Significance
Snake venoms are toxic protein cocktails used for prey capture. To investigate the evolution of these complex biological weapon systems, we sequenced the genome of a venomous snake, the king cobra, and assessed the composition of venom gland expressed genes, small RNAs, and secreted venom proteins. We show that regulatory components of the venom secretory system may have evolved from a pancreatic origin and that venom toxin genes were co-opted by distinct genomic mechanisms. After co-option, toxin genes important for prey capture have massively expanded by gene duplication and evolved under positive selection, resulting in protein neofunctionalization. This diverse and dramatic venom-related genomic response seemingly occurs in response to a coevolutionary arms race between venomous snakes and their prey.
Keywords: genomics, phylogenetics, serpentes
Abstract
Snakes are limbless predators, and many species use venom to help overpower relatively large, agile prey. Snake venoms are complex protein mixtures encoded by several multilocus gene families that function synergistically to cause incapacitation. To examine venom evolution, we sequenced and interrogated the genome of a venomous snake, the king cobra (Ophiophagus hannah), and compared it, together with our unique transcriptome, microRNA, and proteome datasets from this species, with data from other vertebrates. In contrast to the platypus, the only other venomous vertebrate with a sequenced genome, we find that snake toxin genes evolve through several distinct co-option mechanisms and exhibit surprisingly variable levels of gene duplication and directional selection that correlate with their functional importance in prey capture. The enigmatic accessory venom gland shows a very different pattern of toxin gene expression from the main venom gland and seems to have recruited toxin-like lectin genes repeatedly for new nontoxic functions. In addition, tissue-specific microRNA analyses suggested the co-option of core genetic regulatory components of the venom secretory system from a pancreatic origin. Although the king cobra is limbless, we recovered coding sequences for all Hox genes involved in amniote limb development, with the exception of Hoxd12. Our results provide a unique view of the origin and evolution of snake venom and reveal multiple genome-level adaptive responses to natural selection in this complex biological weapon system. More generally, they provide insight into mechanisms of protein evolution under strong selection.
Snake venom contains biologically active proteins (toxins) encoded by several multilocus gene families that each comprise several distinct isoforms (1, 2). Venom is produced in a postorbital venom gland (3) and associated in elapids (cobras and their relatives) and viperids (vipers and pit vipers) with a small downstream accessory gland of unknown function (Fig. 1). Understanding the origin and evolution of the snake venom system is not only of great intrinsic biological interest (3–5), but is also important for drug discovery (1, 2, 6), understanding vertebrate physiological pathways (7, 8), and addressing public health concerns about the enormous number of snake bites suffered in tropical countries (9, 10).
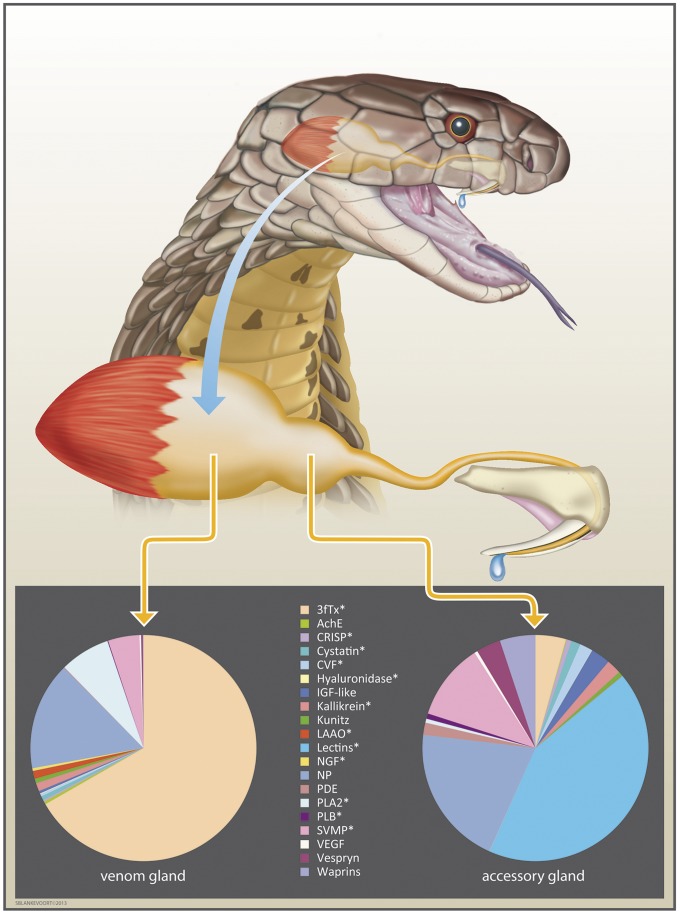
Fig. 1.
