Significance
This work explores the environmental correlates of variations in codon 104 of the p53 gene in three mammalian species: two subterranean mammals, highland- and lowland-dwelling wild zokors (Myospalax baileyi and Myospalax cansus, respectively), and one highland-dwelling aboveground species, the root vole (Microtus oeconomus). In Microtus oeconomus the codon 104E variation in p53 suppresses apoptotic gene reactivation and cell apoptosis. In contrast, in M. baileyi the codon104N variation is responsible for the transactivation of apoptotic genes under three environmental stresses—hypoxia, hypercapnia (acidic stress, high CO2), and cold temperature—that characterize its ecological niche in the Tibet plateau. We conclude that p53 in nature is adapted in structure and function in accordance with specific ecological stresses.
Keywords: ecological stress, evolution, Apaf1, Noxa, Puma
Abstract
Mutational changes in p53 correlate well with tumorigenesis. Remarkably, however, relatively little is known about the role that p53 variations may play in environmental adaptation. Here we report that codon asparagine-104 (104N) and glutamic acid-104 (104E), respectively, of the p53 gene in the wild zokor (Myospalax baileyi) and root vole (Microtus oeconomus) are adaptively variable, meeting the environmental stresses of the Tibetan plateau. They differ from serine-104 (104S) seen in other rodents, including the lowland subterranean zokor Myospalax cansus, and from serine 106 (106S) in humans. Based on site-directed mutational analysis in human cell lines, the codon 104N variation in M. baileyi is responsible for the adaptive balance of the transactivation of apoptotic genes under hypoxia, cold, and acidic stresses. The 104E p53 variant in Microtus oeconomus suppresses apoptotic gene transactivation and cell apoptosis. Neither 104N nor 104E affects the cell-cycle genes. We propose that these variations in p53 codon 104 are an outcome of environmental adaptation and evolutionary selection that enhance cellular strategies for surviving the environmental stresses of hypoxia and cold (in M. baileyi and M. oeconomus) and hypercapnia (in M. baileyi) in the stressful environments of the Qinghai-Tibet plateau.
The regulatory mechanisms of p53 mutation related to tumorigenesis have been widely studied and elucidated (1, 2). Notably, however, p53 evolution and adaptation to environmental stresses have not attracted as much attention. Current studies show that p53 is a master sensor and regulator in response to various stressors, such as DNA damage and hypoxia (3–6). Activation of p53 by stresses results in cell-cycle arrest, DNA repair, senescence, or apoptosis in which a series of p53 target genes are involved to maintain genomic integrity (2). The p53 variations associated with environmental stresses have been described in the Mexican salamander axolotl Ambystoma mexicanum and the Israeli blind subterranean mole rat (Spalax judaei; hereafter, S.j.) (7–9).
For animals existing on high plateaus, hypoxia and cold serve as strong environmental selective pressures generating adaptive complexes to cope with these stresses. Animals that have evolved on plateaus adopt various strategies involving multiple variations to regulate a series of genes (3, 7). The zokor (Myospalax baileyi, Thomas, 1911; hereafter M.b.) and root vole (Microtus oeconomus, Pallas, 1776; hereafter M.o.) are the dominant native mammals living on the alpine meadow of the Qinghai-Tibet Plateau of China at altitudes of 3,000–4,500 m (equivalent to 11.0–13.0% O2 at sea level). M.b. is genetically close to Myospalax cansus (Lyon, 1907; hereafter, M.c.), which lives in subterranean burrows at a lower altitude of about 800 m in the lowland of western China. M.b. and M.c. spend their entire life cycle at 70–250 cm underground with significantly low O2 and high CO2 levels in their burrows (10). Since the collision of the Indian and the Eurasian plates during the Tertiary (40–50 Mya) formed the Tibet plateau (11), small mammals living in this region have been geographically and ecologically isolated from other species and have adapted to the stressful plateau environment, contributing to the East Asian biodiversity (12–15). Our previous work demonstrated that mammals of the Qinghai-Tibet plateau are well adapted to the hypoxic environment (16–19), with particular expression patterns of HIF-1α and IGF-I and its binding protein (IGFBP-1), which mediate protection against hypoxia (20–23). Cells exposed to hypoxia succumb to p53-dependent apoptosis (24–27); thus mutations in p53 are required for cell survival under selective pressures. We examined the hypothesis that plateau mammals are adapted to this environment with p53 alterations linked to hypoxia, hypercapnia (high CO2), and cold.
Here we report that the variations of p53 codon 104 in three rodent species during long-term evolution and adaptation at the Qinghai-Tibet plateau reflect diverse survival strategies. The present study provides insights into the contribution of p53 variations to native mammals’ adaptation to the diverse and extreme environmental stresses of their habitats.
Results
Comparison and Phylogenetic Tree of M.b., M.c., and M.o. p53 Sequences.
