Significance
The molecular basis of morphological and physiological adaptations in snakes is largely unknown. Here, we study these phenotypes using the genome of the Burmese python (Python molurus bivittatus), a model for extreme phenotypic plasticity and metabolic adaptation. We discovered massive rapid changes in gene expression that coordinate major changes in organ size and function after feeding. Many significantly responsive genes are associated with metabolism, development, and mammalian diseases. A striking number of genes experienced positive selection in ancestral snakes. Such genes were related to metabolism, development, lungs, eyes, heart, kidney, and skeletal structure—all highly modified features in snakes. Snake phenotypic novelty seems to be driven by the system-wide coordination of protein adaptation, gene expression, and changes in genome structure.
Keywords: comparative genomics, transposable elements, systems biology, transcriptomics, physiological remodeling
Abstract
Snakes possess many extreme morphological and physiological adaptations. Identification of the molecular basis of these traits can provide novel understanding for vertebrate biology and medicine. Here, we study snake biology using the genome sequence of the Burmese python (Python molurus bivittatus), a model of extreme physiological and metabolic adaptation. We compare the python and king cobra genomes along with genomic samples from other snakes and perform transcriptome analysis to gain insights into the extreme phenotypes of the python. We discovered rapid and massive transcriptional responses in multiple organ systems that occur on feeding and coordinate major changes in organ size and function. Intriguingly, the homologs of these genes in humans are associated with metabolism, development, and pathology. We also found that many snake metabolic genes have undergone positive selection, which together with the rapid evolution of mitochondrial proteins, provides evidence for extensive adaptive redesign of snake metabolic pathways. Additional evidence for molecular adaptation and gene family expansions and contractions is associated with major physiological and phenotypic adaptations in snakes; genes involved are related to cell cycle, development, lungs, eyes, heart, intestine, and skeletal structure, including GRB2-associated binding protein 1, SSH, WNT16, and bone morphogenetic protein 7. Finally, changes in repetitive DNA content, guanine-cytosine isochore structure, and nucleotide substitution rates indicate major shifts in the structure and evolution of snake genomes compared with other amniotes. Phenotypic and physiological novelty in snakes seems to be driven by system-wide coordination of protein adaptation, gene expression, and changes in the structure of the genome.
Biological research increasingly incorporates nontraditional model organisms, particularly model organisms with extreme phenotypes that can provide novel insight into vertebrate and human biology. Snakes are one such model, and they exhibit many extreme phenotypes that can be viewed as innovative evolutionary experiments capable of illuminating key aspects of vertebrate gene function and systems biology (1–5). The evolutionary origin of snakes involved extensive morphological and physiological adaptations, including limb loss, functional loss of one lung, and elongation of the trunk, skeleton, and organs (6). They evolved suites of radical adaptations to consume extremely large prey relative to their body size, including the evolution a kinetic skull and diverse venom proteins (7–9). They also evolved the ability to extensively remodel their organs and physiology on feeding (10, 11), while enduring fluctuations in metabolic rates that are among the most extreme of any vertebrate (11). At the molecular level, previous research has shown that they have undergone an unprecedented degree of evolutionary redesign and accelerated adaptive evolution across multiple mitochondrial proteins (1, 12). We hypothesized that the extreme changes in the mitochondrial proteins were likely to extend to metabolic genes in the nuclear genome. We also hypothesized that other genes and gene networks associated with extreme physiological and phenotypic adaptations in snakes may also have undergone significant evolutionary changes.