The king cobra venom system with venom and accessory gland expression profiles. Pie charts display the normalized percentage abundance of toxin transcripts recovered from each tissue transcriptome. Three-finger toxins are the most abundant toxin family in the venom gland (66.73% of all toxin transcripts and 4.37% in the accessory gland), and they are represented in the genome by at least 21 loci. Lectins are the most abundant toxin family in the accessory gland (42.70% of all toxin transcripts and 0.03% in the venom gland), and they are represented in the genome by at least six loci. Asterisks indicate toxin gene families annotated in the genome. 3FTx, three-finger toxin; AchE, acetylcholinesterase; CRISP, cysteine-rich secretory protein; CVF, cobra venom factor; IGF-like, insulin-like growth factor; kallikrein, kallikrein serine proteases; kunitz, kunitz-type protease inhibitors; LAAO, l-amino acid oxidase; NGF, nerve growth factor; PDE, phosphodiesterase; PLA2, phospholipase A2; PLB, phospholipase-B; SVMP, snake venom metalloproteinase. Drawing made based on a photo by F.J.V.
The birth and death model of gene evolution is the canonical framework used to explain the evolutionary origin of snake venom toxins. Drivers of toxin diversification may include (i) directional selection for toxins that facilitate prey capture, (ii) the need to target a diversity of receptors in different prey, and (iii) the concomitant evolution of venom resistance in some prey as part of an evolutionary arms race (2). The lack of genome sequences for any venomous snake and the consequent dependence on transcriptome data have hampered our understanding of not only the tempo and mode of venom toxin evolution but also, the genomic mechanisms that regulate toxin–gene expression.
To address these issues, we have produced a draft genome of a venomous snake—that of an adult male Indonesian king cobra (Ophiophagus hannah). This iconic species is the longest venomous snake in the world. Native to tropical Asia, it feeds on other snakes, and it is a member of the family Elapidae. We also deep-sequenced transcriptomes and small RNAs of the venom gland, the accessory gland, and a pooled, multitissue archive and characterized the king cobra venom proteome. These unique datasets provide an unprecedented insight into the evolution of venom.
Results and Discussion
King cobra genome sequence data (SI Appendix, Table S1) were first assembled de novo into contigs, which were subsequently oriented and merged into scaffolds. Haploid genome size was estimated by flow cytometry to be 1.36–1.59 Gbp (SI Appendix, Fig. S1). The assembled draft genome has an N50 contig size of 3.98 Kbp and an N50 scaffold size of 226 Kbp. The total contig length is 1.45 Gbp, and the total scaffold length (which contains gaps) is 1.66 Gbp.
As a genome quality check, we examined the Hox cluster, because it is well-characterized in other vertebrates (11). We annotated all 39 Hox genes, which we found clustered at four genomic regions, like in other vertebrates. However, the gene clusters are substantially larger than the Hox clusters observed in mammals (SI Appendix, Fig. S2). Of special interest is the absence of Hoxd12 from the king cobra, the Burmese python (Python molurus bivittatus) (12), and other snake genomes (13) (SI Appendix, Fig. S3). Hoxd12 is important for limb development in tetrapods (11) and thus, may have been lost along with limbs before the snake diversification. We also mapped microRNAs that had been previously located within mammalian and avian Hox clusters (SI Appendix, Fig. S2 and Dataset S1).
We interrogated the king cobra genome and annotated the open reading frames of 12 venom toxin gene families (Fig. 1 and SI Appendix, Fig. S4). Venom toxins are thought to have been co-opted from gene homologs with nontoxic physiological functions that are expressed in tissues other than the venom gland (14, 15). Our analysis of tissue-specific transcriptomic data (12, 16–18) provides genome-scale confirmation that these venom genes have, indeed, been recruited from a wide variety of tissue types (SI Appendix, Table S2). Syntenic comparisons of king cobra genomic architecture with the genomes of other vertebrates revealed that toxin co-option has occurred by two distinct mechanisms: (i) gene hijacking/modification and (ii) duplication of nontoxin genes (SI Appendix, Fig. S5); they were followed in both cases by selective expression in the venom gland.