The p53 mRNAs of the subterranean M.b. and M.c. and the fossorial but above-ground–foraging M.o. of the Tibet plateau were cloned and sequenced. The coding regions of M.b. and M.c. p53 are composed of 1,179 bp, coding a protein of 392 aa. M.b. and M.c. p53 proteins have 98% identity, and both have identities of 85%, 83%, 80%, and 95% with humans, rats, mice, and S.j. p53 protein, respectively. M.o. p53 is composed of 1,176 bp and codes a protein of 391 aa, showing identities of 82%, 83%, 78%, 88%, and 88% to humans, rats, mice, S.j, and M.b. p53 protein, respectively. Phylogenetic trees based on the p53 sequences indicated that in M.b., M.c., M.o., and S.j. p53 evolved adaptively and convergently against hypoxia (Fig. 1 and Fig. S1), although M.o. and M.b. p53 diverges from M.c. p53 in terms of cytochrome b (Fig. 1A).
Fig. 1.

(A) Phylogenetic trees based on the p53 and cytochrome b protein sequences of M. baileyi, M. cansus, and Microtus oeconomus as well as those of other rodents constructed using the neighbor-joining method. (B) Photographs of M.b., M.c., and M.o.
Multiple alignment analysis was performed using Multalin software as described (28). Compared with human p53, three amino acid residues of M.b. p53 and two amino acid residues of M.o. p53 within the DNA-binding domain (DBD) were altered. We found the mutations serine-104-asparagine (S104N), alanine-127-cysteine (A127C), and valine-215-isoleucine (V215I) within the DBD of M.b. p53, as well as serine-104-glutamic acid (S104E) and serine-258-proline (S258P) in M.o. p53 (the corresponding positions in humans are 2 aa greater; i.e., codon 106). In addition to the DBD, there were a leucine insertion at position 322 in the C terminus of M.b. p53, an alanine-86-valine (A86V) mutation in the N terminus, and an arginine-340-serine (R340S) mutation in the C terminus of M.o. p53. The R340S mutation was found to be an evolutionarily positively selected site detected by using PAML 4 software (29) and was tested with the branch-site test as described (30, 31). We examined these sites because the M.o.-specific A86V mutation is close to the core domain and because the S104E mutation (corresponding to codon 106 in humans), which is found only in four fishes (Barbus barbus, Platichthys flesus, Tetraodon miurus, and Xiphophorus hellerii) and the squid Loligo forbesi, living in deep water or at the sandy bottom, resides in the core domain. The 104N of M.b. p53 also exists in the rat, mouse, cattle, sheep, rabbit, and the Mongolian gerbil, but 104S exists in S.j., M.c., and human. The mutations of A127C, V215I, and leucine-322 are specific to M.b. (Fig. S2).
Expression of M.b. and M.o. p53 Under Hypoxia.
p53 mRNA levels under hypoxia were assessed by quantitative real-time RT-PCR. Mimicking the oxygen levels at an altitude of 7 km for 8 h induced significant increases in rat p53 mRNA in liver (32). In contrast, it reduced p53 mRNA in the M.b. liver (Fig. S3A). Remarkably, the p53 protein levels in both M.b. and M.o. livers were decreased in hypoxic conditions, as determined by Western blotting (Fig. S3B).
Codon 104 Variation Is Critical for Transcription of Apoptotic Genes in M.b., M.c., and M.o.
To compare the functional characteristics of M.b., M.c., and M.o. WT p53, a dual-luciferase reporter assay was used in p53-null human non-small cell lung cancer NCI-H1299 cells and in cervical cancer HeLa cells, which have low endogenous p53. The p53 target apoptotic genes IGFBP3, Apoptotic protease activating factor 1 (Apaf1), BCL2-associated X protein (Bax), the Bcl-2 homology 3 (BH3)-only pro-apoptotic protein (Noxa), and P53 upregulated modulator of apoptosis (Puma) and the cell-cycle arrest genes p21 and the human homologue of mouse double minute 2 (Hdm2) were examined (33, 34). All rodent and human WT p53 expression plasmids were cotransfected with reporter plasmids of these target genes, and dual-luciferase reporter assays were performed. We found that M.b. p53 markedly activated IGFBP3, Apaf1, and Bax but suppressed Noxa. M.c. p53 activated IGFBP3, Apaf1, Bax, and also Puma. M.o. p53, however, suppressed all the apoptotic gene transcriptions (Fig. 2 and Fig. S4). Endogenous expression of these apoptotic genes was tested in cells transfected with human, M.b., M.c., and M.o. p53, showing an expression pattern similar to that detected by dual-luciferase reporter assays (Fig. S4). Moreover, p21 and Hdm2 were not affected by human and animal p53, except that p21 was suppressed by M.c. p53 (Fig. S4). Furthermore, M.b. p53 induced high expression of Bcl-2, but human, M.c., and M.o. p53 suppressed Bcl-2 (Fig. 2). These results suggest that codon 104 variations in p53 of M.b., M.c., and M.o. are correlated to distinct transcriptional patterns for apoptotic or antiapoptotic genes (Table S1).
