Significance
A GGGGCC expansion mutation located in intron 1 of chromosome 9 ORF 72 (C9ORF72) was recently described as a common cause of familial amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD). We show that this single mutation results in the accumulation of sense and antisense RNA foci plus six expansion proteins expressed by repeat-associated non-ATG (RAN) translation. RNAs accumulate in nuclear foci and the RAN proteins form cytoplasmic aggregates in neurons that often cluster in affected brain regions. These results indicate that bidirectional transcription and RAN translation are fundamental pathologic features of C9ORF72 ALS/FTD. Additionally these data have broad implications that change our understanding of how microsatellite expansion mutations are expressed in patient cells and how they cause disease.
Keywords: cytoplasmic inclusions, clustered aggregates, noncoding RNA
Abstract
The finding that a GGGGCC (G4C2) hexanucleotide repeat expansion in the chromosome 9 ORF 72 (C9ORF72) gene is a common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) links ALS/FTD to a large group of unstable microsatellite diseases. Previously, we showed that microsatellite expansion mutations can be bidirectionally transcribed and that these mutations express unexpected proteins by a unique mechanism, repeat-associated non-ATG (RAN) translation. In this study, we show that C9ORF72 antisense transcripts are elevated in the brains of C9ORF72 expansion-positive [C9(+)] patients, and antisense GGCCCC (G2C4) repeat-expansion RNAs accumulate in nuclear foci in brain. Additionally, sense and antisense foci accumulate in blood and are potential biomarkers of the disease. Furthermore, we show that RAN translation occurs from both sense and antisense expansion transcripts, resulting in the expression of six RAN proteins (antisense: Pro-Arg, Pro-Ala, Gly-Pro; and sense: Gly-Ala, Gly-Arg, Gly-Pro). These proteins accumulate in cytoplasmic aggregates in affected brain regions, including the frontal and motor cortex, hippocampus, and spinal cord neurons, with some brain regions showing dramatic RAN protein accumulation and clustering. The finding that unique antisense G2C4 RNA foci and three unique antisense RAN proteins accumulate in patient tissues indicates that bidirectional transcription of expanded alleles is a fundamental pathologic feature of C9ORF72 ALS/FTD. Additionally, these findings suggest the need to test therapeutic strategies that target both sense and antisense RNAs and RAN proteins in C9ORF72 ALS/FTD, and to more broadly consider the role of antisense expression and RAN translation across microsatellite expansion diseases.
The chromosome 9p21-linked form of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), the most common cause of familial FTD and ALS identified to date, is caused by an expanded GGGGCC (G4C2) hexanucleotide repeat in intron 1 of chromosome 9 ORF 72 (C9ORF72) (1, 2). The C9ORF72 mutation is found in 40% of familial and 7% of sporadic ALS cases and 21% of familial and 5% of sporadic FTD patients (3). The discovery of the C9ORF72 expansion has generated substantial excitement because it connects ALS and FTD to a large group of disorders caused by microsatellite expansion mutations (4).
Traditionally, microsatellite expansion mutations located in predicted coding and noncoding regions were thought to cause disease by protein gain- or loss-of-function or RNA gain-of-function mechanisms (4). Protein loss-of-function has been proposed to underlie C9ORF72-driven ALS/FTD because the expansion mutation leads to decreased levels of variant 1 transcripts and potential decreases in C9ORF72 protein expression (1, 5). Additionally, because the C9ORF72 G4C2 expansion mutation is located in an intron, several studies have pursued the hypothesis that C9-linked ALS/FTD results from a toxic RNA gain-of-function mechanism in which G4C2 expansion RNAs sequester important cellular factors in nuclear RNA foci. Multiple G4C2 RNA binding proteins have been identified, but so far there is no demonstration that any of these candidates directly bind endogenous expansion transcripts or colocalize with RNA foci observed in patient cells or autopsy tissue (6–9).
Repeat-associated non-ATG (RAN) translation, which we initially discovered in spinocerebellar ataxia type 8 (SCA8) and myotonic dystrophy type 1 (DM1) (10), has also been proposed for C9ORF72 ALS/FTD (11, 12) and Fragile X-associated tremor ataxia syndrome (13). In this mechanism, hairpin-forming microsatellite expansion transcripts express proteins in one or more reading frames without an AUG-initiation codon (10). A variety of names have recently been ascribed to these RAN-translated proteins (e.g., homopolymeric, dipeptide, RANT). We propose that all proteins expressed across microsatellite expansion mutations in the absence of an ATG-initiation codon be referred to as RAN proteins to prevent confusion, because additional expansion mutations that undergo RAN translation are identified.
In C9ORF72 ALS/FTD, RAN proteins expressed from G4C2 sense transcripts were shown to accumulate as protein aggregates in C9(+) autopsy tissue (11, 12). A strength of these studies, which used antibodies directed against RAN-predicted GR, GP, and GA repeat motifs, is that they provide a potential link between the mutation and known pathologic features of C9ORF72 ALS/FTD. A potential weakness is that the antibodies used were directed only against the predicted repeat motifs, which are also found in unrelated endogenous proteins (14–16).
Although it is now clear that a growing number of expansion mutations are bidirectionally transcribed (17–22), nearly all research on these disorders has focused on understanding the impact of expansion mutations expressed from the sense direction. Our earlier studies on SCA8 and DM1 CAG•CTG expansion mutations showed that sense and antisense expansion transcripts are expressed and RAN proteins accumulate in patient tissues (10). Mori et al. also presented evidence by RT-PCR that C9ORF72 antisense transcripts upstream of the expansion mutation are expressed in patient brains (12). Here we show that C9ORF72 ALS/FTD antisense transcripts containing the GGCCCC (G2C4) expansion accumulate in patient brains as nuclear foci. Additionally, we developed a panel of antibodies directed to both the repeat motifs and unique C-terminal regions, and demonstrate that both sense and antisense RAN proteins accumulate in C9ORF72 patient CNS autopsy tissue. The identification of antisense G2C4 RNA foci and three unique antisense RAN proteins in C9ORF72 brains suggests that bidirectional transcription and RAN translation are fundamental pathologic features of C9ORF72 ALS/FTD.
Results
Antisense RNA Foci in C9ORF72-Expansion Patients.
Based on our identification of bidirectional transcription and RAN translation in SCA8 (10, 21), we performed a series of experiments to test the hypotheses that antisense (AS) C9ORF72 expansion transcripts form AS-G2C4 RNA foci and express antisense proteins by RAN translation or from short antisense open-reading frames (AS-ORFs).
First, we confirmed C9ORF72 antisense transcripts are expressed using a linkered (LK) strand-specific RT-PCR strategy to compare expression of the sense and antisense transcripts in intron 1b, 5′ of the AS-G2C4 expansion, and exon 1a. For the antisense strand in intron 1b, strand-specific RT-PCR was performed using a LK–AS-ORF-R primer for the RT reaction and AS-ORF-F and the LK for PCR to specifically amplify antisense cDNAs (Fig. 1A). Similar strategies were used to amplify sense transcripts from the same region of intron 1b and sense and antisense transcripts in exon 1a.
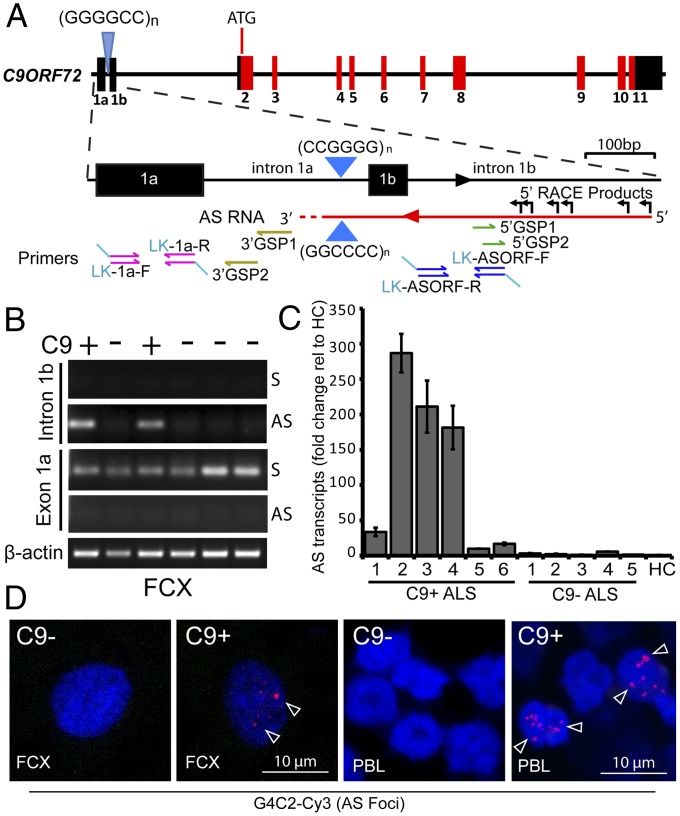
Fig. 1.