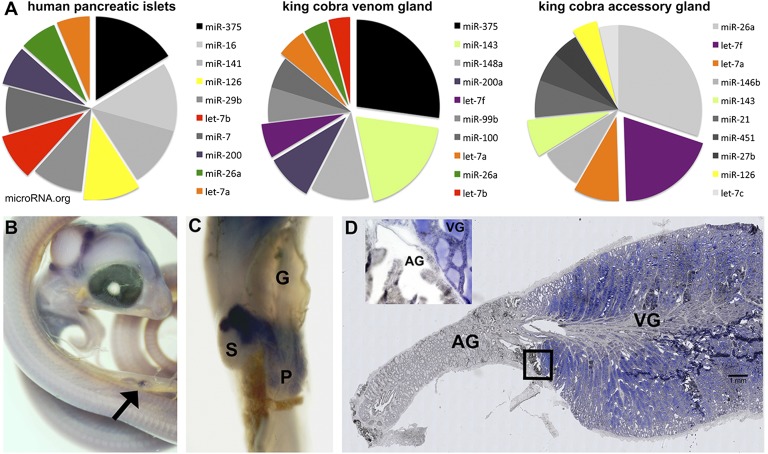
Sequencing and analysis of microRNA (miRNA) libraries made from a range of different tissues showed molecular similarities between the king cobra venom gland and known profiles of human and mouse pancreas (Fig. 2A). The most abundant miRNA in our venom gland library is miR-375, a canonical miRNA in the vertebrate pancreas. In the mouse, chicken, and zebrafish, miR-375 expression is restricted to the pancreas and pituitary gland (19, 20). Here, we detected miR-375 expression in the embryonic pancreas of the copperhead ratsnake (Coelognathus radiatus), the islet cell masses associated with the pancreas and spleen of the spitting cobra (Naja siamensis), and importantly, the venom gland of the king cobra (Fig. 2 B–D and SI Appendix, Fig. S6). In the past, it has been hypothesized that the snake venom gland evolved by evolutionary modification of the pancreatic system (21–23), although this hypothesis has since been abandoned, because little evidence exists that toxins expressed in the venom gland have been co-opted from related proteins expressed in the pancreas (14). However, our results are consistent with miR-375 being part of a core genetic network regulating secretion that has been co-opted during the evolution of the snake venom gland from an ancestral role in the pancreas and foregut secretory cells (24); it highlights an inherent link between these two secretory tissues, a link which was first suggested by Kochva et al. (21–23).
Fig. 2.
MiRNA expression profiles of the king cobra venom gland and accessory gland and miRNA expression patterns by in situ hybridization. (A) The 10 most abundant miRNAs in the venom gland show similarities with the known expression profile of the vertebrate pancreas (shown here for human; microRNA.org). (B) In situ hybridization of miR-375 in a C. radiatus embryo 27 d postoviposition with expression detected in the pancreas (arrow). (C) In situ hybridization of miR-375 in an N. siamensis embryo 32 d postoviposition, showing expression in the islet cell masses of the pancreas and the intrasplenic islet tissue. (D) In situ hybridization of miR-375 in a tissue section of the venom system of an adult O. hannah showing expression in the main venom gland. (Inset) Boundary of the venom gland (expression) and accessory gland (no expression) (SI Appendix, Fig. S6). AG, accessory gland; G, gallbladder; P, pancreas; S, spleen; VG, venom gland.
We identified 20 toxin families in the king cobra venom gland transcriptome (Fig. 1 and Dataset S2), including all toxin families annotated in the genome. Of the transcriptome hits, 14 toxin families were identified in the venom proteome (SI Appendix, Figs. S7–S9 and Tables S3 and S4 and Dataset S3), and nerve growth factor, phospholipase-B, and cobra venom factor have not previously been reported in king cobra venom. We also identified a unique snake venom protein, insulin-like growth factor, which we found selectively expressed in the venom gland and the venom proteome (SI Appendix, Fig. S10 and Table S4). Recent findings have shown adaptive evolution in insulin-like growth factor genes in snakes, although the site of their expression was unknown (25). Evidence of selective venom gland expression combined with adaptive evolution is consistent with a function of these proteins as venom toxins. In addition, we discovered a unique independent recruitment event of l-amino acid oxidase into king cobra venom (SI Appendix, Fig. S11).
Comparisons of toxin expression in the venom gland, accessory gland, and pooled multitissue archive revealed that most toxins are expressed at high levels only in the venom gland (Fig. 1 and SI Appendix, Fig. S10). Our results indicate that toxin gene transcription in the venom gland is regulated independently from its expression in those other tissues. Most toxins observed in the venom gland transcriptome are expressed at low levels in the accessory gland. One exception was the lectin toxin family, with expression that was at least 40 times higher in the accessory gland (SI Appendix, Fig. S10). Our evolutionary analysis of the lectins shows that they have been recruited to the oral secretory glands before the radiation of the advanced snakes, followed by expansion of the gene family (SI Appendix, Figs. S12 and S13). Our king cobra data suggest a model in which venom-like lectin paralogs have then repeatedly become transcriptionally activated in the accessory gland and deactivated in the venom gland (SI Appendix, Figs. S12 and S13).
In situ hybridization showed that the expression of these recruited lectins is concentrated in the serous cells located in the proximal region (26) of the accessory gland (Fig. 3 and SI Appendix, Fig. S14). No lectins were detected in the king cobra venom proteome (SI Appendix, Figs. S7–S9), consistent with their low transcript abundance in the venom gland. These results suggest that lectins do not contribute to king cobra envenoming, which is in contrast to many other venomous snakes (1, 27), and that their repeated recruitment to the accessory gland is associated with the subsequent evolution of unidentified, nontoxic functions (15).