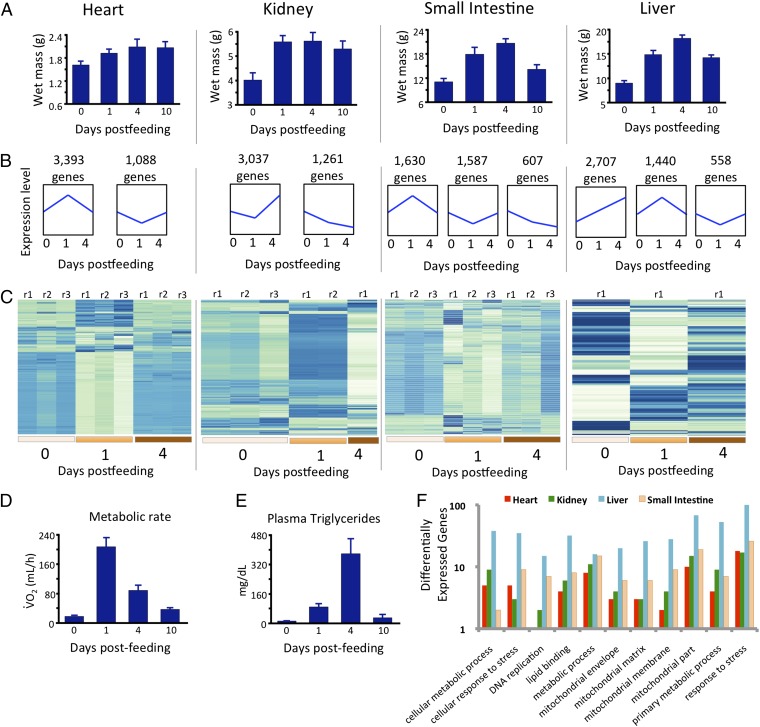
To gain insight into the genomic basis of these phenotypes, we sequenced and annotated the genome of a female Burmese python (Python molurus bivittatus). The python is the most extreme vertebrate model for studying physiological remodeling, rapid organ growth, and metabolic fluctuations after feeding (10, 11). Within 2–3 d after feeding, Burmese pythons can experience 35–100% increases in the mass of major organs, including the heart, liver, small intestine, and kidneys (Fig. 1A) (10, 13). On completion of digestion, each of these phenotypes is reversed; physiological functions are down-regulated, and tissues atrophy in a matter of days (Fig. 1A) (14).
Fig. 1.
Coordinated shifts in physiology and gene expression across organs in pythons after feeding. (A) Rapid increases in organ mass that accompany feeding in the python. (B) Generalized trends in gene expression that are significantly overrepresented in each organ across time points before and after feeding in the python. Results are based on cluster analysis of gene expression profiles and identification of statistically overpopulated profiles. The numbers of genes clustered within each profile are shown above profiles, and trends in the relative magnitude of gene expression for each set of genes are shown in blue. (C) Heat maps of normalized gene expression levels for the top 300 significantly differentially expressed genes across time points shown for different tissues and time points before and after feeding. Low expression levels are indicated by pale colors, and high expression levels are indicated by darker blue. Most time points per tissue have replicates that are indicated at the tops of profiles [labeled by replicate (r) number]; every column per tissue indicates a different individual. (D) Changes in oxygen consumption indicating changes in oxidative metabolism in fasted and fed pythons. (E) Changes in plasma triglyceride levels in fasted and fed pythons. (F) Numbers of significantly differentially expressed genes between 0 and 1 DPF in select GO categories in different tissues.
To complement the genome sequencing results, we also studied transcriptional responses to feeding in four python organs (heart, kidney, liver, and small intestine) for multiple time points. We also evaluated evidence for positive selection in protein coding genes on the lineages leading to the python, cobra, or their common ancestor. In addition, we analyzed changes in functionally important multigene families, such as vemoronasal and olfactory receptors, and opsin genes. Finally, we studied changes in aspects of snake genome structure [repeat content, microsatellite structure, and guanine-cytosine (GC) content] and rates of molecular evolution.
Results and Discussion
The python genome was sequenced using a hybrid approach (combining Illumina and 454 reads), and it is available at National Center for Biotechnology Information: Bioproject PRJNA61234 (GenBank accession no. AEQU00000000). The scaffolded python genome assembly Pmo2.0 is 1.44 Gbp (including gaps), which happened to be the same as the genome size estimated for the related species P. reticulatus (1.44 Gbp) (15). This assembly (Pmo2.0) has an N50 contig size of 10.7 kb and a scaffold size of 207.5 kb (SI Appendix, Tables S1 and S2). Transcriptomic data were used to annotate this genome to provide robust gene models (SI Appendix, Tables S3–S8). For comparative genomic analysis, we analyzed the python genome in conjunction with the genome of the king cobra, Ophiophagus hannah (8). The repetitive contents of the python and king cobra genomes estimated using the de novo P clouds method (16, 17) were similar (python = 59.4%, cobra = 60.4%). Annotation of readily identifiable repeats using a standard consensus repeat element reference library-based approach (18) also found similar repetitive content in the python (31.8%) and cobra (35.2%) genomes (SI Appendix, Table S6). These percentages are only slightly lower than the percentages for humans (∼67% for P clouds and 45% for library-based methods) (16), although the snake genomes are around one-half as large.