G2C4 antisense transcripts accumulate as RNA foci in C9ORF72 patient tissues. (A) Schematic diagram of C9ORF72 gene and antisense transcripts and relative location of primers for strand-specific RT-PCR and RACE primers. (B) Strand-specific RT-PCR of sense (S) and antisense (AS) transcripts (across intron 1b and exon 1a) from FCX of C9(+) and C9(−) ALS patients. (C) Strand-specific qRT-PCR showing elevated antisense mRNA in C9(+) compared with C9(−) ALS patients and a healthy control (HC). (D) FISH with G4C2-Cy3 probe showing G2C4 antisense RNA foci (red) in C9(+)FCX and PBLs which are absent in C9(−) cases. Nuclear foci in FCX are indicated by arrowheads.
Intron 1b antisense transcripts were detected by RT-PCR in the frontal cortex (FCX) from C9(+) ALS/FTD patients but not C9(−) ALS/FTD or normal controls (Fig. 1B), and quantitative RT-PCR (qRT-PCR) shows these transcripts are dramatically increased among six C9(+) ALS/FTD cases (Fig. 1C). In contrast, intron 1b sense transcripts were not detected by RT-PCR in the FCX (Fig. 1B). In blood, both intron 1b sense and antisense transcripts are expressed and no differences in expression levels between C9(+) and C9(−) groups were observed (Fig. S1 A and B). Additional longitudinal studies with larger numbers of patients will be required to determine if changes in sense or antisense transcript levels occur in blood as disease progresses. RACE analysis of FCX samples showed intron 1b antisense transcripts begin at varying sites 251–455 bp upstream of the G2C4 repeat (Fig. 1A, and Fig. S1C). In contrast, 3′ RACE, using primers located 40 and 90 bp 3′ of the G2C4 repeat (3′ GSP1 or 3′ GSP2), did not detect transcripts. These data suggest that the 3′ end of the antisense transcript does not overlap the sense exon 1a region, located 170 bp 3′ of the antisense G2C4 repeat. Consistent with this result, sense but not antisense transcripts are detected by strand-specific LK–RT-PCR using primers overlapping exon 1a (Fig. 1B).
To determine if antisense transcripts include the AS-G2C4 repeat expansion, we performed RNA FISH using a Cy3-labeled (G4C2)4 probe to detect putative AS-G2C4 RNA foci. AS-G2C4 RNA foci accumulate in C9(+) (n = 4) but not C9(−) ALS (n = 3) or healthy control (n = 1) FCX samples (see Fig. 1D and Fig. S1D for examples). The percentage of nuclei that were positive for antisense and sense foci varied from 9% to 19% of nuclei in individual C9(+) samples, with no significant difference in frequency between sense and antisense foci (P = 0.2) (Fig. S1E). Double-labeling shows sense (green) and antisense (red) foci, which are typically found in separate cells, can occasionally be found in the same cell (Fig. S1F).
Because RNA foci in peripheral tissues could provide biomarkers of the disease, we examined peripheral blood leukocytes (PBLs) for RNA foci. Our data show sense and antisense RNA foci are detected in 29% and 7% of cells, respectively, in C9(+) (n = 3 individuals and 1,513 cells) but not C9(−) controls (n = 8) (see Fig. 1D and Fig. S2 A–C for examples). A striking difference in the pattern of foci accumulation is observed in blood, with 63% of antisense-positive cells having >five nuclear foci, whereas PBLs positive for sense foci typically have only one focus per cell and no cells were observed with five or more foci. As expected, RNA-foci in C9(+) PBLs were not detected with a CAGG-Cy3 control probe (Fig. S2D) and FISH signal from the Cy3-G4C2 AS-probe can be competed with excess unlabeled G4C2 probe (Fig. S2E). Additionally, these foci were resistant to DNase I and sensitive to RNase I digestion (Fig. S2F).
Taking these data together, we show C9ORF72 antisense transcripts are elevated in the FCX in C9(+) ALS but not C9(−) ALS or normal controls. We also demonstrate that unique antisense transcripts containing the AS-G2C4 expansion mutation are expressed and accumulate in nuclear RNA foci in the C9(+) FCX. Additionally, we show sense and antisense foci accumulate in blood, providing potential biomarkers of C9ORF72 ALS/FTD in a readily accessible tissue.
Dual Immunological Strategy to Detect RAN Proteins.
When we first described RAN translation in SCA8 and DM1, multiple antibodies directed against epitope tags, the repeat motif, and the C-terminal regions of these proteins were used (10) to demonstrate their expression. Although two groups recently reported evidence that RAN proteins are expressed in C9ORF72 ALS/FTD patients (11, 12), both groups relied on a single antibody for each of the GP, GR, or GA repeat motifs. Because these repeat motifs are found in a number of other proteins (14–16), we used a dual immunological strategy and developed antibodies that recognize the predicted repeat motifs or their corresponding unique C-terminal regions.
A schematic diagram showing eight putative C9ORF72 RAN proteins expressed across sense G4C2 (S-G4C2) and AS-G2C4 transcripts is shown in Fig. 2A and Fig. S3. Predicted proteins include six putative RAN proteins, three sense (GPS, GRS, and GAS) and three antisense (PAAS, PRAS, and GPAS), and additional putative ATG-initiated antisense proteins in the PR and GP frames (ATG-PRAS, and ATG-GPAS). RAN proteins with the GP repeat motif are predicted in both the sense and antisense directions (GPS and GPAS) and unique C-terminal regions are predicted in five of the six reading frames. To test for the accumulation of these proteins in vivo, we developed a series of polyclonal antibodies against the predicted repeat motifs or available corresponding C-terminal regions are shown in Fig. 2A and further described in Fig. S3 and Table S1. Antibodies to test for putative antisense proteins (rabbit α-PAAS, α-PA-CTAS, α-PRAS, α-PR-CTAS, α-GPS/AS, α-GP-CTS, and mouse α-GPS/AS) were generated and their specificities demonstrated in cells transfected with constructs expressing epitope-tagged recombinant protein by Western blot and immunofluoresence (IF) detection (Fig. 2B and Fig. S4). Additional antibodies detecting repeat and C-terminal regions expressed in the sense direction are characterized in Fig. S5. In summary, we generated 10 unique polyclonal antibodies against antisense and sense repeat motifs and unique C-terminal regions.
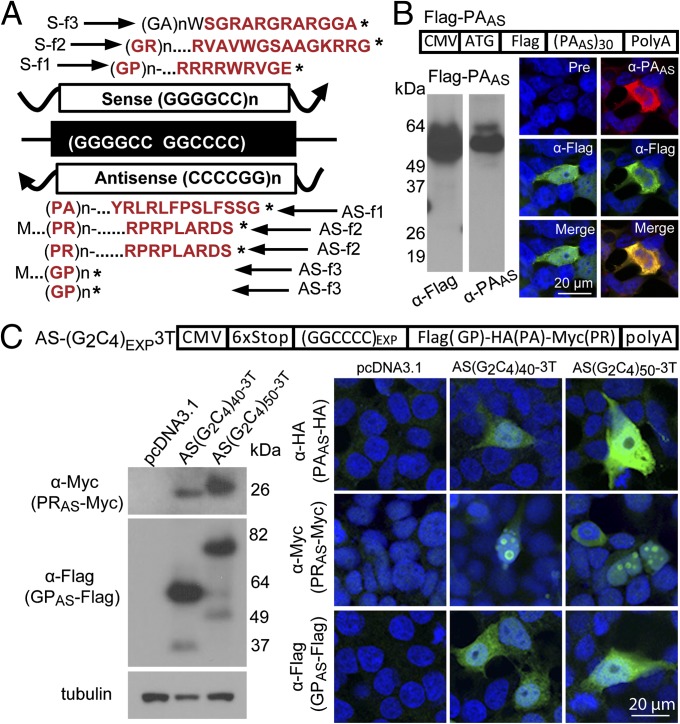
Fig. 2.
In vitro evidence for RAN translation of antisense G2C4 expansion and dual immunological detection strategy. (A) Diagram of putative proteins translated from sense and antisense transcripts. Sequences highlighted in red were used to generate antibodies. CT, C-terminal; f1–3, reading frames 1–3; *, stop codon. (B) Abbreviated example of validation of α-PAAS rabbit polyclonal antibody. Immunoblots (Lower Left) and IF staining (Lower Right) of HEK293T cells transfected with Flag-PAAS construct (Upper) and probed with α-Flag and α-PAAS antibodies. See Figs. S4 and S5 for additional controls and validation of eight additional antibodies generated against RAN repeat motifs and CT regions. (C) Immunoblots (Lower Left) and IF staining (Lower Right) of HEK293T cells 48 h posttransfection with the AS-(G2C4)EXP-3T construct (Upper). PRAS and GPAS expansion proteins were detected by Western blot and PAAS, PRAS and GPAS proteins were detected by IF in transfected cells.
RAN Translation of Antisense GGCCCC Repeat Expansions in Vitro.