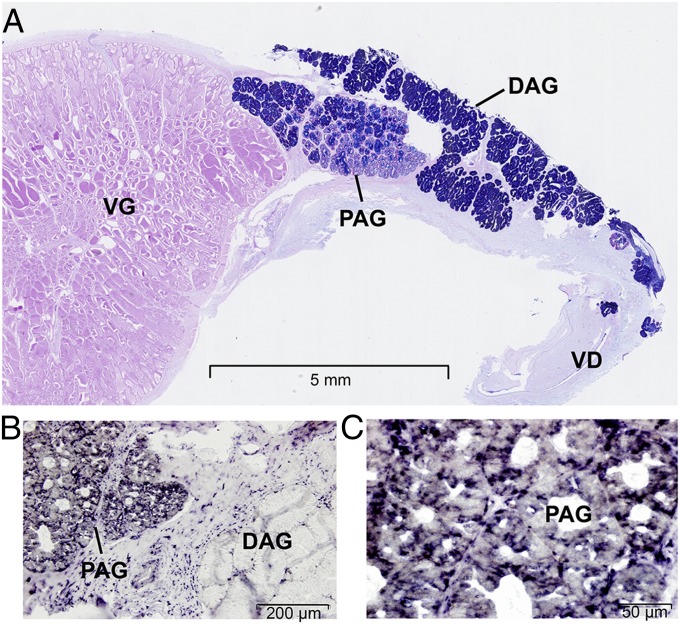
Fig. 3.
Histological section of the complete venom apparatus of the king cobra and spatial expression of lectin genes in the accessory gland. (A) Longitudinal section of the venom system reveals the two regions of the accessory gland: the proximal portion (PAG) and the distal portion (DAG; consistent with a previous morphological study) (26). The venom system is stained by alcian blue and periodic acid–Schiff, in which the secretory epithelial cells and secretion of the venom gland are periodic acid–Schiff-positive and the seromucous acini of the PAG and the mucous acini comprising the DAG are stained with alcian blue. (B) In situ hybridization of lectin gene Oh-516 (genome ID s8808 gene 2) shows that lectin expression is restricted to the PAG. DAG shows no staining. (C) Detail of the PAG shown in B showing strong granular staining in the epithelium of the PAG (SI Appendix, Fig. S14). VD, venom duct; VG, venom gland.
The venom gland transcriptome and venom proteome revealed multiple related venom isoforms for many different toxin families. To investigate the role of gene duplication in driving the genomic expansion of venom genes, we examined the evolutionary history of nine different toxin families by comparing gene orthologs and paralogs from other venomous snakes and the Burmese python, and green anole lizard (Anolis carolinensis) genomes and tissue transcriptomes (12, 16–18) (SI Appendix, Figs. S11 and S15–S22). We then used these data to perform tests of directional selection. Our results reveal multiple distinct patterns of gene duplication and sequence evolution under positive selection in different protein-coding gene families both before and after their recruitment into venom-producing pathways (Fig. 4 A and B and SI Appendix, Table S5). Significantly, we found evidence of higher rates of duplication and selection in the most highly expressed, proteomically abundant, and functionally important (28) gene families analyzed. The major lethal toxin family of the king cobra, the three-finger toxins (28, 29), is the most abundantly represented and isomerically diverse toxin family found in the venom gland transcriptome and venom proteome (Fig. 1 and SI Appendix, Fig. S7). This family has undergone massive expansion and shows high levels of positive selection and gene duplication (Fig. 4 B and C). In addition, phospholipase A2, snake venom metalloproteinase, and kallikrein toxin families also exhibit substantial gene duplication (Fig. 4D), and evidence of positive selection was identified in two of these gene families (Fig. 4B).
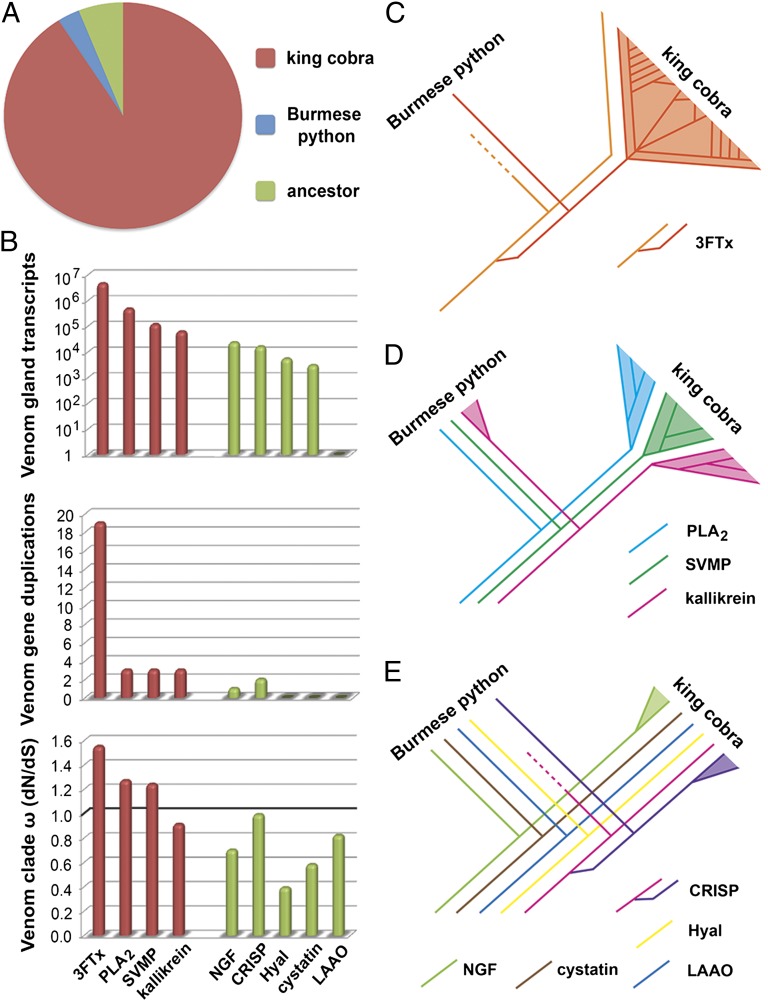
Fig. 4.