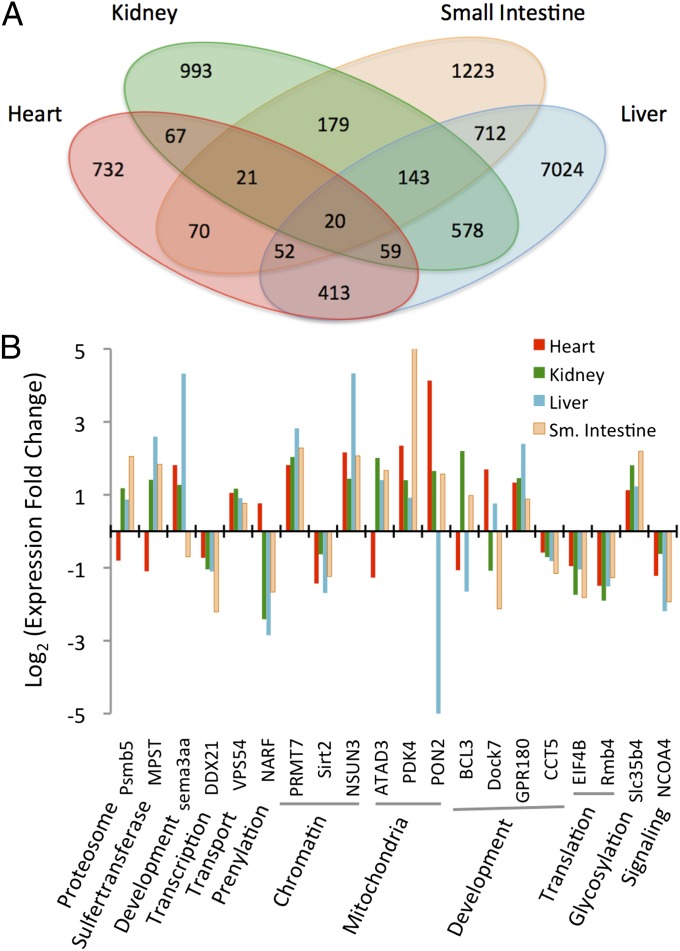
We studied transcriptional responses to feeding in four organs (heart, kidney, liver, and small intestine) for multiple time points before [0 d postfed (DPF)] and after (1 and 4 DPF) feeding (Fig. 1 A–D). These responses involve thousands of genes and large changes in gene expression that are tightly coordinated with the extreme and rapid changes in organ size and performance after feeding (Fig. 1 and SI Appendix, Figs. S1–S17 and Tables S9 and S10). The genes with significant expression responses are functionally diverse and involved in metabolism, chromatin remodeling, growth and development, and human pathologies (Dataset S1). Given the postfeeding increases in oxidative metabolism (Fig. 1D), plasma lipid levels (Fig. 1E), and organ size (Fig. 1A) (10, 11, 14) in the python, we expected genes associated with metabolism, lipids, mitochondria, and DNA replication to be highly regulated between 0 and 1 DPF. As predicted, many differentially expressed genes are associated with these functional groups [based on gene ontology (GO) (19) term analyses], with the one exception being the lack of genes in the heart associated with DNA replication (Fig. 1F). This lack is consistent with previous findings that the python heart experiences hypertrophy (cell growth) rather than hyperplasia (cell division) during postfeeding growth (20, 21). To elucidate core shared aspects of this response, we identified 20 genes that are significantly differentially expressed in all four organs between 0 and 1 DPF (Fig. 2A). Functions of these genes include chromatin remodeling, mitochondrial function, development, translation, and glycosylation, and there are organ-specific patterns in the direction and magnitude of expression changes, with the heart tending to be the most unique (Fig. 2B and Dataset S1).
Fig. 2.
Differentially expressed genes between fasting and 1 DPF across tissues in the python. (A) Numbers of genes significantly differentially expressed between 0 and 1 DPF in four python tissues and the overlap in these gene sets among tissues. (B) Fold expression changes between 0 and 1 DPF for 20 genes differentially expressed in all four tissues and broad functional classifications of these genes.