Fig. 2.
Comparison of transactivation of apoptotic genes by WT p53 and 104 mutants in M.b., M.c., and M.o. Results shown are the mean ± SD of three independent transfection assays performed in duplicate. **P < 0.01, ***P < 0.001, WT M.b. and WT M.c. compared with human WT p53 (unpaired t test). #P < 0.05, M.b. WT compared with M.b. 104 mutant. +P < 0.05, ++P < 0.01, +++P < 0.001, M.o. WT compared with M.o. 104 mutant. &P < 0.05, &&&P < 0.001 M.c. WT compared with M.c. 104 mutant.
Site-Directed Mutagenesis.
We conducted site-directed mutagenesis to highlight the nature and functions of the mutations found in these codons in natural populations. Site-directed mutagenesis of p53 revealed that variation in codon 104 is a key factor in the p53 target gene transactivations seen in M.b. and M.o as compared with the other variations within the core domain (Fig. S5). Amino acid replacements in p53 codon 104 showed that the M.b. p53 N104S mutation down-regulated the expression of IGFBP3 and Apaf1; however, the M.c. p53 S104N mutation down-regulated the expression of Apaf1, Bax, Noxa, and Puma but up-regulated the expression of IGFBP3. Humanization of M.o. p53 (E104S) up-regulated the transactivation of all apoptotic genes from a suppressed status (Fig. 2). These reversed effects suggest that the p53 codon 104 variations in highland-dwelling M.b. and M.o. and in lowland-dwelling M.c. correlate to their distinct transactivational patterns for apoptotic genes and are likely to have a causal relationship for some apoptotic genes. It has been known that apoptotic activities of p53 can be transcriptionally repressed (35–37). Bcl-2 can be either activated to protect cells from apoptosis (38) or transcriptionally repressed by p53 (39–41). Similar to human p53, M.c., and M.o. p53 transcriptionally repressed Bcl-2, whereas M.b. p53 strongly activated Bcl-2 (Fig. 2 and Table S1).
p53 Induces Different Cell Fates in M.b. and M.o. Because of Codon 104 Variation.
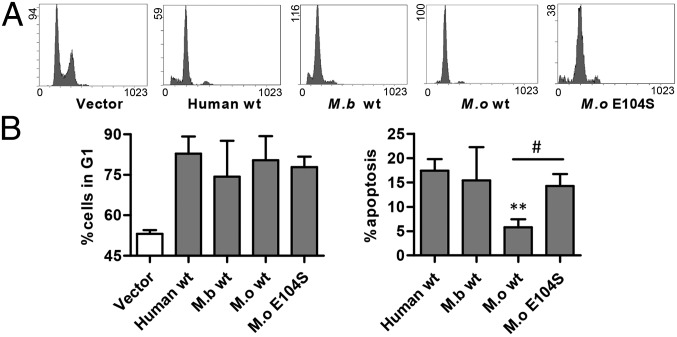
To determine whether the differential transactivation of apoptotic targets by M.b. and M.o. p53 altered their ability to induce apoptosis, we performed flow cytometry on HeLa cells transfected with human, M.b., and M.o. WT p53. All p53 fragments were tagged with GFP, forming p53–GFP fusion plasmids. GFP-fused p53 also induces apoptosis and cell-cycle arrest (42, 43). The DNA content of cells stained with propidium iodide was analyzed using flow cytometry. WT human, M.b., and M.o. p53 induced similar degrees of G1-phase arrest (Fig. 3). However, the functional apoptotic activity of M.b. p53 was similar to that of human p53 (sub-G1 DNA content), whereas M.o. p53 was defective in inducing apoptosis. Humanization of codon 104 restored its apoptotic ability (Fig. 3). Apoptosis detected using double stained method showed similar results (Fig. 4). The apoptotic function of M.o. p53 was closely correlated with its down-regulated transactivation of the apoptotic targets IGFBP3, Apaf1, Bax, Puma, and Noxa (Fig. 2), suggesting that the evolutionary development of the codon 104 variation resulted in the down-regulation of the apoptotic function of M.o. p53 (Table S1). This development allows M.o. to escape from hypoxia-induced apoptosis.
Fig. 3.
Flow cytometry showing cell-cycle arrest and apoptosis induced by WT and mutated p53 of humans, M.b., and M.o. (A) Cell-cycle arrest and apoptosis induced by empty vector and WT p53 from human, M.b., and M.o. and the p53 E104S mutation of M.o. (B) Histograms representing the G1 phase arrest and apoptotic rate induced by each p53. The apoptotic rate for cells transfected with WT p53 was calculated as the percentage of cells with sub-G1 DNA content minus the percentage that transfected with empty vector. Results shown are the mean ± SD of four independent transfection assays, **P < 0.01, compared with control (unpaired t test). #P < 0.05, M.o. E104S mutant compared with WT p53.
