Abstract
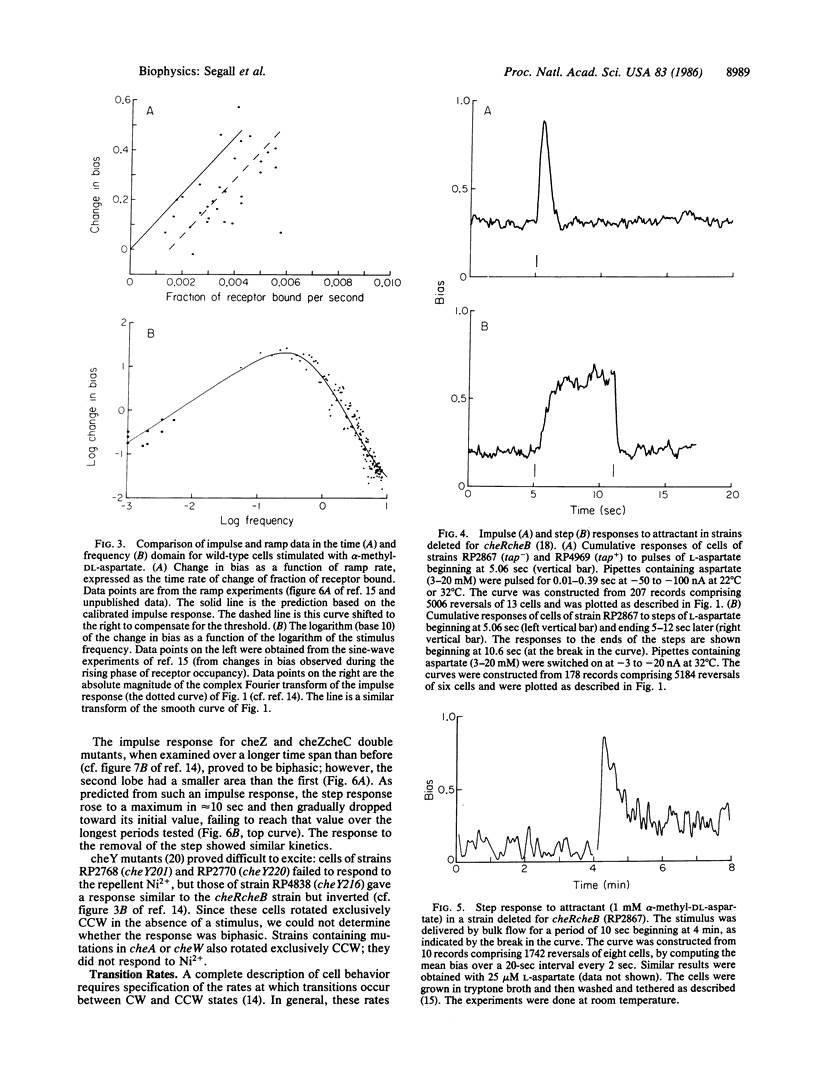
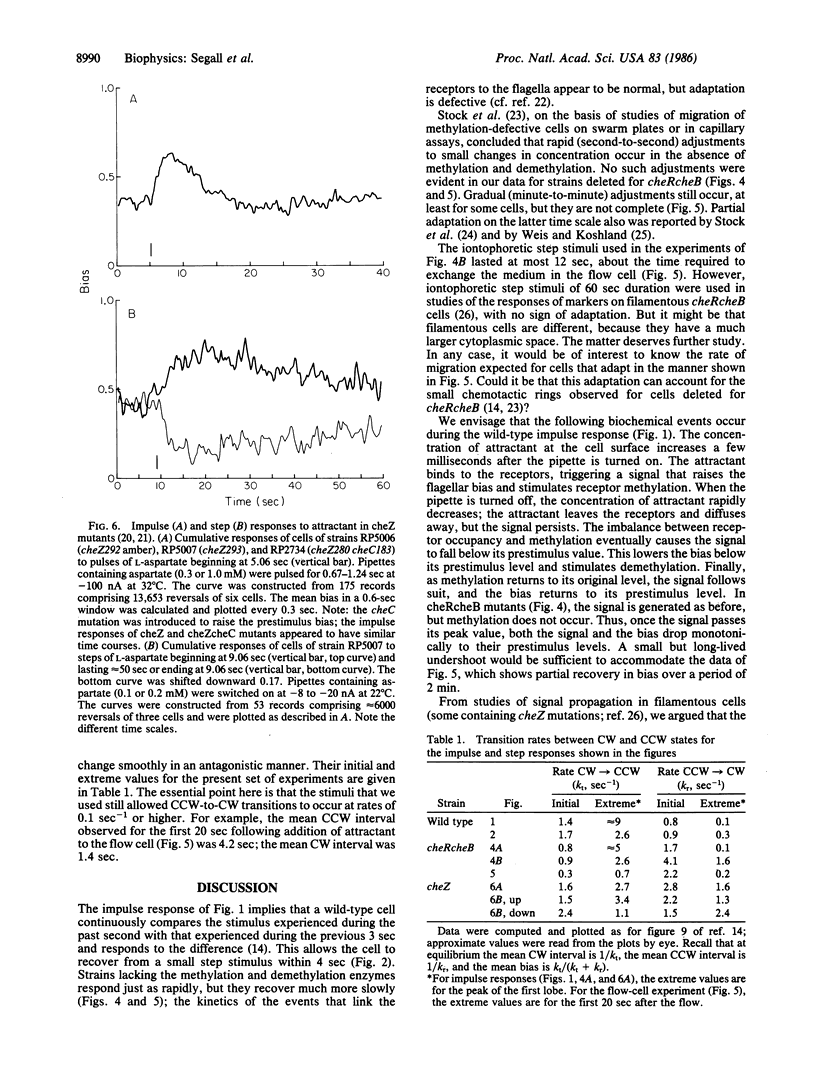
Responses of tethered cells of Escherichia coli to impulse, step, exponential-ramp or exponentiated sine-wave stimuli are internally consistent, provided that allowance is made for the nonlinear effect of thresholds. This result confirms that wild-type cells exposed to stimuli in the physiological range make short-term temporal comparisons extending 4 sec into the past: the past second is given a positive weighting, the previous 3 sec are given a negative weighting, and the cells respond to the difference. cheRcheB mutants (defective in methylation and demethylation) weight the past second in a manner similar to the wild type, but they do not make short-term temporal comparisons. When exposed to small steps delivered iontophoretically, they fail to adapt over periods of up to 12 sec; when exposed to longer steps in a flow cell, they partially adapt, but with a decay time of greater than 30 sec. cheZ mutants use a weighting that extends at least 40 sec into the past. The gain of the chemotactic system is large: the change in occupancy of one receptor molecule produces a significant response.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973 Jan;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemoreceptors in bacteria. Science. 1969 Dec 26;166(3913):1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in bacteria. Science. 1966 Aug 12;153(3737):708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Block S. M. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol. 1984 Nov;130(11):2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Brown D. A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972 Oct 27;239(5374):500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Tedesco P. M. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block S. M., Segall J. E., Berg H. C. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983 Apr;154(1):312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block S. M., Segall J. E., Berg H. C. Impulse responses in bacterial chemotaxis. Cell. 1982 Nov;31(1):215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Berg H. C. Temporal stimulation of chemotaxis in Escherichia coli. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1388–1392. doi: 10.1073/pnas.71.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S., Koshland D. E., Jr Membrane receptors for aspartate and serine in bacterial chemotaxis. J Biol Chem. 1979 Oct 10;254(19):9695–9702. [PubMed] [Google Scholar]