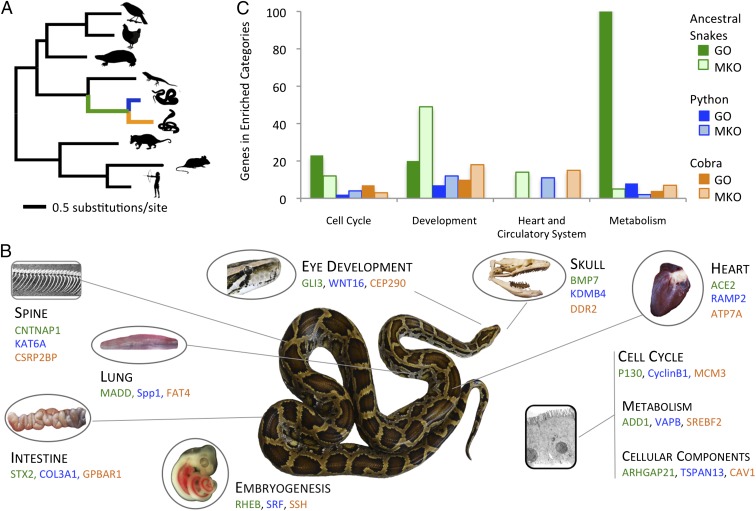
Based on previous work showing extraordinary selection in snake mitochondrial genomes (1, 22, 23), we hypothesized that many nuclear-encoded metabolic genes might show evidence of positive selection (particularly on the ancestral snake branch), and we were curious if positive selection might partially explain other unique physiological and phenotypic features of snakes. To detect selection on protein coding genes, we assembled orthologous gene alignments for 7,442 genes from the python and cobra along with all other tetrapod species in Ensembl (24). We used branch site codon models to detect genes that experienced positive selection on the lineages leading to the python, cobra, or their common ancestor (Fig. 3). We inferred positive selection in 516 genes on the ancestral snake lineage, 174 genes on the cobra, and 82 genes on the python at a P value < 0.001 (Dataset S2). To link these gene sets to phenotypes, we identified mouse KO phenotypes (25) and GO (19) terms that were statistically enriched on different snake lineages (Dataset S3 and SI Appendix, Supplementary Methods). A number of functionally enriched categories of positively selected genes in snakes is readily interpretable in light of the unique aspects of snake physiology and morphology (Fig. 3, Dataset S3, and SI Appendix, Fig. S17). Genes under positive selection include genes that are functionally related to development, the cardiovascular system, signaling pathways, cell cycle control, and lipid and protein metabolism (Fig. 3). We also find a high level of correspondence between enriched categories of differentially expressed genes involved in organ remodeling (Figs. 1 and 2) and genes that have experienced positive selection in snakes, including genes involved in the cell cycle, development, the heart and circulatory system, and metabolism (Fig. 3B).
Fig. 3.
Functional categories of genes under positive selection related to the extreme biology of snakes. (A) Phylogenetic tree of amniotes with branch lengths estimated by maximum likelihood analysis of aligned Ensembl 1:1 orthologs from a subset of taxa analyzed for positive selection. Branches representing snake lineages are colored green for ancestral snake lineage, blue for ancestral python lineage, and orange for ancestral cobra lineage. (B) Examples of genes that have experienced positive selection (P < 0.001) on snake lineages and are related to prominent phenotypic or cellular traits of snakes (colors correspond to the branches in A). Genes are grouped with phenotypic characteristics based on GO and mouse KO phenotype (MKO) terms associated with these genes (Datasets S2 and S3), although no claim is made that genes listed directly explain the snake phenotypes that they are associated with per se or that specific genes shown were selected based on their relative prominence in other literature. (C) Functional clusters of GO and MKO terms that are statistically enriched (P < 0.05) for positively selected genes (P < 0.001). Numbers on the y axis represent the combined numbers of genes in clustered enriched categories.