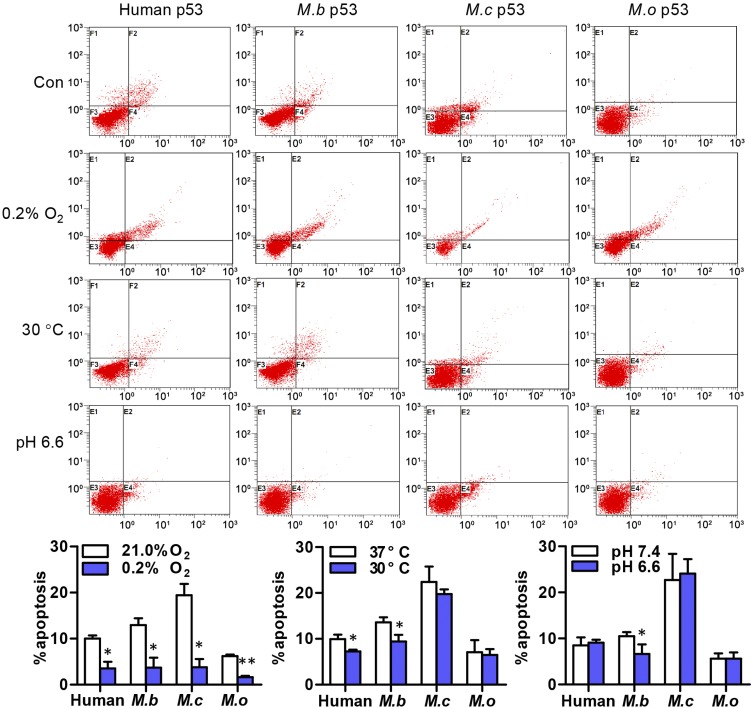
Fig. 4.
M.b., M.c., and M.o. have different apoptotic p53 variants in response to hypoxia, cold, and acidic stresses. Apoptosis induced by these p53 genes under each stress was determined by using flow cytometry to detect GFP-p53+ cells stained with Annexin V- phycoerythrin and 7-amino-actinomycin. Results shown are the mean ± SD of three independent transfection assays. *P < 0.05, **P < 0.01, stress compared with control (unpaired t test).
Variation in Codon 104 Contributes to Different p53 Transactivation Patterns in Response to Hypoxia, Cold, and Acidic Stress in M.b., M.c., and M.o.
M.b. and M.o. evolved adaptive resistance to hypoxic and hypercapnic stresses concomitant with the geological uplift of the Qinghai-Tibet plateau. Therefore, we investigated the response of p53 target apoptotic genes to these stresses in H1299 cells transfected with WT p53 or codon 104 mutants such as N104S for M.b., S104N for M.c., and E104S for M.o.
Under strong hypoxia stress (0.2% O2), both WT and 104-mutant p53 in the three mammals suppressed Bax and Puma but activated Noxa. However, M.b. WT p53 (104N) up-regulated Apaf1, but its N104S mutant up-regulated IGFBP3; M.c. WT p53 (104S) up-regulated IGFBP3, but its S104N mutant up-regulated Apaf1. Furthermore, M.o. WT p53 up-regulated IGFBP3 and Apaf1, but these genes were down-regulated after E104S mutation (Fig. 5, Fig. S6A, and Table S2). These data suggest that the variations in p53 codon 104 in the three mammals correlate with their distinct transactivation of apoptotic genes and exhibit a functional and causal relationship in the responses of plateau-dwelling M.b. and lowland-dwelling M.c. to hypoxic stress. Down-regulation of Bax and Puma in the wild mammals benefits cell survival against apoptosis.
Fig. 5.
Up-regulation and down-regulation of the transcription of p53 target genes by WT p53 and of p53 104 mutants under hypoxia and cold stress. ↑, up-regulation; ↓, down-regulation; ―, unchanged.
Under cold (30 °C) stress, both M.b. WT p53 and its N104S mutant up-regulated Bax, Noxa, and Puma and down-regulated IGFBP3; M.c. WT p53 up-regulated all apoptotic genes, but IGFBP3 became down-regulated after S104N mutation. In contrast to M.o. WT p53, its E104S mutant induced down-regulation of Puma, IGFBP3, and Apaf1 in response to cold stress (Fig. 5, Fig. S6B, and Table S2), suggesting that variations in the mammals’ WT p53 codon 104 are linked to their distinct transcription pattern for apoptotic genes in response to cold.