To test if the AS-G2C4 expansions can undergo RAN translation, we performed a series of experiments in transfected HEK293T cells. First, we generated triply tagged AS-G2C4 minigene constructs, AS-(G2C4)EXP-3T. These constructs have a 6xStop codon cassette (two stops in each frame) upstream of an AS-(G2C4)EXP and three different C-terminal epitope tags to monitor protein expression in all reading frames [i.e., AS-(G2C4)EXP-3T transcripts translated in three frames result in antisense Pro-Ala (PAAS-HA), Pro-Arg (PRAS-Myc), and Gly-Pro (GPAS-Flag) RAN proteins] (Fig. 2C). Immunoblotting of AS-(G2C4)40-3T and AS-(G2C4)50-3T lysates detected two epitope-tagged RAN proteins, PRAS-Myc and GPAS-Flag, but not PAAS-HA (Fig. 2C). The (PRAS)40-Myc and (PRAS)50-Myc proteins migrated at approximately their predicted sizes of 20 and 27 kDa, respectively. In contrast, but similar to previously described RAN proteins (10), the (GPAS)40-Flag and (GPAS)50-Flag proteins migrated substantially higher than their predicted sizes (10–15 kDa) at 50 and 75 kDa, respectively (Fig. 2C). The faint lower molecular-weight bands on this blot may result from repeat contractions seen during bacterial culture or differences in translational start site. IF shows antisense RAN proteins are expressed in all three reading frames (Fig. 2C). The detection of PAAS-HA by IF but not Western blotting may be caused by a lower frequency of cells expressing RAN PAAS-HA from these constructs. Additionally, recombinant GPAS-Flag and PAAS-HA proteins have a cytoplasmic localization, whereas PRAS-Myc proteins are distributed in both the nucleus and cytoplasm. These localization differences may result from different properties of the repeat motifs or the C-terminal flanking sequences found in this epitope-tagged construct. In an additional series of experiments, we also show sense S-G4C2-expansion constructs containing 30, 60, and 120 repeats express GPS-Flag, GRS-HA and GAS-Myc RAN proteins (Fig. S6 A–C). In summary, these data show that recombinant AS-G2C4 and S-G4C2 expansion transcripts express RAN proteins in all six reading frames.
Antisense G2C4 RAN Proteins Accumulate in the Brain.
Because C9ORF72 FTD/ALS affects the brain and spinal cord, we used several approaches to determine if novel antisense proteins are expressed in C9ORF72 expansion-positive autopsy tissue. Aggregated proteins are difficult to isolate from the human brain; therefore, we used a sequential protein-extraction protocol (23) on frozen C9(+) and C9(−) ALS FCX autopsy samples. Antisense PAAS and PRAS proteins were detected with α-PAAS, α-PA-CTAS, α-PRAS, and α-PR-CTAS on immuno-dot blots of 1% (vol/vol) Triton-X100 insoluble, 2% SDS soluble extracts from a subset of C9(+) but not C9(−) ALS patients (Fig. 3A). Additional immuno-dot blots showing evidence for sense-RAN protein (GPS, GRS, GAS) accumulation in C9(+) ALS/FTD FCX are shown in Fig. S6D. α-GPS/AS, α-PAAS, and α-PRAS antibodies also detected high molecular-weight smears in 2% SDS insoluble fractions from C9(+) ALS FCX samples after resuspending the pellets in sample buffer containing 8% SDS (23) (Fig. 3B). The differences in migration pattern seen for the recombinant proteins (Fig. 2 B and C), which migrate as one or more bands, and the smears observed in patient tissue extracts (Fig. 3B) likely reflect differences in the RAN proteins because of much longer repeat tracts in patient samples and their extraction from highly insoluble aggregates. By immunohistochemistry (IHC), we demonstrated next that protein aggregates were detectable in the perikaryon of hippocampal neurons from C9(+) ALS/FTD autopsy tissue (Fig. 3C) but not in C9(−) ALS patients or control subjects (Fig. S7) using antibodies against the repeat motifs (α-PAAS, α-PRAS, α-GPS/AS) as well as antibodies directed to predicted C-terminal sequences beyond the PA and PR repeat tracts (α-PA-CTAS and α-PR-CTAS). None of these antibodies recognizes the 1C2-positive polyGln aggregates found in HD autopsy tissue (Fig. S8).
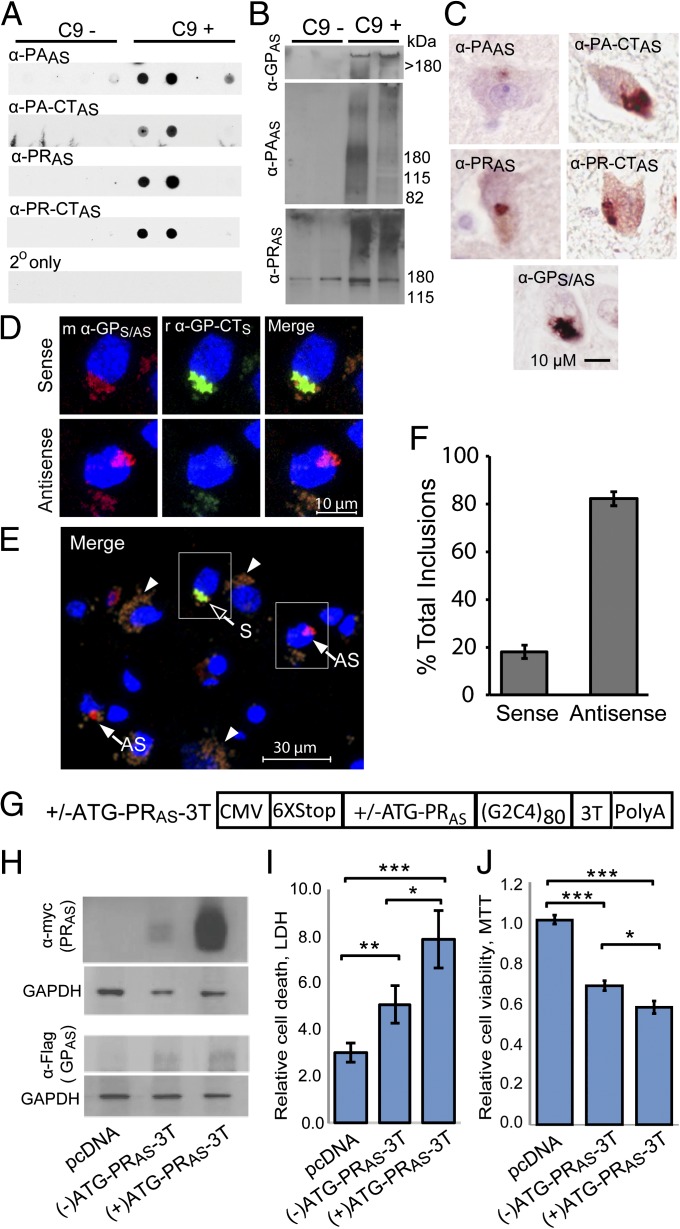
Fig. 3.
In vivo evidence for RAN-translation of the G2C4 AS repeat and toxicity studies. (A) Dot blots containing frontal cortex lysates from four different C9(−) and four different C9(+) ALS/FTD patients were probed with α-PAAS, α-PA-CTAS, α-PRAS, and α-PR-CTAS antibodies. Results show evidence of RAN protein accumulation with all four antibodies in two of four C9(+) samples and weaker positive staining in one sample with α-PAAS. (B) Immunoblots of C9(+) and C9(−) ALS frontal cortex 2% SDS insoluble fractions with α-PRAS, α-PAAS, and α-GPS/AS. (C) IHC detection of PAAS, PRAS, and GPS/AS protein aggregates in hippocampal neurons from C9(+) ALS patients detected with α-PAAS, α-PA-CTAS, α-PRAS, α-PR-CTAS, and α-GPS/AS antibodies. (D) IF staining with mouse α-GPS/AS (Cy3, red) and rabbit α-GP-CTS (Alexa Fluor 488, green) of C9(+)hippocampal tissue with sense inclusions positive for both antibodies (Upper) and antisense inclusions positive for only GP repeat antibody (Lower). Colocalization of α-GPS/AS and α-GP-CTS staining was confirmed by confocal z-stacks. (E) IF staining of larger region. Arrowheads indicate background lipofuscin autofluroescense, which can be distinguished from the real signal because it fluoresces in all three channels and when merged appears as brownish-orange signal that is distinct from the Cy3 and Alexa Fluor 488 signals shown (31). (F) Percentages of double- (sense) and single- (antisense) labeled aggregates were quantitated among 1,389 cells from three different tiled areas and a total of 88 aggregates. (G–J) RAN and PRAS toxicity studies. (G) G2C4 expansion constructs (+/−ATG-PR-3T) with or without an ATG initiation codon in PRAS frame and 3′ epitope tags. (H) Western blots showing levels of PRAS and GPAS in cells transfected with constructs in G. (I) LDH and (J) MTT assays of transfected HEK293T cells. *P ≤ 0.05, **P ≤ 0.01), ***P ≤ 0.001, n = 6 independent experiments).