Contrasting evolutionary histories of king cobra toxin gene families. (A) The vast majority of toxin family gene duplication events occurred in the king cobra lineage compared with the Burmese python and their common ancestor. (B) Comparisons of venom gland expression, venom-related gene duplication events, and rate of evolution of main toxin families (red) and ancillary toxin families (green). (C) Massive expansion of the three-finger toxin gene family and (D) moderate expansion of other pathogenic toxin families by duplication of venom-expressed genes after the split of the Burmese python from the advanced snakes. (E) Ancillary toxin families show reduced evidence of gene duplication. Colored lines indicate gene loci, with line splits representing gene duplication events and dotted lines indicating gene loss. Venom gene duplications are defined as duplications that occurred after the split of the Burmese python from the advanced snakes (king cobra). ω represents the dN/dS ratio identified for venomous gene clades. The boundary for directional selection is indicated by a bold line. Note the logarithmic scale in the normalized venom gland expression graph. 3FTx, three-finger toxin; CRISP, cysteine-rich secretory protein; Hyal, hyaluronidase; kallikrein, kallikrein serine proteases; LAAO, l-amino acid oxidase; NGF, nerve growth factor; PLA2, phospholipase A2; SVMP, snake venom metalloproteinase.
Gene duplication coupled with positive selection is the mechanism underlying venom protein neofunctionalization (30–34). Our results are, therefore, consistent with a prominent role for prey-driven natural selection in generating the genetic diversity of the most pathogenic toxin families (28). By contrast, toxin families with ancillary functions show lower levels of gene expression, little to no evidence of gene duplication, and no evidence of directional selection (Fig. 4 B and E). For example, hyaluronidase, which possibly functions to break down prey tissue at the envenomation site (35), is not under positive selection. These results suggest that ancillary venom genes are less likely to generate resistance in prey and therefore, likely to experience lower selection pressures. These gene families likely have conserved functional activities and do not participate in the evolutionary arms race seen in the more toxic venom protein families.
In conclusion, this study highlights the diversity of genomic responses to extrinsic selective factors (the imperative to overpower prey quickly). These responses include function-modulated patterns of transcript abundance, gene duplication, and protein evolution in different toxin families. In contrast with the only other venomous vertebrate genome sequenced to date [the platypus (36, 37)], gene duplication is apparently of fundamental importance in the adaptive evolution of the king cobra venom system. This distinction likely reflects the differences in selective pressures relating to the very different biological role of venom in these organisms. Platypus venom is implicated in male–male combat, and its evolution is driven by sexual selection, whereas snake venom is primarily used for predatory purposes. The requirement of snake venom to rapidly immobilize prey coupled with the concomitant evolution of resistance in some prey species apparently results in an evolutionary arms race that drives a diverse and dramatic genomic response in venomous snakes. Our study provides unique genome-wide perspectives on the adaptive evolution of such venom systems as well as to protein evolution in general, and thus, it contributes an essential foundation for understanding and comparing evolutionary genomic processes in venomous organisms.
Methods
SI Appendix, SI Materials and Methods has additional information relating to the methodologies described below.
Tissue Acquisition and Processing.
All animal procedures complied with local ethical guidelines. Genome sequencing was undertaken on a blood sample obtained from an adult male king cobra that originated in Bali, Indonesia. Venom was extracted, and 4 d later (to maximize mRNA production), the venom gland, accessory gland, and other tissue samples were sourced from a second Indonesian adult male specimen and stored in RNAlater.
Genome Sequencing.