Under acidic stress, the WT and p53 104 mutants in the three mammals showed a similar alteration in the apoptotic target genes. However, in response to pH 6.6, WT p53 induced activated Bcl-2 transcription in M.b., suppressed Bcl-2 transcription in M.c., and did not change Bcl-2 transcription in M.o. Importantly, although Bcl-2 transcription was up-regulated by M.b. WT p53, it was down-regulated by N104S mutation. No change in response to pH 6.6 was seen with the S104N mutation in M.c. p53 or the E104S mutation in M.o. p53 (Fig. 6, Fig. S6C, and Table S2). This result suggests that WT p53 in these species share a similar transcriptional regulation for apoptotic genes but divergent regulation for antiapoptotic genes. M.b. WT 104N seems to be linked specifically to Bcl-2 transactivation under low pH, which may play protective effects against apoptosis under acidic conditions.
Fig. 6.
Comparison of the transcription of p53 target genes by WT p53 and 104 of p53 mutants under acidic stress. ↑, up-regulation; ↓, down-regulation; ―, unchanged.
p53 Induces Different Apoptosis Rates Under Hypoxia, Cold, and Acidic Stress in M.b., M.c., and M.o.
To assess WT p53’s induced responses of cell apoptosis, cells transfected with M.b., M.c., and M.o. WT p53 expression plasmids were tested under hypoxic, cold, or acidic stresses using flow cytometry. The contributions of WT p53 to apoptosis were confirmed by detecting GFP–p53+ cells. All WT p53 induced lower levels of apoptosis under hypoxia of 0.2% O2 (Fig. 4). This reduction may correlate with the decreased Bax and Puma transcription (Fig. 5). M.c. and M.o. WT p53 failed to reduce apoptosis, but M.b. WT p53 induced down-regulation of cell apoptosis under 30 °C cold stress (Fig. 4), possibly through suppressed IGFBP3 (Fig. 5). Furthermore, M.c. and M.o. WT p53 failed to induce a reduction in cell apoptosis, but M.b. WT p53 induced reduced apoptosis (Fig. 4) under pH 6.6 acidic stress, possibly because of up-regulated Bcl-2 (Fig. 6). These data suggest that in the two highland-dwelling species M.b. and M.o. and in the lowland-dwelling species M.c. p53 has distinct adaptive apoptotic target genes and/or antiapoptotic Bcl-2 gene transactivation (Tables S1 and S2).
Discussion
Overview of Results.
Our results suggest that the variation of p53 codon 104 in M.b. and M.o. is the result of adaptation to the stressful environment of the Qinghai-Tibet plateau. The rising of the Qinghai-Tibet plateau started by the collision of the Indian and Eurasian plates 40–50 Mya (11). M.b. and M.o. were isolated geographically and ecologically and encountered lower temperatures and O2 levels as the plateau rose (44). In addition to other allelic changes, we found that codon 104 differs with 104N in M.b. and 104E in M.o. We propose that these changes are adaptively related to cold and hypoxic environments. Interestingly, other than the rodent M.o., only the squid Loligo forbesi and four fishes (Barbus barbus, Platichthys flesus, Tetraodon miurus, and Xiphophorus hellerii) have 104E (Fig. S2). Among these species, both X. hellerii and L. forbesi live in hypoxic aquatic environments (7), and T. miurus lives on sandy bottoms in deep water (45). P. flesus is also known to be moderately tolerant of hypoxia (46, 47). The homoplasy of this site shared by these species strongly suggests that the 104E in M.o. p53 is related to hypoxia tolerance. In M.b., however, 104N could be an adaptation coupled to hypoxia and hypercapnia at the high altitudes (∼3,000–4,500 m) which M.b. inhabits. The two 104 variations may correlate with distinct adaptive strategies to the different ecological niches of the two plateau species. This view is supported by the present results showing that 104N in Tibetan M.b. has a high transactivation to apoptotic target genes and to antiapoptotic Bcl-2. This effect differs from the suppressed apoptotic genes and unchanged Bcl-2 induced by M.o. p53 104E. Likewise, it differs from activated apoptotic genes and suppressed Bcl-2 induced by p53 104S in M.c., which inhabits burrows at an altitude of <800 m in west China. It also differs from the action of p53 in S.j. in Israel (7). M.c. is a lowland zokor and is a relative of M.b. However, its p53 transactivation of apoptotic genes is stronger than that of M.b., and this increased activation has been demonstrated to correlate with the codon 104S variation (Fig. 2 and Fig. S4). Therefore, the 104E and 104N variations in Qinghai-Tibet plateau’s mammals appear to be associated with hypoxic and cold environments; in particular, the 104N variant seems to be associated with hypoxia and high CO2 (Table S2).
Adaptive Environmental Correlates of p53.
