA, Zhou S, Eves R, Shen L, Jia L, Mak AS. Phosphorylation of cortactin by p21-activated kinase. Arch Biochem Biophys. 2006;456:183–193. doi: 10.1016/j.abb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Welches WR, Brosnihan KB, Ferrario CM. A comparison of the properties and enzymatic activities of three angiotensin processing enzymes: angiotensin-converting enzyme, prolyl endopeptidase and neutral endopeptidase 24.11. Life Sci. 1993;52:1461–1480. doi: 10.1016/0024-3205(93)90108-f. [DOI] [PubMed] [Google Scholar]
- Zhang MM, Yu K, Xiao C, Ruan DY. The influence of developmental periods of sodium valproate exposure on synaptic plasticity in the CA1 region of rat hippocampus. Neurosci Lett. 2003;351:165–168. doi: 10.1016/j.neulet.2003.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.