Previous studies using antibodies directed against the GP repeat motif detected aggregates, which were assumed to be expressed from the sense strand (11, 12). It is important to note, however, that GP repeat-containing proteins are predicted to be expressed from both sense and antisense transcripts (Fig. 2A). In the sense direction the predicted RAN GPS protein contains a unique C-terminal (CT) sequence. In contrast, the antisense GPAS protein has a stop codon immediately after the repeat. To distinguish sense GPS RAN proteins from antisense GPAS proteins, we performed double-label IF experiments on C9(+) human hippocampal autopsy sections using rabbit α-GP-CTS to detect the CT region of the sense-GP protein and mouse α-GPS/AS to detect both sense and antisense GP expansion proteins. Double-labeling showed two types of inclusions: (i) putative sense inclusions double-labeled with mouse α-GPS/AS (red) and rabbit α-GP-CTS (green), and (ii) putative antisense inclusions singly labeled with mouse α-GPS/AS (red) (Fig. 3D). Approximately 18% of inclusions showed the sense pattern with double-labeling and 82% of inclusions showed the antisense pattern and were positive for α-GPS/AS and negative for α-GP-CTS (Fig. 3 E and F). These data independently confirm that α-GPS/AS antibodies recognize GP proteins containing the endogenous sense C-terminal sequence. Additionally, it appears that the majority of GP-repeat inclusions are expressed from the antisense RNA. Although a potential caveat is that if premature termination exists, both antibodies may underestimate the amount of these proteins. Protein blot analysis, however, argues against premature translational termination. As seen in Fig. S4E (far Right) both the N-terminal α-V5 and C-terminal α-GP-CTs antibodies recognize the same size, single band in lysates expressing recombinant ATG-V5-(GP)120-CT protein. If there were substantial premature termination across the GP expansion then the N-terminal α-V5 antibody would be expected to recognize multiple bands or a smear. Although it is possible that premature termination occurs across longer RAN-initiated repeat tracts, our data suggest that premature termination across GP-encoding expansions containing (G2C4)120 repeats, if it occurs at all, is infrequent.
Taken together, these results indicate that insoluble, aggregate-forming antisense RAN proteins are expressed from all three antisense reading frames and demonstrate the importance of characterizing protein aggregates with both repeat and C-terminal antibodies.
Antisense G2C4 Expansions and RAN Proteins Are Toxic to Cells.
In addition to GPAS and PRAS RAN proteins expressed by RAN translation, two of the antisense reading frames have upstream ATG initiation codons that may result in both ATG-initiated GPAS and PRAS proteins (ATG-GPAS and ATG-PRAS) (Fig. 2A and Fig. S3). We previously showed that the presence of an ATG-initiation codon does not prevent RAN translation from also occurring in all three reading frames (10). Therefore, GPAS and PRAS proteins may be expressed by both AUG-initiated or RAN translation. To explore the effects that an ATG-initiation codon has on RAN protein expression for the G2C4 expansion, we generated an additional minigene construct by placing an ATG-initiation codon in front of the AS-G2C4 repeat (Fig. 3G). We selected the PRAS frame for analysis because an ATG initiation codon naturally occurs in this reading frame. Western blotting shows that HEK293T cells transfected with (+)ATG-PR-3T express substantially higher levels of PRAS protein compared with (−)ATG-PRAS-3T–transfected cells (Fig. 3H). In contrast, qRT-PCR and protein blotting showed transcript levels (Fig. S9A) and levels of RAN-translated GPAS (Fig. 3H) were comparable. Similar to Fig. 2C, RAN-translated PAAS was not detectable by protein blot. We then tested the effects of these constructs on cell viability using complementary assays; lactate dehydrogenase (LDH) detection and methylthiazol tetrazolium (MTT). The LDH assay measures the amount of LDH released into the medium and is an indicator of lost membrane integrity and cell death. For the LDH assay, cells transfected with the (−)ATG-PRAS-3T or (+)ATG-PRAS-3T constructs showed 1.9- and 2.9-fold increases in cell death compared with vector control cells (P = 0.008 and 0.001), respectively (Fig. 3I). Additionally, (+)ATG-PRAS-3T–transfected cells, which express elevated levels of PRAS protein, show a 1.5-fold increase in cell death compared with cells transfected with the (–)ATG-PRAS-3T construct (P = 0.034) (Fig. 3I). The MTT assay, which measures cellular metabolic activity of NAD(P)H-dependent oxidoreductase enzymes and reflects the number of viable cells present, showed similar results. Cells transfected with (–)ATG-PRAS-3T and (+)ATG-PRAS-3T constructs showed dramatic decreases in the number of metabolically active cells [33% (P < 0.00001) and 43% (P < 0.00001), respectively], compared with untreated cells or empty vector controls (Fig. 3J). Additionally, elevated PRAS expression in cells transfected with (+)ATG-PRAS-3T had significantly lower levels of metabolic activity compared with (–)ATG-PRAS-3T cells (P < 0.05). By light microscopy, cell detachment and changes in cell morphology were evident in (–)ATG-PRAS-3T compared with control cells. These phenotypes were worse in (+)ATG-PRAS-3T cells, which express elevated levels of PRAS (Fig. S9 B–D). A similar study using (+/−)ATG-GPs-3T–transfected cells demonstrates that GPs proteins are also toxic to cells (Fig. S9 E–I).
Taken together, these data demonstrate that: (i) the G2C4 and G4C2 expansion mutations are toxic to cells; (ii) increased PRAS and GPS proteins expressed in cells transfected with the (+)ATG-PRAS-3T or (+)ATG-GPS-3T constructs increase cell toxicity and death above levels caused by any DNA, RNA, and RAN protein effects of the (−)ATG-PRAS-3T or (−)ATG-GPS-3T constructs. Therefore, the PRAS and GPS proteins are intrinsically toxic to cells.
All Six RAN Proteins Form Aggregates in the Brain.
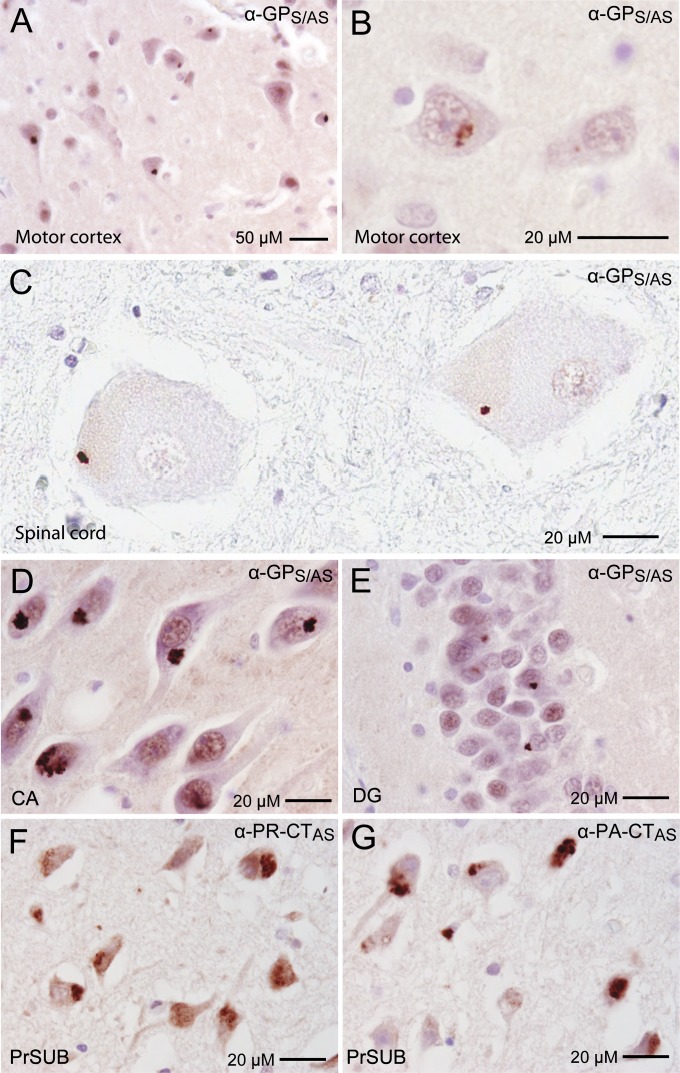
To determine if all six RAN proteins from both sense and antisense RNA strands are expressed in C9(+) ALS patients, IHC staining was performed on sections of paraffin-embedded brain tissues using nine polyclonal antibodies against repeat-expansion or C-terminal sequences of these proteins. In C9(+) cases, there were abundant globular and irregular-shaped neuronal cytoplasmic inclusions in the hippocampus, the majority of which were in the dentate gyrus and in pyramidal cells in the cornu ammonis (CA) regions (Fig. 4). These RAN inclusions were also detected in C9(+) motor cortex (Fig. 4). GPS/AS-positive inclusions were detected in all examined C9(+) cases but not in C9(−) cases or normal control sections. In the CA regions of the hippocampus and in the motor cortex, clusters of GPS/AS aggregates were frequently found in C9(+) cases in the hippocampus, with >20% of neurons showing α-GPS/AS inclusions (Table 1). Fewer aggregates were detected with the α-GP-CT sense antibody. These data are consistent with our separate double-labeling experiments (Fig. 3 D–F), which suggest most GP aggregates are translated from C9ORF72 antisense strand.