The WT p53 variations in the two Tibetan species M.b. and M.o. are associated with cellular apoptosis and antiapoptosis induced by hypoxia. Several studies have demonstrated that the p53 DBD is conserved and important for its functions as a tumor suppressor (2, 26). Only a few studies report that the p53 variation is associated with environmental adaptation. In tumorigenesis, the most frequently mutated sites in the DBD are found at codons 175R, 248R, and 273R of p53, which are referred to as “hotspots” (48). As expected, these changes are not found in M.b. or M.o. The mutated codon 104 residing in the DBD appears to be associated with environmental fitness. In human cancers, a germ-line mutation in this codon was reported in a patient with multiple primary cancers (49). Using site-directed mutagenesis, we found that the codon 104N mutation in M.b. and the 104E mutation in M.o. p53 are required for divergent responses of IGFBP3 and Apaf1 to hypoxia and cold stresses. This observation led us to analyze the 3D structure of p53 tetramer binding to DNA and highlighting the location of codon 104. Although located in the core domain, codon 104 does not interact directly with DNA (Fig. S7); thus, it should not induce universal changes in transactivation by p53. The transcriptional repression of Bcl-2 is associated with a competitive binding of p53 against a Bcl-2 transcriptional activator Brn-3a (40). The codon 104 mutation of M.b. p53 did not affect the activation of Bcl-2 (Fig. 2) but did affect the activation of Bcl-2 under acidic stress (Fig. 6). Other sites of variation in M.b. p53, especially those within the mSin3A-binding domain, also seemed to contribute to Bcl-2 activation.
The codon 104 variations in the two Tibetan species also appear to be related to the specific local environment of their niches. The high CO2 stress in the underground burrows of M.b. results in a significantly higher partial pressure of CO2 in the mammal’s arterial (51.97 mm Hg) and venous (76.86 mm Hg) blood, challenging the blood-buffering system. In rats the CO2 pressure is lower (33.68 mm Hg in arterial blood and 40.05 mm Hg in venous blood) (10, 50). Our study demonstrated that under acidic stress of pH 6.0, M.b. p53 transactivation of the apoptotic target gene IGFBP3 was decreased and that of the antiapoptotic gene Bcl-2 was increased dramatically. However, the N104S mutation in M.b. resulted in enhanced IGFBP3 and reduced Bcl-2 activation (Fig. 6 and Fig. S6C). Therefore, the 104N variation is involved in the regulation of blood acidity. These data suggest that the 104N mutation in M.b. contributes to the animal’s tolerance of a high-CO2 environment. In addition to hypoxia and hypercapnia, the low annual mean temperature on the Tibet plateau provides an additional stress. Evidence shows that genotypic variation of p53 is related to the average temperature of the population’s environment (51). Although M.b. and M.o. live at the same altitude, p53 in these two rodents showed markedly different responses to cold stress (Fig. 5). This difference could be explained by the two mammals’ different niches. M.o. forages above ground, subjected to extremely low (average, −14.8 °C) ambient temperatures in winter and an average ambient temperature of 9.8 °C in summer. The temperature in the underground burrows of M.b. is milder and is relatively stable. This difference may have contributed to the differential evolution of codon 104 in the two plateau-dwelling rodents. The holarctic distribution of M.o. also suggests the species’ adaptation to cold environments (52). In M.b. the 104N mutation of p53 contributes to the decreased IGFBP3 transactivation under cold and acidic stresses (Figs. 5 and 6), suggesting an elaborate modulation of p53 to cope with multiple stresses to underground life.
Conclusions
Our study has shown an evolutionary adaptive variation in codon p53 104 in the two Qinghai-Tibet plateau mammals, the underground-dwelling M.b. (104N) and ground-dwelling M.o. (104E). These variations affect hypoxia, cold, and pH regulation, indicating that the p53 variations are adaptively associated with the variable plateau niches below and above ground. Generally, these results suggest that M.b. and M.o. are fitting model organisms for investigating the adaptive molecular evolutionary mechanisms of the interaction between gene and environment.
Materials and Methods
Myospalax baileyi (250–350 g, Fig. 1B) and Microtus oeconomus (18–25 g, Fig. 1B) were captured from the field near Haibei Research Station of the Alpine Meadow Ecosystem, Chinese Academy of Sciences (37°39′N, 101°19′E) in Qinghai, China. M. cansus (250–300 g, Fig. 1B) were captured from the field near Yan'an, north Shaanxi (36°36′N, 109°31′E) of China. Adult male Sprague-Dawley rats (150–200 g; Certification No. SCXK20080033) were purchased from the Experimental Animal Center, Zhejiang Academy of Medical Science. Animal protocols were approved by the IACUC of the School of Medicine, Zhejiang University and followed National Institutes of Health laboratory animal care guidelines. The methods used in this study (animal breeding, molecular cloning, sequence analysis, cell culture, plasmid construction, mutagenesis, transient transfection, dual-luciferase reporter assay, quantitative real-time RT-PCR, Western blotting, and flow cytometry) are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. I. C. Bruce at the School of Medicine, Zhejiang University, and Robin Permut at the Institute of Evolution, University of Haifa, for editing the paper and Greg Cole of the University of California, Los Angeles, for helpful comments on the manuscript. This work was supported by Grants 2012CB518200 and 2006CB504100 from the National Basic Research Program (973) of the Ministry of Science and Technology of China and by Grants 31071047 and 30870300 from the National Natural Science Foundation of China. E.N. received financial support from the Ancell-Teicher Research Foundation for Genetics and Molecular Evolution.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320369110/-/DCSupplemental.