The ancestral snake lineage shows significant enrichment of positively selected genes in GO and mouse knock out (KO) phenotype categories related to metabolism, eye structure, spine and skull shape, and embryonic patterning mechanisms contributing to somite formation and left–right asymmetry (Fig. 3 and Dataset S3). The high number of positively selected nuclear genes associated with metabolism on this lineage corresponds well with previous studies indicating substantial mitochondrial protein selection also occurring early in snake evolution (1, 12). Other enriched categories correspond well with the major phenotypic shifts, including limb loss, trunk elongation and skeletal changes, associated with the shift to a fossorial lifestyle. On the cobra lineage, we found enrichment of categories related to heart, lung, and neuronal development (Fig. 3 and Dataset S3). This lineage includes the ancestor of colubroid snakes, many of which (like the cobra) are highly active foragers, and categories of positively selected genes may be related to this shift in natural history. The python lineage showed enrichment in categories regulating heart and blood vessel development, hematopoiesis, and cell cycle regulation, which correspond well with the python’s extreme postfeeding response, and potentially, the use of constriction to subdue prey in members of this lineage. These enriched categories in the python include the angiogenic PDGF pathway, signaling through the cytoskeletal regulator ρ-GTPase, the TGF-β/bone morphogenetic protein signaling pathway, and categories associated with bone strength and growth (Dataset S3). Many of the positively selected genes also have prominent medical significance. For example, angiotensin 1 converting enzyme and endothelin 1 are important therapeutic targets for cardiovascular disease (26, 27). Similarly, GRB2-associated binding protein 1, which integrates receptor tyrosine kinase signaling, is known to contribute to breast cancer, melanomas, and childhood leukemia (28).
The extreme phenotypes that characterize snakes can also be linked to changes in multigene families. A prominent feature of snakes is a long forked tongue used to enhance chemoreception. Genes encoding vomeronasal receptors, olfactory receptors, and ephrin-like receptors all show major expansions in the ancestral snake lineage as well as the cobra and python, indicating that expansions in these gene families may also contribute to enhanced chemoreception in snakes (SI Appendix, Figs. S18–S20). It is hypothesized that ancestral snakes were fossorial (6), which reduced selection for light perception. Supporting this fossorial snake ancestor hypothesis, we found that 10 visual and nonvisual opsin genes were lost in snakes but are otherwise present in squamates, including RH2, SWS2, PIN, PPIN, PARIE, MEL1, NEUR2, NEUR3, TMT2, and TMTa (SI Appendix, Fig. S21 and Table S11). The absence of these genes was verified using tBlastx searches for these opsins in the cobra and python annotated gene sets as well as genome assemblies and cDNA assemblies.
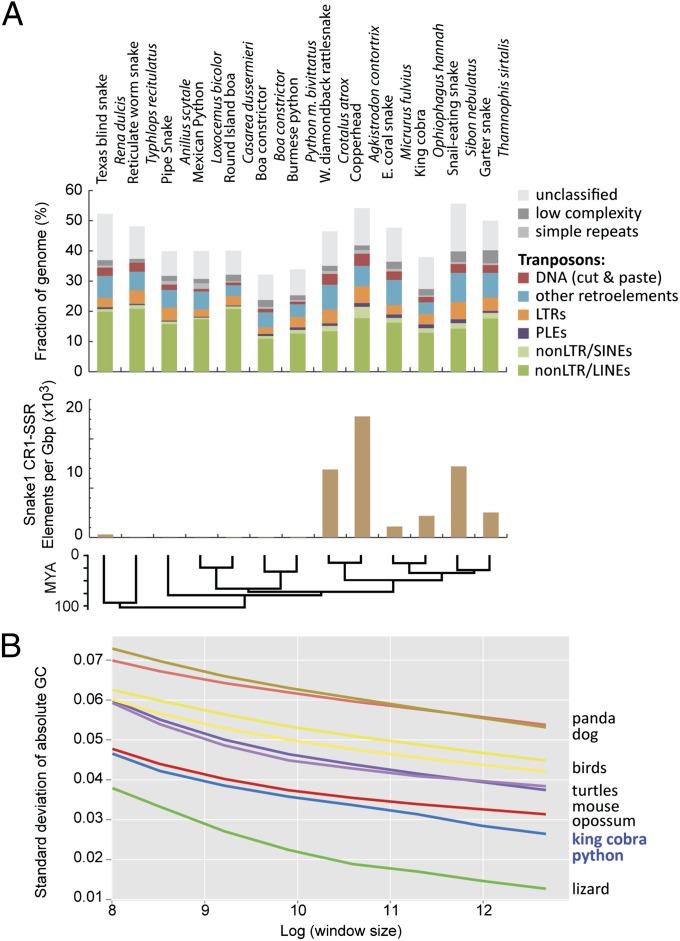
As with the mitochondrial genome (1, 22, 23), snake nuclear genomes have evolved substantial changes in structure and patterns of molecular evolution relative to other vertebrate genomes. To compare repeat element content among partially sequenced and fully assembled snake genomes, we used a single multispecies combined repeat element library for analysis of repeat content (SI Appendix, Supplementary Methods). Despite low variance in snake genome size (SI Appendix, Fig. S21), repeat library-based annotations of genome samples from 10 additional snake species show surprisingly high variance in genomic repeat content but a relatively constant diversity of repeat types (Fig. 4 and SI Appendix, Tables S5–S8) (29). Exceptions are the peculiar families of snake1 (L3) CR1 LINEs that tend to contain microsatellite repeats at their 3′ end (30), which we find to have expanded almost exclusively in colubroid snakes, such as the King Cobra, the Western Diamondback Rattlesnake and Copperhead, and the Garter snake (Fig. 4B). An unexpected result from the analysis of the Anolis lizard genome was that it lacked the GC isochore structure present in mammalian and avian genomes (31, 32). The GC isochore structures of snakes, however, are intermediate between the lack of isochores in Anolis and the clear isochore structure in turtles, birds, and mammals (Fig. 4). These differences in isochore structure raise the intriguing possibility that snakes reevolved GC