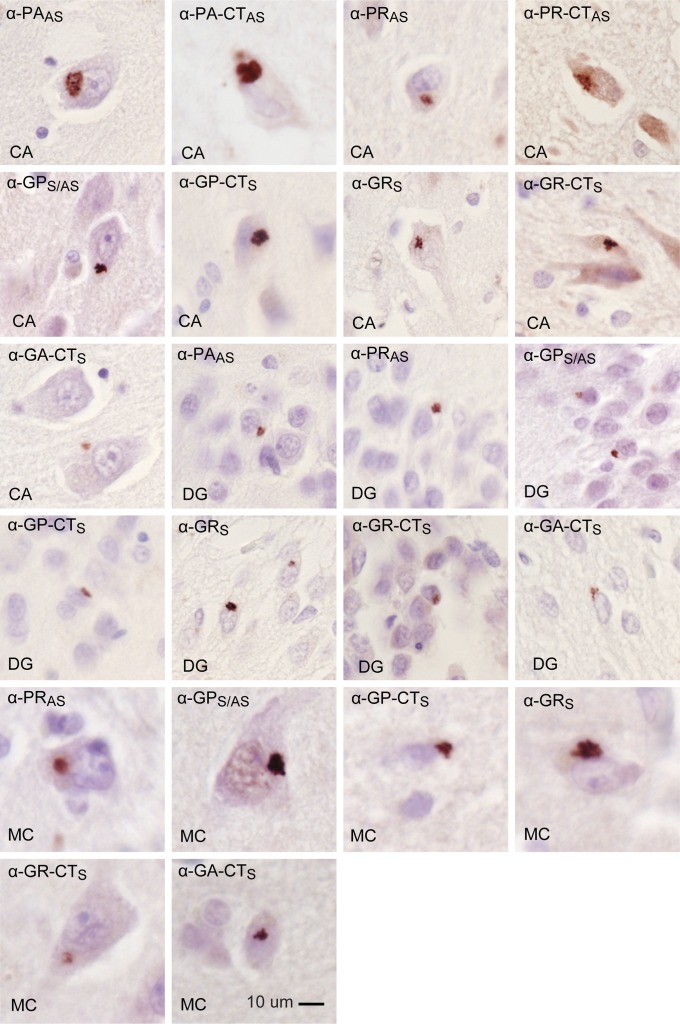
Fig. 4.
In vivo evidence for RAN translation in both antisense and sense directions of C9ORF72. Cytoplasmic inclusions detected by IHC using antibodies against sense (α-GRS, α-GR-CTS, α-GA-CTS, α-GP-CTS) and antisense (α-PAAS, α-PA-CTAS, α-PRAS, α-PR-CTAS), and α-GPS/AS, which recognizes GP proteins made in both the sense and antisense directions. Aggregates are found in neurons of the CA and DG regions of the hippocampus and the motor cortex (MC) of C9(+) ALS autopsy tissue.
Table 1.
Summary of histopathological findings in C9ORF72-positive ALS/FTD cases and controls
| Region and case | Case information |
RAN inclusions |
||||||||
| C9 EXP | Age | Sex/race | PMD | DX | GP | PA | PR | GR | GA | |
| Hippocampus | ||||||||||
| 1 | + | 59 | F/W | 4 | ALS | +++ | + | + | ++ | ++ |
| 2 | + | 42 | M/W | 10 | A/F | +++ | + | NA | NA | NA |
| 3 | + | 74 | F/W | 16 | FTD | +++ | +++* | +++* | ++ | + |
| 4 | + | 45 | M/W | 3 | ALS | +++ | — | + | ++ | + |
| 5 | + | 82 | F/W | 17 | FTD | +++ | + | + | ++ | + |
| 6 | + | 86 | F/W | 10 | A/F | ++ | — | + | ++ | — |
| 7 | — | 76 | M/W | 7 | ALS | — | — | — | — | — |
| 8 | — | 55 | M/W | 7.5 | ALS | — | — | NA | NA | NA |
| 9 | — | 60 | M/W | 16 | CON | — | — | NA | NA | NA |
| 10 | — | 81 | M/W | 6 | FTD | — | NA | NA | — | — |
| 11 | — | 83 | M/W | 17 | FTD | — | — | — | — | — |
| 12 | — | 77 | M/W | 16 | CON | — | — | — | — | — |
| Motor cortex | ||||||||||
| 1 | + | 59 | F/W | 4 | ALS | +++ | — | — | ++ | ++ |
| 2 | + | 42 | M/W | 10 | A/F | +++ | + | + | + | — |
| 7 | — | 76 | M/W | 7 | ALS | — | — | — | — | — |
| 8 | — | 55 | M/W | 7.5 | ALS | — | — | — | — | — |
| 9 | — | 60 | M/W | 16 | CON | — | — | — | — | — |
| Spinal cord | ||||||||||
| 2 | + | 42 | M/W | 6 | A/F | + | — | — | — | — |
| 13 | + | 53 | M/W | 10 | ALS | + | — | — | — | — |
| 14 | + | 55 | F/W | ? | A/F | + | — | — | — | — |
| 7 | — | 76 | M/W | 7 | ALS | — | — | — | — | — |
| 8 | — | 55 | M/W | 7.5 | ALS | — | — | — | — | — |
| 9 | — | 60 | M/W | 16 | CON | — | — | — | — | — |
| 15 | — | 64 | F/W | 0 | ALS | — | — | — | — | — |
| 16 | — | 79 | M/W | 33 | ALS | — | — | — | — | — |
| 17 | — | 79 | M/W | 10 | ALS | — | — | — | — | — |
—, no inclusions; +, occasional; ++, moderate; +++, numerous inclusions; *, variable staining from section to section; A/F, ALS/FTD; CON, control; DX, diagnosis; F, female; M, male; NA, not available; PMD, postmortem interval; W, white/Caucasian. The apparent differences in the frequencies of the various inclusions may reflect differences in protein conformation and epitope availability or differences in the affinities of these antibodies.
PAAS inclusions were detected in hippocampus in four of six C9(+) cases tested and in one of two motor cortex samples (Table 1). In C9(+) cases, the frequency of PAAS inclusions is strikingly lower in the hippocampus and motor cortex compared with GPS/AS inclusions, but high-intensity regional staining with extremely large PAAS inclusions in >50% of neurons was found in one patient (Table 1). PRAS-positive inclusions were also seen in hippocampus in all C9(+) cases examined and in the motor cortex in one of two C9(+) cases tested. Similar to the PAAS staining, PRAS inclusions are less frequent but intense regional staining was occasionally observed.
In the sense direction, GRS-positive inclusions were found in the hippocampus and motor cortex in all C9(+) cases examined, but these inclusions appeared less frequent than the GPS/AS aggregates. GAS inclusions were only occasionally detected by IHC as small perinuclear inclusions in the hippocampus and in motor cortex (Fig. 4 and Table 1).
The apparent differences in the frequency of various types of aggregates may result from differences in protein conformation and epitope availability or differences in the affinities of these antibodies, which were designed to different epitopes. Differences in methods, antibody affinity, and epitope availability may also explain why previously reported GA aggregates detected with α-GA antibodies (12) were the most frequent type of sense aggregate, in contrast to our findings that α-GPS/AS aggregates are abundant and α-GA-CTS aggregates are rare. Taken together, our data show that all six RAN proteins form aggregates in the C9(+) autopsy brains.
Inclusions of RAN Proteins in Upper and Lower Motor Neurons.
A central feature of ALS is the gradual degeneration and death of upper motor neurons in the motor cortex and lower motor neurons in the brainstem and spinal cord. To test if RAN proteins accumulate in upper and lower motor neurons, we performed IHC using all nine antibodies against predicted proteins in both sense and antisense directions. In C9(+) cases, abundant GPS/AS neuronal cytoplasmic inclusions were seen in all layers of the motor cortex, with frequent GPS/AS aggregates in pyramidal neurons of layer III and throughout layer V (Fig. 5A and Fig. S10). Although cell death and atrophy made motor neurons in layer V difficult to identify, GPS/AS inclusions in he remaining upper motor neurons were found (Fig. 5B). Additionally, PAAS-, PRAS-, GRS-, and GAS-positive inclusions were also found in the motor cortex (Fig. 4 and Table 1). In a similar series of experiments performed in spinal cord sections, we detected GPS/AS aggregates in all three cases examined and aggregates in lower motor neurons in two of three C9(+) patients but not in C9(−) ALS cases or normal controls (Fig. 5C and Table 1). Although GPS/AS aggregates seen in most cells have a perinuclar location, the cytoplasmic aggregates in the lower motor neurons were located away from the nucleus in the periphery. Surprisingly, PAAS, PRAS, GRS, and GAS proteins were not detected in the three C9(+) spinal cord samples we examined; this may result from lower overall RAN protein expression or less-frequent aggregation in these cells. Alternatively, it is possible that RAN protein aggregates are less-frequently detected in spinal cord neurons because they are more vulnerable and RAN protein toxicity causes the substantial motor neuron loss in C9(+) ALS patients. This report of RAN protein accumulation in lower motor neurons is unique. The identification of GP-aggregates in both upper and lower motor neurons links C9 RAN-protein accumulation to the neurons that are selectively vulnerable in ALS.