References
- 1.Asker C, Wiman KG, Selivanova G. p53-induced apoptosis as a safeguard against cancer. Biochem Biophys Res Commun. 1999;265(1):1–6. doi: 10.1006/bbrc.1999.1446. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Chen XQ, Du JZ. Cellular adaptation to hypoxia and p53 transcription regulation. J Zhejiang Univ Sci B. 2009;10(5):404–410. doi: 10.1631/jzus.B0820293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci USA. 2003;100(17):9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resnick MA, Tomso D, Inga A, Menendez D, Bell D. Functional diversity in the gene network controlled by the master regulator p53 in humans. Cell Cycle. 2005;4(8):1026–1029. doi: 10.4161/cc.4.8.1904. [DOI] [PubMed] [Google Scholar]
- 5.Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. Proc Natl Acad Sci USA. 2008;105(3):944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol Cell Biol. 2006;26(6):2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashur-Fabian O, et al. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci USA. 2004;101(33):12236–12241. doi: 10.1073/pnas.0404998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soussi T, May P. Structural aspects of the p53 protein in relation to gene evolution: A second look. J Mol Biol. 1996;260(5):623–637. doi: 10.1006/jmbi.1996.0425. [DOI] [PubMed] [Google Scholar]
- 9.Villiard E, et al. Urodele p53 tolerates amino acid changes found in p53 variants linked to human cancer. BMC Evol Biol. 2007;7:180. doi: 10.1186/1471-2148-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei DB, Wei L, Zhang JM, Yu HY. Blood-gas properties of plateau zokor (Myospalax baileyi) Comp Biochem Physiol A Mol Integr Physiol. 2006;145(3):372–375. doi: 10.1016/j.cbpa.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Harrison TM, Copeland P, Kidd WS, Yin A. Raising tibet. Science. 1992;255(5052):1663–1670. doi: 10.1126/science.255.5052.1663. [DOI] [PubMed] [Google Scholar]
- 12.Zhisheng A, Kutzbach JE, Prell WL, Porter SC. Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature. 2001;411(6833):62–66. doi: 10.1038/35075035. [DOI] [PubMed] [Google Scholar]
- 13.Guo ZT, et al. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature. 2002;416(6877):159–163. doi: 10.1038/416159a. [DOI] [PubMed] [Google Scholar]
- 14.Cun YZ, Wang XQ. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol Phylogenet Evol. 2010;56(3):972–982. doi: 10.1016/j.ympev.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Nei M, Maruyama T, Wu CI. Models of evolution of reproductive isolation. Genetics. 1983;103(3):557–579. doi: 10.1093/genetics/103.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du JZ, Li QF, Chen XG. The changing of corticosterone level in native Ochotona curzoniae. Acta Theriologica Sinica. 1983;3(1):47–52. [Google Scholar]
- 17.Du JZ, Li QF, Chen XG. Effect of simulated altitude on liver of Ochotona curzoniae and rats. Acta Zool Fenn. 1984;171:201–203. [Google Scholar]
- 18.Li QF, Du JZ. Effect of chronic hypoxia on liver of Ochotona curzoniae and rat. Acta Theriologica Sinica. 1986;6(4):261–266. [Google Scholar]
- 19.Li QF, Chen XG, You ZB, Du JZ. A comparative study on effects of acute hypoxia upon livers of three small mammals. Acta Theriologica Sinica. 1987;7(1):51–57. [Google Scholar]
- 20.Chen XQ, Wang SJ, Du JZ, Chen XC. Diversities in hepatic HIF-1, IGF-I/IGFBP-1, LDH/ICD, and their mRNA expressions induced by CoCl(2) in Qinghai-Tibetan plateau mammals and sea level mice. Am J Physiol Regul Integr Comp Physiol. 2007;292(1):R516–R526. doi: 10.1152/ajpregu.00397.2006. [DOI] [PubMed] [Google Scholar]
- 21.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75(1):73–82. [PubMed] [Google Scholar]
- 22.Schmid C. Insulin-like growth factors. Cell Biol Int. 1995;19(5):445–457. doi: 10.1006/cbir.1995.1088. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, et al. Distinct post-transcriptional regulation of Igfbp1 gene by hypoxia in lowland mouse and Qinghai-Tibet plateau root vole Microtus oeconomus. Mol Cell Endocrinol. 2013;376(1-2):33–42. doi: 10.1016/j.mce.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Hammond EM, Giaccia AJ. The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun. 2005;331(3):718–725. doi: 10.1016/j.bbrc.2005.03.154. [DOI] [PubMed] [Google Scholar]
- 25.Le QT, Denko NC, Giaccia AJ. Hypoxic gene expression and metastasis. Cancer Metastasis Rev. 2004;23(3-4):293–310. doi: 10.1023/B:CANC.0000031768.89246.d7. [DOI] [PubMed] [Google Scholar]
- 26.Zhao K, Chai X, Johnston K, Clements A, Marmorstein R. Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution. J Biol Chem. 2001;276(15):12120–12127. doi: 10.1074/jbc.M011644200. [DOI] [PubMed] [Google Scholar]