isochore structure or that the Anolis (or an ancestral squamate) lineage lost GC isochore structure. Trends in GC content at third codon position across amniotes indicate a shift to higher AT content in snakes and based on equilibrium GC content at third codon position calculations, a continued erosion of GC content in king cobra and an increase of GC content in python (SI Appendix, Fig. S24). We also used whole-genome pairwise alignments among Anolis, python, and king cobra to examine how GC content varies among squamates, and we found that both snake species had similar GC profiles to each other but lower GC content than lizard when comparing across aligned genomic sites (SI Appendix, Fig. S25). This inference of dynamic GC content evolution is consistent with a shift in isochore structure in reptile genomes.
Fig. 4.
Variation and uniqueness of snake genome content and structure. (A) Amounts and types of readily identified repeat elements in snake complete and sampled genomes. Estimates in Top and Middle show abundance of genomic repeat elements across 10 snake species based on sampled genome sequences, except for the python and cobra, which are based on complete genomes. Bottom shows genomic density of snake1 CR1 LINE elements for subfamilies that tend to contain microsatellite repeats at their 3′ tails in snake genomes and genome samples. (B) Evidence for shifts in genomic GC isochore structure in squamate reptiles. The y axis is the standard variation of GC content when examining the genome at nonoverlapping window sizes (from 5- to 320-kb windows log-transformed on the x axis). Larger values indicate greater among-window GC content heterogeneity. For example, at all spatial scales, mammals have the greatest GC heterogeneity, and squamate reptiles have the least GC heterogeneity. The right side of the graph (GC SD at a window size of 320 kb) shows GC heterogeneity at a spatial scale on the order of isochore structure; the low GC heterogeneity of squamate reptiles indicates a reduced representation of GC-rich isochores compared with the other taxa. LTR, long terminal repeat; PLE, Penelope-like element; SINE, short interspersed nuclear element.
Rates of molecular evolution also differ substantially across reptile lineages (33). Although turtle genes evolve slowly compared with other sequenced amniotes (34), we find snake genes have evolved rapidly compared to other amniotes (Fig. 3A). Based on a subset of 10,000 aligned codons sampled from orthologous gene alignments, snake lineages experienced relatively high rates of evolution compared with the turtle and other amniote lineages (SI Appendix, Fig. S26). Analysis of the full set of 62,817 fourfold degenerate third codon positions from the orthologous gene set indicates that snake neutral substitution rates are also accelerated relative to other reptilian lineages (SI Appendix, Fig. S27). Additional analyses of 44 nuclear genes for >150 squamate reptile species (from a previous phylogenetic study) (35) indicate accelerated neutral evolution in the ancestral lineages of squamate reptiles, snakes, and colubroid snakes (SI Appendix, Fig. S28).
The comparative systems genomics approach that we have taken to study snake genomes has provided hundreds of candidate genes to study the process by which genes and gene networks coevolve to produce phenotypic diversity in vertebrates. The degree to which the physiological, morphological, and metabolic changes in the snakes coincide with molecular changes is remarkable. It has been hypothesized that major morphological changes are primarily driven by changes in gene expression (36). Snakes provide an alternative vertebrate model system, in which the extensive system-wide evolutionary coordination of protein adaptation, gene expression, and changes in the structure and organization of the genome itself seem to have driven phenotypic novelty. Although there are examples of such types of changes in other vertebrates (37), we expect that the genomic changes seen in snakes and documented here are exceptional in their number and magnitude. There have been sufficient vertebrate mitochondrial genomes sampled to make a convincing case that the adaptive changes observed in snake mitochondrial genomes are truly exceptional (1, 22, 23), and among the 60+ currently published vertebrate nuclear genomes, the degree of change observed in snakes is exceptional as well. The python genome, together with the genome of the king cobra (8), will accelerate understanding of the genomic features that underlie the phenotypic uniqueness of snakes.