Fig. 5.
Clustered RAN-positive cells in hippocampus and motor cortex and RAN aggregates in motor neurons. IHC showing cytoplasmic α-GPS/AS aggregates in: (A) layer III of motor cortex; (B) upper motor neuron in layer V of the motor cortex; (C) lower motor neurons in the lumbar spinal cord (LSC); (D) in CA and (E) DG regions of the hippocampus. (F and G) Abundant PA-CTAS and PR-CTAS cytoplasmic inclusions in the presubiculum (PrSub) from one patient.
High-Density Clustering of Cells with RAN-Protein Aggregates.
Both sense and antisense proteins accumulate in neurons of C9ORF72 autopsy brains. In general, two types of aggregation patterns were observed: (i) isolated cells with cytoplasmic aggregates and (ii) high-density clusters of cells with cytoplasmic aggregates in which ∼10 to more than 50% of neurons are positive. Clustered aggregates were most frequently detected for GP and were found in the CA1–4 and dentate gyrus (DG) of the hippocampus (Fig. 5 D and E). The clustered GP aggregates in the DG were smaller and less frequent than the large cytoplasmic aggregates in CA regions. Additional clustered GP aggregates were frequently found in subiculum and presubiculum of the hippocampus as well as the motor cortex. Immunostaining of serial sections showed that multiple RAN proteins are often found in the same region. For example, intense clustered staining for PA, PR, GP, GA, and GR proteins is found in the same region of the presubiculum in serial sections from one C9(+) patient (Fig. 5 F and G).
Immunostaining for PA showed that some brain regions have abundant aggregates, whereas other regions in the same section are relatively spared. For example, Fig. 6A illustrates a gradient of PA inclusions (presubiculum > subiculum > CA1) across hippocampal regions in a single section in one patient. PA inclusions in this patient were numerous (>50% of neurons) in the presubiculum (I), moderate in subiculum (II), and rare in CA1 hippocampal regions (III and IV). Consistent with the focal regional staining seen in this section, PA staining was not detected in sections from a separate block of hippocampal tissue taken from the same patient. These data suggest expression of the PA RAN protein is variable from cell to cell or that aggregation of PA in one cell triggers aggregation in neighboring cells, as has been proposed in Parkinson and Alzheimer’s disease (24–30).
Fig. 6.
Clustered staining of RAN proteins. (A) Low-power image of IHC staining with α-PA-CTAS shows variations in staining intensity (red is positive) in regions I–IV, with Insets showing higher-power images. (B) Examples of aggregates from region I show immunoreactivity against all nine antibodies with similar staining for antibodies against repeat and unique C-terminal epitopes.
Next, we used serial sections from this C9(+) case to show that antibodies directed against both the repeat motifs (α-PA, α-PR, α-GP, α-GR) and corresponding C-terminal regions (α-PA-CT, α-PR-CT, α-GP-CT, α-GR-CT, α-GA-CT) detect aggregates in the same densely staining region of the presubiculum (region I) (Fig. 6B). These results show both sense and antisense RAN protein aggregates accumulate in this region. The detection of similar aggregates using antibodies that recognize either the repeat motifs or specific C-terminal regions adds substantial confidence that these antibodies are truly recognizing proteins expressed across both the antisense G2C4 and sense G4C2 expansion transcripts, and provides new tools to understand the biological impact of RAN translation in C9ORF72 ALS/FTD.
Discussion
There has been much excitement about the finding that an intronic microsatellite expansion mutation in C9ORF72 causes a common form of both familial and sporadic ALS/FTD (1, 2). The three major pathological mechanisms being considered for this disease include haploinsufficiency (1, 2), RNA gain-of-function (6–9), and RAN translation (10–13). To date, efforts to understand the molecular mechanisms of this disease have focused exclusively on understanding the consequences of the C9ORF72 expansion mutation in the sense direction. In this article, we demonstrate that the C9ORF72 expansion mutation is also expressed in the antisense direction and provide evidence that antisense RNA foci and antisense RAN proteins contribute to C9ORF72 ALS/FTD. We show (i) antisense C9ORF72 but not sense transcripts are elevated in C9(+) autopsy tissue; (ii) antisense G2C4 expansion transcripts form RNA foci that accumulate in C9(+) brain and blood; (iii) RAN translation occurs across antisense G2C4 expansion constructs in cell culture; (iv) sense and antisense RAN proteins accumulate in C9(+) autopsy brains using a dual immunological approach with both repeat and C-terminal antibodies; and (v) RAN protein aggregates accumulate in upper and lower motor neurons, linking RAN translation directly to the key pathologic feature of ALS.
Since the initial report that G4C2 RNA foci accumulate in C9ORF72 ALS/FTD patient tissues (1), a leading hypothesis is that G4C2 sense transcripts sequester and dysregulate RNA binding proteins similar to the sequestration of muscleblind (MBNL) proteins in DM1, DM2, and SCA8 (4). Several groups have already reported G4C2 binding proteins and are testing their potential role in disease (6–9). Our finding that antisense G2C4 foci also accumulate in patient cells suggests that G2C4 antisense RNAs and binding proteins may play a role. Additionally, our identification of sense and antisense foci in C9(+) peripheral blood may prove useful as an easily accessible biomarker of C9ORF72 ALS/FTD. Biomarkers that monitor both sense and antisense transcripts may be particularly important because therapies that decrease expression of one strand may turn out to increase expression of the other strand. It has been previously shown that decreasing antisense transcripts leads to epigenetic changes and increases in sense transcripts in SCA7 (17). Moving forward, it will be important to examine the role that sense and antisense transcripts, their respective binding proteins, as well as any potential interactions between these factors, might play in C9ORF72 ALS/FTD.
Using a dual immunological approach, we provide evidence that G2C4 antisense transcripts express novel antisense proteins (PAAS, PRAS, GPAS) by RAN translation or from two short ORFs (ATG-PRAS and ATG-GPAS). These data add three unique RAN proteins, plus two putative ATG-initiated antisense proteins, to the previous reports of three sense RAN proteins (11, 12). It is tempting to speculate, based upon the presence of RAN protein aggregates in affected tissues and RAN protein toxicity in vitro data, that these eight C9ORF72 proteins create a toxic cellular environment. However, many important questions remain. First, are RAN proteins toxic in vivo? This question may take a long time to sort out, but based on the polyglutamine disease research, RAN protein aggregates may be protective and it may be that soluble or microaggregated RAN proteins cause problems (4). Second, what tissues and cell types express RAN proteins? In SCA8 and C9ORF72 ALS/FTD, RAN proteins have been reported in neurons but RAN proteins have also been found in peripheral tissues, for example, in DM1 RAN proteins are detected in blood (10), which raises the question of whether RAN protein expression is a systemic problem. Third, why are microsatellite expansion disorders typically adult onset diseases? Could aging or stress increase RAN protein expression or accumulation? Fourth, how do the repeat motifs and flanking sequences impact disease? Additionally, RAN translation across CAG expansions has been shown to occur even in the context of an ATG-initiated ORF (10). This raises the question of whether RAN proteins contribute to disorders, such as Huntington disease, which are currently considered to be caused by effects of polyglutamine-expanded mutant huntingtin protein (4). Fifth, are ATG-initiated antisense GP and PR proteins expressed, and if so, are they expressed at higher levels than their respective RAN proteins? Although additional studies will be required to answer these questions for C9ORF72 ALS/FTD, previous studies of RAN translation in SCA8 show that flanking sequences can affect RAN protein levels (10). Additionally, where RAN translation starts is likely to depend on reading frame. For CAG repeats, RAN translation initiation in the polyGln frame occurs close to or at the beginning of the repeat tract, regardless of an upstream ATG initiation codon. In contrast, RAN in the polyAla frame initiates at sites throughout the length of the repeat (10). Repeat length can also affect the relative levels of RAN protein expression in the various reading frames (10). Sixth, do RAN proteins have a normal function and could loss of this function contribute to disease? In this and other studies (10, 13), it has been shown that RAN translation can occur across relatively short repeats. Do the CAG repeats of 30 and 40 that are normally found in spinocerebellar ataxia type 1 (SCA1) and TATA binding protein (TBP) genes normally undergo RAN translation, and if so, what do these proteins do? Understanding how repeat length and flanking sequence affect RAN translation in C9ORF72 ALS/FTD may provide insights essential for the development of therapies that control or block RAN protein expression.
One of the most puzzling features of ALS/FTD is why C9ORF72 patients, who have been apparently healthy for decades, undergo rapid neurological decline and variable phenotypes. It will be important to determine if C9ORF72 antisense transcript levels increase with age or correlate with disease onset. Additionally, considerable variations in aggregation frequency and patterns, and RAN protein accumulation, are seen within and between patients. Understanding if these differences are driven by variability of RNA or RAN protein expression or aggregation propensity may provide insight into the triggers of disease onset, phenotypic variability (FTD vs. ALS), and progression.