- 27.Graeber TG, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379(6560):88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 28.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 31.Cao YB, Chen XQ, Wang S, Wang YX, Du JZ. Evolution and regulation of the downstream gene of hypoxia-inducible factor-1alpha in naked carp (Gymnocypris przewalskii) from Lake Qinghai, China. J Mol Evol. 2008;67(5):570–580. doi: 10.1007/s00239-008-9175-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Wang MY, Hao K, Chen XQ, Du JZ. CRHR1 mediates p53 transcription induced by high altitude hypoxia through ERK 1/2 signaling in rat hepatic cells. Peptides. 2013;44:8–14. doi: 10.1016/j.peptides.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 33.Moroni MC, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3(6):552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 34.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9(4):428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 35.Sabbatini P, Chiou SK, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15(2):1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91(19):8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan KM, Vousden KH. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18(7):3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang E, Korsmeyer SJ. Molecular thanatopsis: A discourse on the BCL2 family and cell death. Blood. 1996;88(2):386–401. [PubMed] [Google Scholar]
- 39.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16(10):5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budhram-Mahadeo V, et al. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem. 1999;274(21):15237–15244. doi: 10.1074/jbc.274.21.15237. [DOI] [PubMed] [Google Scholar]
- 41.Scian MJ, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27(18):2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 42.Norris PS, Haas M. A fluorescent p53GFP fusion protein facilitates its detection in mammalian cells while retaining the properties of wild-type p53. Oncogene. 1997;15(18):2241–2247. doi: 10.1038/sj.onc.1201406. [DOI] [PubMed] [Google Scholar]
- 43.Wahlfors J, Loimas S, Pasanen T, Hakkarainen T. Green fluorescent protein (GFP) fusion constructs in gene therapy research. Histochem Cell Biol. 2001;115(1):59–65. doi: 10.1007/s004180000219. [DOI] [PubMed] [Google Scholar]
- 44.Tang L, et al. Allopatric divergence and phylogeographic structure of the plateau zokor (Eospalax baileyi), a fossorial rodent endemic to the Qinghai-Tibetan Plateau. J Biogeogr. 2010;37(4):657–668. [Google Scholar]
- 45.Bhaskaran A, May D, Rand-Weaver M, Tyler CR. Fish p53 as a possible biomarker for genotoxins in the aquatic environment. Environ Mol Mutagen. 1999;33(3):177–184. [PubMed] [Google Scholar]
- 46.Lundgreen K, Kiilerich P, Tipsmark CK, Madsen SS, Jensen FB. Physiological response in the European flounder (Platichthys flesus) to variable salinity and oxygen conditions. J Comp Physiol B. 2008;178(7):909–915. doi: 10.1007/s00360-008-0281-9. [DOI] [PubMed] [Google Scholar]
- 47.Jørgensen JB, Mustafa T. The effect of hypoxia on carbohydrate metabolism in flounder (Platichthys flesus L.)—I. Utilization of glycogen and accumulation of glycolytic end products in various tissues. Comp Biochem Physiol B. 1980;67:243–248. [Google Scholar]
- 48.Glazko GV, Koonin EV, Rogozin IB. Mutation hotspots in the p53 gene in tumors of different origin: Correlation with evolutionary conservation and signs of positive selection. Biochim Biophys Acta. 2004;1679(2):95–106. doi: 10.1016/j.bbaexp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Kimura K, et al. Germline p53 mutation in a patient with multiple primary cancers. Jpn J Clin Oncol. 2001;31(7):349–351. doi: 10.1093/jjco/hye070. [DOI] [PubMed] [Google Scholar]
- 50.Shams I, Avivi A, Nevo E. Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic-hypercapnic stresses. Comp Biochem Physiol A Mol Integr Physiol. 2005;142(3):376–382. doi: 10.1016/j.cbpa.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Shi H, et al. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet. 2009;84(4):534–541. doi: 10.1016/j.ajhg.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunhoff C, Galbreath KE, Fedorov VB, Cook JA, Jaarola M. Holarctic phylogeography of the root vole (Microtus oeconomus): Implications for late Quaternary biogeography of high latitudes. Mol Ecol. 2003;12(4):957–968. doi: 10.1046/j.1365-294x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.