- Clegg D. O., Koshland D. E., Jr The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist F. W., Elwell R. A., Lovely P. S. Studies of bacterial chemotaxis in defined concentration gradients. A model for chemotaxis toward L-serine. J Supramol Struct. 1976;4(3):329–342. doi: 10.1002/jss.400040304. [DOI] [PubMed] [Google Scholar]
- Dahlquist F. W., Lovely P., Koshland D. E., Jr Quantitative analysis of bacterial migration in chemotaxis. Nat New Biol. 1972 Mar 29;236(65):120–123. doi: 10.1038/newbio236120a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Sensory transduction in bacterial chemotaxis. Int Rev Cytol. 1983;81:33–70. doi: 10.1016/s0074-7696(08)62334-7. [DOI] [PubMed] [Google Scholar]
- Larsen S. H., Reader R. W., Kort E. N., Tso W. W., Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974 May 3;249(452):74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Koshland D. E., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M., Ornston M. K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977 May 5;112(1):1–30. doi: 10.1016/s0022-2836(77)80153-8. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978 Jul;135(1):45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R. Interaction of the cheC and cheZ gene products is required for chemotactic behavior in Escherichia coli. Proc Natl Acad Sci U S A. 1979 May;76(5):2390–2394. doi: 10.1073/pnas.76.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S., Eisenbach M. Direction of flagellar rotation in bacterial cell envelopes. J Bacteriol. 1984 Apr;158(1):222–230. doi: 10.1128/jb.158.1.222-230.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E., Ishihara A., Berg H. C. Chemotactic signaling in filamentous cells of Escherichia coli. J Bacteriol. 1985 Jan;161(1):51–59. doi: 10.1128/jb.161.1.51-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E., Manson M. D., Berg H. C. Signal processing times in bacterial chemotaxis. Nature. 1982 Apr 29;296(5860):855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Koshland D. E., Jr Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Feb;72(2):710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J., Borczuk A., Chiou F., Burchenal J. E. Compensatory mutations in receptor function: a reevaluation of the role of methylation in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8364–8368. doi: 10.1073/pnas.82.24.8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J., Kersulis G., Koshland D. E., Jr Neither methylating nor demethylating enzymes are required for bacterial chemotaxis. Cell. 1985 Sep;42(2):683–690. doi: 10.1016/0092-8674(85)90125-4. [DOI] [PubMed] [Google Scholar]