This study, when combined with other studies (10–13, 21, 22), makes a strong case that bidirectional transcription and RAN translation are likely to be involved in many microsatellite disorders. Remarkably, RAN translation has now been reported in all four diseases (SCA8, DM1, ALS/FTD, and Fragile X-associated tremor ataxia syndrome) in which researchers have investigated whether RAN proteins accumulate in patient tissues (10–13). Although it may be possible that antisense oligo or RNAi approaches aimed at reducing expression of expansion transcripts will provide effective therapies, it is also possible that these approaches will be ineffective. Therefore, understanding the roles that sense and antisense RNAs and RAN proteins play in microsatellite expansion diseases is essential for developing additional therapeutic targets. Finally, the idea that eight proteins and two mutant RNAs can be expressed from an intronic mutation has broad implications for understanding fundamental mechanisms of protein synthesis and translational control that should now be considered for a broad category of neurological disorders.
Materials and Methods
cDNA Constructs.
CCCGGGGCC(GGGGCC)2GGGGCCC and CCCGGGGCC(GGGGCC)28GGGGCCC fragments that contain upstream 6xStop codons were synthesized and cloned into pIDTSmart vector by Integrated DNA Technologies. Underlined sequences generate SmaI (CCCGGG) and PspOM1 (GGGCCC) restriction enzyme sites. The 6xStops-(GGGGCC)4-3T and 6xStops-(GGGGCC)30-3T constructs were generated by subcloning NheI/XhoI fragment into a pcDNA3.1 vector containing triple epitopes. To expand the size of the GGGGCC repeats, a SmaI/XhoI fragment was subcloned into PspOMI blunted with T4 DNA polymerase/XhoI of pcDNA-6xStops-(GGGGCC)EXP-3T. To reverse the orientation of GGGGCC repeats in pcDNA-6xStop-3T construct, a SmalI/ClaI fragment was subcloned into pBluescript SK+ to generate pBluescript-(GGGGCC)EXP. The AfeI/XhoI fragment pBluescript-(GGGGCC)EXP was subcloned into pcDNA-6xStop-3T to make pcDNA-6xStop-(GGCCCC)EXP-3T construct.
RT-PCR.
Strand-specific RT-PCR in autopsy tissues.
Total RNA was isolated from FCX autopsy tissues and PBL of ALS patients and healthy controls with TRIzol (Invitrogen). To detect transcripts from both strands, cDNA was generated from 0.25 μg of total RNA using the SuperScript III system (Invitrogen) with LK strand-specific reverse primers and PCR with strand-specific forward and LK primers. The PCR reactions were done as follows: 94 °C for 3 min, then 35 cycles of 94 °C for 45 s, 58 °C for 45 s, and 72 °C for 1 min, followed by 6 min at 72 °C. Bands were cloned and sequence to verify their specificity of the PCR amplification.
RT-PCR for toxicity assay in 293T cells.
Total RNA from cells was extracted using miRNeasy Mini kit (Qiagen) according to the manufacturer's protocol. Total RNA was reverse-transcribed using the SuperScript III RT kit (Invitrogen) and random-hexamer primers. The expression of the different G4C2-3xTag constructs were analyzed by RT-PCR and qPCR using primer set: 3xTag-Fw and 3xTag-Rv. β-Actin expression was used as a reference gene amplified with primer set ACTB3 and ACTB4. Primer sequences are listed in Table S2.
Real time RT-PCR.
Two-step quantitative RT-PCR was performed on a MyCycler Thermal Cycler system (Bio-Rad) using SYBR Green PCR Master Mix (Bio-Rad) and ASORF strand-specific cDNA and primer sets. Control reactions were performed with human β-actin primers ACTB3 and ACTB4 using oligo dT synthesized total cDNA as template. Quantitative RT-PCR was performed for 40 cycles (95 °C 30 s, 60 °C 30 s) in an optical 96-well plate with each sample cDNA/primer pair done in triplicate. The relative fold-changes were generated by first normalizing each experimental Ct value to their β-actin Ct value and then normalized to the healthy control antisense ΔΔCt. Primer sequences are listed in Table S2.
Rapid Ampliciation of 5′ and 3′ cDNA Ends (5′ and 3′ RACE).
Four micrograms of total RNA from 2 C9(+) ALS patients and 2 C9(−) ALS patients FCX autopsy tissues were used for 5′ and 3′ RACE (5′ RACE systems and 3′ RACE; Life Technologies). In 5′ RACE, Primer ASORF R was used for gene-specific first-strand cDNA synthesis and nested reverse primers are 5′ GSP1 and 5′ GSP2. In 3′ RACE, nested forward primers are 3′ GSP1 and 3′ GSP2. The 3′ RACE and 5′ RACE products were gel-extracted, cloned with TOPO TA Cloning (Invitrogen) and sequenced. Primer sequences are listed in Table S2.
Production of Polyclonal Antibodies.
The polyclonal rabbit antibodies were generated by New England Peptide and the polyclonal mouse antibody was generated by the Interdisciplinary Center for Biotechnology Research at the University of Florida. In sense strand (GGGGCC), antisera were raised against synthetic poly(GP), poly(GR) peptides and C-terminal regions of predicted GP, GR, and GA RAN proteins (Fig. S3). In the antisense strand (GGCCCC), antisera were raised against synthetic poly(PA), poly(PR) peptides and the C-terminal regions of predicted PA and PR RAN proteins. Peptides used to generate antibodies to both antisense and sense proteins and their use for Western blot, IF, and IHC is summarized in Table S1.
Cell Culture and Transfection.
HEK293T cells were cultured in DMEM supplemented with 10% FBS and incubated at 37 °C in a humid atmosphere containing 5% CO2. DNA transfections were performed using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions.
Human Samples.
Frozen frontal cortex tissue samples for biochemical and histological analysis included samples from seven C9(+) ALS, five C9(−) ALS controls, and one normal control were used in this research. RNA quality was sufficient in six of the seven samples for qRT-PCR. Additionally, paraffin-embedded fixed tissues from eight C9(+) ALS/FTD and seven C9(−) ALS/FTD cases, as well as two normal controls, were used for IHC analysis. Blood samples from three C9(+) cases and eight controls were used for the blood studies. PBL were isolated from the buffy coat of freshly collected whole blood following brief centrifugation at 2,000 × g. Red blood cells were preferentially lysed and removed using RBC Lysis Buffer (Roche); PBLs were centrifuged, washed once with PBS, and dried on slides. This study was conducted in compliance with the Declaration of Helsinki. Institutional review boards of the University of Florida, The Johns Hopkins University, and Washington University in St. Louis approved the study. Written, informed consent was obtained from participants or relevant parties at the time of enrollment.
Immunofluorescence.
The subcellular distribution of polymeric proteins was assessed in transfected HEK293T cells by IF. Cells were plated on eight-well tissue-culture chambers and transfected with plasmids the next day. Forty-eight hours posttransfection, cells were fixed in 4% PFA in PBS for 30 min and permeabilized in 0.5% triton X-100 in PBS for 15 min on ice. The cells were blocked in 1% normal goat serum (NGS) in PBS for 30 min. After blocking, the cells were incubated for 1 h at room temperature in blocking solution containing the rabbit anti-Myc (Abcam), mouse anti-HA (Covance), mouse anti-Flag (Sigma), rabbit α-GR, and rabbit α-GR-CT primary antibodies at a dilution of 1:400. The slides were washed three times in PBS and incubated for 1 h at room temperature in blocking solution containing goat anti-rabbit conjugated to Cy3 (Jackson ImmunoResearch) and goat anti-mouse conjugated to Alexa Fluor 488 (Invitrogen) secondary antibodies at a dilution of 1:200. The slides were washed three times in PBS and mounted with mounting medium containing DAPI (Invitrogen). IF in patient hippocampal tissue was performed on 6-µm fresh-frozen sections. A similar protocol was used as in transfected cells except 2% NGS was used as blocking buffer and higher dilution of antibodies was used (mouse α-GP 1:1,000 and rabbit α-GP-CT 1:5,000).
RNA-FISH on Brain Sections.
Slides with FCX tissue were fixed in 4% PFA in PBS for 10 min and incubated in prechilled 70% ethanol for 30 min on ice. Following rehydration in 40% formamide in 2× SSC for 10 min, the slides were blocked with hybridization solution (40% formamide, 2× SSC, 20 µg/mL BSA, 100 mg/mL dextran sulfate, and 10 µg/mL yeast tRNA, 2 mM Vanadyl Sulfate Ribonucleosides) for 10 min at 55 °C and then incubated with 400 ng/mL of denatured RNA probe in hybridization solution at 55 °C for 2 h. After hybridization the slides were washed three times with 40% formaminde in 2× SSC and briefly washed one time in PBS. If double-labeling was performed, then denatured G2C4-Cy-5 probe was added, incubated, and processed as above. Autofluorescence of lipofuscin was quenched by 0.25% of Sudan Black B in 70% ethanol and the slides were mounted with mounting medium containing DAPI (Invitrogen).