Materials and Methods
Burmese Python Genome Sequencing.
All animal procedures were conducted with registered Institutional Animal Care and Use Committee (University of Texas, Arlington, TX) protocols. Python genome sequencing libraries were constructed from a single P. molurus bivittatus female obtained commercially. We created and sequenced multiple whole-genome shotgun libraries on an Illumina Genome Analyzer IIx, an Illumina HiSeq 2000, and a 454 FLX sequencer. In total, we generated 73.8 Gbp (∼49× genome coverage) for python genome assembly and scaffolding, which included data generated for a previous draft assembly (38). The genome was assembled using SOAPdenovo (39) and Newbler (Roche), and alternate assemblies were merged (40). Details are provided in SI Appendix, Supplementary Methods.
Annotation and Gene Prediction.
The genome assembly was annotated using the MAKER annotation pipeline (41). All available RNAseq data from multiple organs and fasted/fed time points were used in developing gene model predictions, and repeat element libraries from both complete snake genomes and the snake genome sampling were used to annotate repeat elements in the python (SI Appendix, Supplementary Methods and SI Appendix, Tables S5–S10). The annotated genome assembly is available under the National Center for Biotechnology Information Bioproject PRJNA61234 (GenBank accession no. AEQU00000000).
Comparative and Evolutionary Analyses.
Ortholog sets were assembled by addition of our python and cobra gene coding DNA sequences (CDSs) sets to the Ensembl Compara v70 1:1 vertebrate ortholog set (24, 42). Analyses included filtering likely gene duplicates in snakes, quality control steps, and alignment. Alignments were analyzed using a maximum likelihood branch site test for sites that were uniquely positively selected on the python, cobra, or ancestral snake lineages (SI Appendix, Supplementary Methods).
Supplementary Material
Acknowledgments
We thank Carl Franklin for donation of the specimen used for genome sequencing. We thank Roche 454 for contributing 454 sequencing used in python genome assembly, Roger Winer for running Newbler assemblies, WestGrid and Compute/Calcul Canada (A.P.J.d.K.) for providing extra computing resources, and two anonymous reviews for constructive comments. The project was supported by setup funds from the University of Texas (to T.A.C.) and the University of Colorado School of Medicine (to D.D.P.); Roche-454 Proof of Concept grants (to T.A.C. and D.D.P.); National Science Foundation Grants IOS 0922528 (to A.M.B.) and IOS 0466139 (to S.M.S.); and National Institutes of Health Grants R01GM083127 (to D.D.P.) and R01GM097251 (to D.D.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AEQU00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314475110/-/DCSupplemental.
References
- 1.Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS One. 2008;3(5):e2201. doi: 10.1371/journal.pone.0002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez C, et al. Control of segment number in vertebrate embryos. Nature. 2008;454(7202):335–339. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- 3.Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399(6735):474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- 4.Di-Poï N, et al. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature. 2010;464(7285):99–103. doi: 10.1038/nature08789. [DOI] [PubMed] [Google Scholar]
- 5.Vonk FJ, Richardson MK. Developmental biology: Serpent clocks tick faster. Nature. 2008;454(7202):282–283. doi: 10.1038/454282a. [DOI] [PubMed] [Google Scholar]
- 6.Vidal N, Hedges SB. Molecular evidence for a terrestrial origin of snakes. Proc Biol Sci. 2004;271(Suppl 4):S226–S229. doi: 10.1098/rsbl.2003.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casewell NR, Huttley GA, Wüster W. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 2012;3:1066. doi: 10.1038/ncomms2065. [DOI] [PubMed] [Google Scholar]
- 8.Vonk FJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackessy SP. Biochemistry and pharmacology of colubrid snake venoms. J Toxicol. 2002;21(1-2):43–83. [Google Scholar]
- 10.Secor SM, Diamond J. Adaptive responses to feeding in Burmese pythons: Pay before pumping. J Exp Biol. 1995;198(Pt 6):1313–1325. doi: 10.1242/jeb.198.6.1313. [DOI] [PubMed] [Google Scholar]
- 11.Secor SM, Diamond J. A vertebrate model of extreme physiological regulation. Nature. 1998;395(6703):659–662. doi: 10.1038/27131. [DOI] [PubMed] [Google Scholar]
- 12.Castoe TA, et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci USA. 2009;106(22):8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox CL, Secor SM. Matched regulation of gastrointestinal performance in the Burmese python, Python molurus. J Exp Biol. 2008;211(Pt 7):1131–1140. doi: 10.1242/jeb.015313. [DOI] [PubMed] [Google Scholar]