We used a whole-genome shotgun sequencing strategy and Illumina sequencing technology. Genomic DNA was isolated from blood using the Qiagen Blood and Tissue DNeasyKit and paired-end libraries prepared from 5 µg isolated gDNA using the Illumina Paired-End Sequencing Sample Prep Kit. Either a 200- or 500-bp band was cut from the gel (library PE200 or PE500, respectively) (SI Appendix, Table S1). Similarly, mate pair libraries were prepared from 10 µg isolated gDNA using the Illumina Mate Pair 2–5 Kb Sample Prep Kit and bands from 2 to 15 Kbp cut from the gel (MP2K, MP7K, MP10K, and MP15K libraries) (SI Appendix, Table S1). After circularization, shearing, isolation of biotinylated fragments, and amplification, the 400- to 600-bp fraction of the resulting fragments was isolated from the gel. Genomic libraries were paired-end sequenced with a read length of 36–151 nt on an Illumina GAIIx instrument.
Genome Assembly.
For genome assembly, we largely followed the strategy pioneered in the work by Li et al. (38) for the assembly of the giant panda genome. Sequencing reads from both paired-end libraries were first used for building initial contigs. Both sets were preprocessed to eliminate low-quality reads and nucleotides as well as adapter contamination. For initial contig assembly, we used the CLC Assembly Cell De Novo Assembler (version 3.2; CLC Bio, Aarhus, Denmark), which implements a De Bruijn graph-based assembler. A run with a minimum-required contig size of 100 bp and a k-mer length of 31 nt resulted in an assembly with a total length of 1.45 Gbp and a contig N50 of 3,982 bp [i.e., 50% of the assembly (725 Mbp) is in contigs of at least this length]. Initial contigs were subsequently oriented into larger supercontigs (scaffolds) using SSPACE (39). SSPACE aligns paired reads to the contigs using Bowtie (40). SSPACE was used to scaffold contigs in a hierarchical fashion using first links obtained from the PE500 library to generate intermediate supercontigs, which were then used as the input for subsequent runs, with links from individual mate-pair libraries increasing in size. At each stage, a minimum of three nonredundant links was required to join two contigs. This procedure resulted in a final scaffold set with a total length of 1.66 Gbp and an N50 of 225,511 bp.
Genome Annotation.
Automated gene prediction was undertaken using the automated annotation pipeline MAKER (41, 42). Gene annotations were made using a protein database combining the Uniprot/Swiss-Prot protein database and all king cobra and green anole (A. carolinensis) sequences from the National Center for Biotechnology Information protein database. Ab initio gene predictions were created by MAKER using the programs SNAP (43) and Augustus (44). Gene models were further improved by providing MAKER with all king cobra mRNAseq data generated in this study, which were combined to generate a joint assembly of transcripts using Trinity (45). A total of three iterative runs of MAKER was used to produce the final gene set. Additional extensive manual annotation was performed to establish the intron–exon boundaries of members of venom toxin gene families.
mRNA-Seq and Small RNA Libraries.
King cobra tissue sequencing libraries were prepared for the venom gland, accessory gland, and a pooled multitissue archive (heart, lung, spleen, brain, testes, gall bladder, pancreas, small intestine, kidney, liver, eye, tongue, and stomach). Total RNA was isolated from each tissue using the Qiagen miRNeasy Kit. Transcriptome libraries were subsequently prepared from 10 µg total RNA (using equal amounts of RNA isolated from each tissue for the pooled multitissue archive) using the Illumina mRNA-Seq Sample Preparation Kit. Total RNA from the same samples was used to prepare the small RNA libraries using the Illumina small RNA v1.5 Sample Preparation Kit. RNAseq and small RNA libraries were sequenced on the Illumina GAIIx sequencing platform.
Transcriptome Assembly.
Reads for the venom gland, accessory gland, and pooled multitissue archive were coassembled with Abyss (46, 47) with various k values (every even number from 50 to 96). The resulting assemblies were joined by an iterative BLAST and cap3 assembler (48). Coding sequences were extracted using an automated pipeline based on similarities to known proteins or by obtaining coding sequences from the larger ORF of the contigs containing a signal peptide. To map the raw Illumina reads to the coding sequences and determine their tissue bias, raw reads from each library were blasted to the coding sequences using blastn with a word size of 25 (−W 25 switch) and allowing recovery of up to three matches. The three matches were used if they had less than two gaps and their scores were equal to the best score. The resulting blast file was used to compile the number of reads each coding DNA sequence received from each library.
miRNA Profiles and in Situ Hybridizations.