Quantitation of sense and antisense RNA foci in the FCX was done on three different C9(+) and two C9(−) ALS FCX samples. Six random pictures were taken by confocal microscopy under 60× magnification and 10 z-stacks (0.6 μm between stacks) were analyzed; 300–400 cells were counted for each patient. Total nuclei were detected by DAPI staining and the foci were detected by Cy3 probes.
RNA-FISH in Peripheral Blood Leukocytes.
All solutions were made with DEPC-treated water. Slides were fixed in cold 4% PFA for 10 min on ice and washed three times in 1× PBS for 5 min each wash. Slides were immersed in ice cold 70% ethanol for 15 min and then rehydrated in 40% formamide/2× SSC for 10 min. After being prehybridized for 30 min at 60 °C, slides were rinsed briefly in 40% formamide/2× SSC and denatured G4C2-Cy3 probe was added hot (75 °C) and incubated in humidified chamber at 60 °C for 90 min. Slides were then washed at 55 °C 3 × 10 min. If double-labeling was performed, then denatured G2C4-Cy5 probe was added, incubated, and processed as above. Prolong Antifade Gold with DAPI (Invitrogen) was added and coverslips mounted. Slides were viewed on Leica confocal using a 100× oil objective for PBLs foci and for brain a 63× water objective. Probes were at a concentration of 0.8 ng/μL and 50 μL were added per slide. Prehybridization and hybridizations solutions are as described above.
The specificity of the RNA foci was determined by treating cells before FISH detection with either RNase (100 μg/mL in 2× SSC), DNase (1 U/μL in DNaseI buffer) or Protease K (120 μg/mL in 2 mM CaCl2, 20 mM Tris, pH 7.5). Treated cells were incubated at 37 °C for 30 min, washed three times in corresponding incubation buffer and rehydrated in 40% formamide/2× SSC. Subsequent FISH detection was performed as described above. Antisense probe specificity was tested by hybridizing slides with 10-fold excess unlabeled (G4C2)4 oligo followed by hybridization with either (G4C2)4 -Cy3 (antisense probe) or (G2C4)4-Cy3 (sense probe). Subsequent treatment and detection were performed as described above.
Western Blotting.
Transfected cells in each well of a six-well tissue-culture plate were rinsed with PBS and lysed in 300 μL RIPA buffer with protease inhibitor mixture for 45 min on ice. DNA was sheared by passage through a 21-gauge needle. The cell lysates were centrifuged at 16,000 × g for 15 min at 4 °C, and the supernatant was collected. The protein concentration of the cell lysate was determined using the protein assay dye reagent (Bio-Rad). Twenty micrograms of protein were separated in a 4–12% NuPAGE Bis⋅Tris gel (Invitrogen) and transferred to a nitrocellulose membrane (Amersham). The membrane was blocked in 5% dry milk in PBS containing 0.05% Tween-20 (PBS-T) and probed with the anti-Flag (1:2,000), anti-Myc (1:1,000), anti-HA (1:1,000), or rabbit polyclonal antibodies (1:1,000) in blocking solution. After the membrane was incubated with anti-rabbit or anti-mouse HRP-conjugated secondary antibody (Amersham), bands were visualized by the ECL plus Western Blotting Detection System (Amersham).
Sequential extraction of patient FCX autopsy tissue was performed as follows: tissue was homogenized in PBS containing 1% Triton-X100, 15 mM MgCl2, 0.2 mg/mL DNase I and protease inhibitor mixture and centrifuged at 16,000 × g for 15 min at 4 °C. The supernatant was collected. The pellet was resuspended in 2% SDS and incubated at room temperature for 1 h, then centrifuged at 16,000 × g for 15 min at 4 °C. The supernatant was collected and the 2% SDS insoluble pellet was resuspended in 8% SDS, 62.5 mM Tris⋅HCl pH 6.8, 10% glycerol and 20% 2-Mercaptoethanol for protein blotting (23).
Protein Dot Blot.
For protein dot blot, 1% Triton-X100 soluble fraction and 2% SDS soluble fraction from the sequential extraction was immobilized onto nitrocellulose membranes with Bio-Dot 96-well microfiltration system (Bio-Rad) under vacuum. The membranes were washed in PBST and blotted with each rabbit polyclonal antibody (1:2,000) using the same protocol as Western blotting.
Immunohistochemistry.
Ten-micrometer sections were deparaffinized in xylene and rehydrated through graded alcohol, incubated with 95–100% formic acid for 5 min, and washed with distilled water for 10 min. Heat-induced epitope retrieval was performed by steaming sections in citrate buffer, pH 6.0, at 90 °C for 30 min. To block nonspecific Ig binding, a serum-free block (Biocare Medical) was applied for 30 min. Rabbit polyclonal antibodies were applied at a dilution of from 1:5,000–1:15,000 in serum-free block (Biocare Medical) and incubated overnight at 4 °C. Linking reagent (streptavidin or alkaline phosphatase; Covance) was applied for 30 min at room temperature. These sections were incubated in 3% H2O2 for 15 min to bleach endogenous peroxidase activity. Then labeling reagent (HRP, Covance) was applied for 30 min at room temperature. Peroxidase activity was developed with NovaRed substrate (Vector) and sections were counterstained with hematoxylin.
Cell-Toxicity Assays.
All of the transfection experiments were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions and at a 60% cell confluence. Next, 500 ng of each vector was transfected in 35-mm wells (35 ng per well in 96 well plates). Cell death was determined by measuring LDH cell release, using CytoTox 96 nonradioactive cytotoxicity assay (Promega) according to the manufacturer’s instructions. Absorbance was recorded at 490 nm and total LDH release was measured by lysing the cells with 1% Triton X-100. In each experiment, determinations were performed in quintuplicates for each experimental condition and average data calculated. Statistical significance was determined using the two-tailed unpaired Student t test for single comparisons (P < 0.05) and the ANOVA when multiple pair-wise conditions were compared.
Cell Viability Assays.
HeK293T cells were transfected in 96-well plates and cell viability was determined 42 h posttransfection with the 3-(4,5- dimethythiazol-. 2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. MTT was added to cell culture media at 0.5 mg/mL final concentration and incubated for 45 min at 37 °C. Cells were then lysed with 100 µL of DMSO upon medium removal and absorbance was measured at 595 nm. In each experiment, determinations were performed in quintuplicates. Statistical significance was determined using Student t test (P < 0.05).
Supplementary Material
Acknowledgments
We thank Drs. M. Swanson and J. Cleary for helpful suggestions and discussions; Dr. P. Rabins for contributions to the studies of frontotemporal dementia at The Johns Hopkins University. This work was funded in part by the Keck Foundation (L.P.W.R.); Target ALS (L.P.W.R.); the Amyotrophic Lateral Sclerosis Association (L.P.W.R.); the Muscular Dystrophy Association (L.P.W.R.); the University of Florida (L.P.W.R.); Instituto de Salud Carlos III (M.B.-C.); The Johns Hopkins Alzheimer’s Disease Research Center Grant NIH P50AG05146 (to J.C.T.); the Samuel I. Newhouse Foundation (J.C.T.); and the Packard Center (J.D.R.).
Footnotes
Conflict of interest statement: T.Z. and L.P.W.R. are listed as inventors on pending patents on RAN proteins.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315438110/-/DCSupplemental.
References
- 1.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, et al. ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majounie E, et al. Chromosome 9-ALS/FTD Consortium French Research Network on FTLD/FTLD/ALS ITALSGEN Consortium Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012;11(4):323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DL, Orr HT, Warren ST. The unstable repeats—Three evolving faces of neurological disease. Neuron. 2013;77(5):825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 6.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288(14):9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125(3):413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 8.Xu Z, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110(19):7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108(1):260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 13.Todd PK, et al. CGG repeat-associated translation mediates neurodegeneration in Fragile X tremor ataxia syndrome. Neuron. 2013;78(3):440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strausberg RL, et al. Mammalian Gene Collection Program Team Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venter JC, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 16.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 17.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70(6):1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet. 2011;20(17):3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilburn B, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington’s disease-like 2 mice. Neuron. 2011;70(3):427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd PD, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16(24):3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 21.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38(7):758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 22.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20(3):483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Wyman T, Yu ZX, Li SH, Li XJ. Abnormal association of mutant huntingtin with synaptic vesicles inhibits glutamate release. Hum Mol Genet. 2003;12(16):2021–2030. doi: 10.1093/hmg/ddg218. [DOI] [PubMed] [Google Scholar]
- 24.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106(31):13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14(5):504–506. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 27.Li JY, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14(5):501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 28.de Calignon A, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73(4):685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, et al. Trans-synaptic spread of tau pathology in vivo. PLoS ONE. 2012;7(2):e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polymenidou M, Cleveland DW. Prion-like spread of protein aggregates in neurodegeneration. J Exp Med. 2012;209(5):889–893. doi: 10.1084/jem.20120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13(24):3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.