- 14.Secor SM. Digestive physiology of the Burmese python: Broad regulation of integrated performance. J Exp Biol. 2008;211(Pt 24):3767–3774. doi: 10.1242/jeb.023754. [DOI] [PubMed] [Google Scholar]
- 15.De Smet WHO. The nuclear Feulgen-DNA content of the vertebrates (especially reptiles), as measured by fluorescence cytophotometry, with notes on the cell and chromosome size. Acta Zool et Pathologica Antverpiensia. 1981;76(1):119–167. [Google Scholar]
- 16.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu W, Castoe TA, Hedges DJ, Batzer MA, Pollock DD. Identification of repeat structure in large genomes using repeat probability clouds. Anal Biochem. 2008;380(1):77–83. doi: 10.1016/j.ab.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smit AFA, Hubley R, Green P (2004) RepeatMasker Open-3.0. Available at http://www.repeatmasker.org. Accessed December 1, 2012.
- 19.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen JB, Rourke BC, Caiozzo VJ, Bennett AF, Hicks JW. Physiology: Postprandial cardiac hypertrophy in pythons. Nature. 2005;434(7029):37–38. doi: 10.1038/434037a. [DOI] [PubMed] [Google Scholar]
- 21.Riquelme CA, et al. Fatty acids identified in the Burmese python promote beneficial cardiac growth. Science. 2011;334(6055):528–531. doi: 10.1126/science.1210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang ZJ, et al. Comparative mitochondrial genomics of snakes: Extraordinary substitution rate dynamics and functionality of the duplicate control region. BMC Evol Biol. 2007;7:123. doi: 10.1186/1471-2148-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castoe TA, et al. Dynamic nucleotide mutation gradients and control region usage in squamate reptile mitochondrial genomes. Cytogenet Genome Res. 2009;127(2–4):112–127. doi: 10.1159/000295342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database Issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eppig JT, et al. The Mouse Genome Database (MGD): Comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40(Database Issue):D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang CC, Struthers AD. Targeting the renin-angiotensin-aldosterone system in heart failure. Nat Rev Cardiol. 2013;10(3):125–134. doi: 10.1038/nrcardio.2012.196. [DOI] [PubMed] [Google Scholar]
- 27.Kaoukis A, et al. The role of endothelin system in cardiovascular disease and the potential therapeutic perspectives of its inhibition. Curr Top Med Chem. 2013;13(2):95–114. doi: 10.2174/1568026611313020003. [DOI] [PubMed] [Google Scholar]
- 28.Ortiz-Padilla C, et al. Functional characterization of cancer-associated Gab1 mutations. Oncogene. 2013;32(21):2696–2702. doi: 10.1038/onc.2012.271. [DOI] [PubMed] [Google Scholar]
- 29.Shedlock AM, et al. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Natl Acad Sci USA. 2007;104(8):2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castoe TA, et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol Evol. 2011;3:641–653. doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita MK, Edwards SV, Ponting CP. The Anolis lizard genome: An amniote genome without isochores. Genome Biol Evol. 2011;3:974–984. doi: 10.1093/gbe/evr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477(7366):587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzika AC, Helaers R, Schramm G, Milinkovitch MC. Reptilian-transcriptome v1.0, a glimpse in the brain transcriptome of five divergent Sauropsida lineages and the phylogenetic position of turtles. Evodevo. 2011;2(1):19. doi: 10.1186/2041-9139-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaffer HB, et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013;14(3):R28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiens JJ, et al. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol Lett. 2012;8(6):1043–1046. doi: 10.1098/rsbl.2012.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3(7):e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan QH, et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013;23(9):1091–1105. doi: 10.1038/cr.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castoe TA, et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12(7):406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao G, et al. Graph accordance of next-generation sequence assemblies. Bioinformatics. 2012;28(1):13–16. doi: 10.1093/bioinformatics/btr588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vilella AJ, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19(2):327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.