The small RNA sequences were analyzed using CLC Bio Genome Workbench. Briefly, small RNA sequences were filtered for quality and size, and reads of low quality and lengths less than 17 or greater than 26 nt were discarded. The remaining pool of small RNAs was compared with miRBase release 18 (http://www.miRBase.org) to extract orthologous mature miRNA sequences from each king cobra RNA sample. These miRNAs were subsequently mapped to the king cobra genome, with 70 bp upstream and downstream of the mature sequence extracted as the potential precursor miRNA sequence using PHP scripts and blast (49) 2.2.26+. The expression level of each miRNA was assessed using CLC Bio and compared with data available at the miRNA targets and expression database (http://www.microRNA.org; release August 2010) for the expression profiles of orthologous miRNA genes in mouse and human (e.g., miR-375). Whole-mount in situ hybridizations for miR-375 detection were performed using 5′ digoxigenin-labeled locked nucleic acid (LNA; Exiqon) probes following the protocol in the work by Darnell et al. (19). The standard tissue section in situ protocol in the work by Jostarndt et al. (50) for paraffin-embedded tissues was followed for miR-375 detection in the adult king cobra venom gland. For whole-mount in situ hybridizations in late-stage snake embryos (27 d postoviposition or older), embryos were skinned, and the abdominal wall was cut open followed by an extended probe hybridization for ∼36 h. All miR-375 LNA in situ hybridizations were carried out at 57 °C (22°C below the calculated probe melting temperature of 79°C) along with a no-probe control. miR-196 LNA in situ was carried out at 47 °C as an additional negative control in the adult venom gland.
Venom Proteomics.
We used king cobra venom extracted from the same animal used for transcriptomics. The venom was reduced, alkylated, digested with trypsin, separated by column chromatography, and analyzed by ESI-ion trap tandem MS. The peptide fragments created by collision-induced dissociation were compared against the assembled king cobra venom gland and accessory gland transcriptomes and a Lepidosaurian (National Center for Biotechnology Information) database using Sequest and Mascot software with a false discovery rate of 0.01.
Evolutionary Analyses.
King cobra sequences exhibiting homology to toxin families were identified through (i) annotation in the genome or transcriptome and (ii) blast searching the king cobra genome and transcriptome datasets in CLC Main Workbench with representative templates of toxin and nontoxin gene homologs. Coding regions of identified toxin gene loci were aligned using the MUSCLE algorithm (51) with putative paralogs and orthologs from selected vertebrates, including other venomous snakes and the P. molurus bivittatus and A. carolinensis genomes and transcriptomes (12, 16–18). These sequences were obtained by mining GenBank for blast hits and using the datasets in work by Casewell et al. (15).
DNA gene trees for each toxin family were reconstructed using Bayesian inference in MrBayes v3.2 (52) incorporating optimized models of sequence evolution selected by MrModelTest v2.3 (53). Each dataset was run in duplicate using four chains for 5 × 106 generations, sampling every 500th cycle from the chain, and using default settings in regards to priors. Tracer v1.4 (54) was used to estimate effective sample sizes for all parameters and verify the point of convergence (burnin), with trees generated before the completion of burnin discarded. The locations of gene expression of snake sequences determined by transcriptomics were mapped on the gene trees to visualize relative expression in different tissue types. Toxin family gene duplication events were inferred by pruning the gene trees to only contain king cobra and Burmese python genes along with a single outgroup sequence. The ensuing gene trees were analyzed using the duplication and loss criterion in iGTP (55) with the following species tree: [outgroup (king cobra, Burmese python)]. For tests of directional selection, we inferred fully resolved maximum likelihood trees from each of the toxin family datasets using the BEST tree-searching algorithm in PHYML (56). The most parsimonious points of recruitment into venom-producing pathways were then reconstructed on these trees, thereby classifying tree branches into venomous and nonvenomous. The method of Yang and Nielsen (57) was implemented in the PAML software package to estimate ωvenomous and ωnonvenomous for each toxin family.
Supplementary Material
Acknowledgments
We thank the following persons who helped us or contributed material used in this study: Austin Hughes, Nathan Dunstan, Daniëlle de Wijze, and Youri Lammers. We thank Bas Blankevoort for constructing Fig. 1. This work received funding from the following sources: internal funding from the Naturalis Biodiversity Center (F.J.V., J.W.A., and M.K.R.), a Rubicon Grant from the Netherlands Organization for Scientific Research (to F.J.V.), a research fellowship from the United Kingdom Natural Environment Research Council (to N.R.C.), an Netherlands Organization for Scientific Research Visitor’s Travel Grant from Nederlandse Organisatie voor Wetenschappelijk Onderzoek (to R.J.R.M., R.M.K., and M.K.R.), a studentship from the United Kingdom Biotechnology and Biological Sciences Research Council (to R.B.C.), and a Smart Mix Grant from the Dutch Government (to M.K.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The king cobra genome assembly and reads reported in this paper have been deposited in the GenBank database (bioproject no. PRJNA201683). The transcriptome sequences reported in this paper have been deposited in the GenBank Short Read Archive database (bioproject no. PRJNA222479). The microRNA sequences reported in this paper have been deposited in miRBase, www.mirbase.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314702110/-/DCSupplemental.
References
- 1.Vonk FJ, et al. Snake venom: From fieldwork to the clinic: Recent insights into snake biology, together with new technology allowing high-throughput screening of venom, bring new hope for drug discovery. Bioessays. 2011;33(4):269–279. doi: 10.1002/bies.201000117. [DOI] [PubMed] [Google Scholar]
- 2.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol Evol. 2013;28(4):219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Vonk FJ, et al. Evolutionary origin and development of snake fangs. Nature. 2008;454(7204):630–633. doi: 10.1038/nature07178. [DOI] [PubMed] [Google Scholar]
- 4.Fry BG, et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439(7076):584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- 5.Saviola AJ, Chiszar D, Busch C, Mackessy SP. Molecular basis for prey relocation in viperid snakes. BMC Biol. 2013;11:20. doi: 10.1186/1741-7007-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2(10):790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 7.Bohlen CJ, et al. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479(7373):410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diochot S, et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature. 2012;490(7421):552–555. doi: 10.1038/nature11494. [DOI] [PubMed] [Google Scholar]
- 9.Kasturiratne A, et al. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5(11):e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohapatra B, et al. Snakebite mortality in India: A nationally representative mortality survey. PLoS Negl Trop Dis. 2011;5(4):e1018. doi: 10.1371/journal.pntd.0001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zákány J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304(5677):1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 12.Castoe TA, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA. 110:20645–20650. doi: 10.1073/pnas.1314475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di-Poï N, et al. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature. 2010;464(7285):99–103. doi: 10.1038/nature08789. [DOI] [PubMed] [Google Scholar]
- 14.Fry BG. From genome to “venome:” Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15(3):403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casewell NR, Huttley GA, Wüster W. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 2012;3:1066. doi: 10.1038/ncomms2065. [DOI] [PubMed] [Google Scholar]
- 16.Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477(7366):587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castoe TA, et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12(7):406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckalbar WL, et al. Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes. BMC Genomics. 2013;14:49. doi: 10.1186/1471-2164-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darnell DK, et al. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235(11):3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 20.Lynn FC, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 21.Kochva E. In: Biology of the Reptilia. Gans C, Gans KA, editors. London: Academic; 1978. [Google Scholar]
- 22.Kochva E, Nakar O, Ovadia M. Venom toxins: Plausible evolution from digestive enzymes. Amer Zool. 1983;23(2):427–430. [Google Scholar]
- 23.Kochva E. The origin of snakes and evolution of the venom apparatus. Toxicon. 1987;25(1):65–106. doi: 10.1016/0041-0101(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 24.Christodoulou F, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463(7284):1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sparkman AM, et al. Rates of molecular evolution vary in vertebrates for insulin-like growth factor-1 (IGF-1), a pleiotropic locus that regulates life history traits. Gen Comp Endocrinol. 2012;178(1):164–173. doi: 10.1016/j.ygcen.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 26.Mackessy SP. Morphology and ultrastructure of the venom glands of the northern pacific rattlesnake Crotalus viridis oreganus. J Morphol. 1991;208:109–128. doi: 10.1002/jmor.1052080106. [DOI] [PubMed] [Google Scholar]
- 27.Morita T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon. 2005;45(8):1099–1114. doi: 10.1016/j.toxicon.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 28.Mebs D, Claus I. In: Snake Toxins. Harvey AL, editor. New York: Pergamon; 1991. pp. 425–447. [Google Scholar]
- 29.Kini RM, Doley R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon. 2010;56(6):855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Kini RM, Chan YM. Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J Mol Evol. 1999;48(2):125–132. doi: 10.1007/pl00006450. [DOI] [PubMed] [Google Scholar]
- 31.Fry BG, et al. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57(1):110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 32.Lynch VJ. Inventing an arsenal: Adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wüster W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 2011;28(9):2637–2649. doi: 10.1093/molbev/msr091. [DOI] [PubMed] [Google Scholar]
- 34.Sunagar K, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol Biol Evol. 2012;29(7):1807–1822. doi: 10.1093/molbev/mss058. [DOI] [PubMed] [Google Scholar]
- 35.Fox JW. A brief review of the scientific history of several lesser-known snake venom proteins: l-Amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon. 2013;62:75–82. doi: 10.1016/j.toxicon.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453(7192):175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong ES, Papenfuss AT, Whittington CM, Warren WC, Belov K. A limited role for gene duplications in the evolution of platypus venom. Mol Biol Evol. 2012;29(1):167–177. doi: 10.1093/molbev/msr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt C, Yandell M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491–505. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59–68. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birol I, et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25(21):2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 47.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6(12):e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Jostarndt K, Puntschart A, Hoppeler H, Billeter R. The use of 33P-labelled riboprobes for in situ hybridizations: Localization of myosin alkali light-chain mRNAs in adult human skeletal muscle. Histochem J. 1994;26(1):32–40. [PubMed] [Google Scholar]
- 51.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nylander JAA. MrModeltest v2. Finland: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 54.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaudhary R, Bansal MS, Wehe A, Fernández-Baca D, Eulenstein O. iGTP: A software package for large-scale gene tree parsimony analysis. BMC Bioinformatics. 2010;11:574. doi: 10.1186/1471-2105-11-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19(6):908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.