Significance
Serine protease inhibitors (serpins) are a protein superfamily whose members are involved in many diseases and are thus attractive drug targets. In addition to protease inhibition, serpins also bind a variety a of other biological molecules, including extracellular matrix components and cell surface receptors. The inhibitory mechanism of serpins requires a conformational change that can also alter their affinity to non-protease ligands. Here a surprising allosteric mechanism of action is revealed for a small molecule inhibitor of the serpin, plasminogen activator inhibitor 1 (PAI-1). Compound binding prevents PAI-1 interaction with both proteases and with its cofactor even though the binding sites are located 40 Å apart. These results suggest the potential for the identification other therapeutically useful serpin inhibitors.
Keywords: fibrinolysis, thrombolysis, fibrosis, cancer
Abstract
Plasminogen activator inhibitor type-1 (PAI-1) is a member of the serine protease inhibitor (serpin) family. Excessive PAI-1 activity is associated with human disease, making it an attractive pharmaceutical target. However, like other serpins, PAI-1 has a labile structure, making it a difficult target for the development of small molecule inhibitors, and to date, there are no US Food and Drug Administration–approved small molecule inactivators of any serpins. Here we describe the mechanistic and structural characterization of a high affinity inactivator of PAI-1. This molecule binds to PAI-1 reversibly and acts through an allosteric mechanism that inhibits PAI-1 binding to proteases and to its cofactor vitronectin. The binding site is identified by X-ray crystallography and mutagenesis as a pocket at the interface of β-sheets B and C and α-helix H. A similar pocket is present on other serpins, suggesting that this site could be a common target in this structurally conserved protein family.
Plasminogen activator inhibitor type 1 (PAI-1) is a serine protease inhibitor (serpin) implicated in numerous pathological processes, including coronary heart disease, chronic fibrotic and inflammatory diseases, and tumor invasion and metastasis (1–6). These associations have made PAI-1 an attractive pharmaceutical target. However, despite extensive studies, only a few small molecule inhibitors have been identified thus far (7–16), and the majority of these are poor pharmaceutical candidates as they have relatively low affinity for PAI-1 and are unable to inactivate PAI-1 bound to its plasma cofactor vitronectin.
PAI-1 is the most potent physiologic inhibitor of tissue-type and urokinase-type plasminogen activators (tPA and uPA, respectively) (17). Like other serpins, PAI-1 has a solvent-exposed, reactive center loop (RCL) that contains an amino acid sequence that confers protease target specificity. The serpin inhibitory mechanism is a multistep process of coordinated conformational changes that are necessary to trap a target protease (18). The first step is the formation of a noncovalent Michaelis complex, followed by the initial steps of a typical serine protease catalytic attack leading to the covalent acyl-enzyme complex (19). However, before the protease can dissociate from the serpin, a dramatic conformational change occurs [termed the stressed to relaxed (S to R) transition], in which the cleaved RCL inserts into the central β-sheet A, translocating the covalently-bound protease 70 Å to the base of β-sheet A (20). This conformational change results in distortion of the protease active site (21, 22) and thereby prevents the deacylation reaction, inactivating the protease. Should this conformational change be interrupted, the protease can complete its cleavage of the serpin RCL, remaining active and leaving the serpin in a cleaved, inactive, loop-inserted state (23).
PAI-1 is unique among serpins in that it readily autoinactivates into a so-called latent form, where the PAI-1 RCL inserts into the central A-sheet without cleavage (24). In the latent form, the scissile bond is inaccessible to target proteases and PAI-1’s affinity for vitronectin is greatly reduced (25). Under physiological conditions, the latency transition is irreversible; however, denaturant-induced refolding can reconstitute PAI-1 in the native, active form (26). Several latency-inducing agents have been described, including amphipathic nonionic detergents (27, 28) and monoclonal antibodies or peptides (29–31). These latter agents are thought to stabilize a prelatent intermediate and thereby shift the structural equilibrium toward the latent conformation. None of these agents have been further developed as therapeutics.
Such a complex antiproteolytic mechanism, requiring global conformational changes in the serpin structure, as well as PAI-1’s conformational instability, suggests that it should be possible to inhibit the serpin mechanism in multiple ways. Indeed, it is anticipated that minor disruptions in the conformational change may be sufficient to prevent trapping of the protease. This complexity is unlike an enzyme with a single active site, or a receptor with a single ligand binding site, and it may be that the structurally dynamic nature of serpins predisposes anti-PAI-1 high-throughput screens to identifying relatively weak affinity or nonspecific small molecules. Furthermore, the transient nature of the native conformation in the absence of the cofactor vitronectin complicates in vitro screening and therapeutic development (32).
We report here the biochemical and structural characterization of a unique high-affinity synthetic PAI-1 inactivator, CDE-096, that is based on our previously described class of polyphenolic PAI-1 inhibitors (33). These inactivators are the first known serpin antagonists with a mechanism of action that involves reversible modulation of the RCL conformation to block Michaelis complex formation. Significantly, CDE-096 is active against both free PAI-1 and vitronectin-bound PAI-1. CDE-096 binds to PAI-1 with nanomolar affinity and induces conformational changes that prevent binding to both proteases and vitronectin. This allosteric mechanism is characterized by reciprocal communication between the high-affinity compound-binding and vitronectin-binding sites. Remarkably, our structural studies suggest a binding site for CDE-096 at the interface between the B and C β-sheets (termed the sB/sC pocket). Although the overall fold of serpins are generally conserved, the sB/sC pocket exhibits considerable structural and sequence variability across the serpin superfamily, thus providing a means of target specificity. Taken together, these data establish a unique mechanistic class of PAI-1 antagonists and provide a distinct binding site for the rational design of future anti-serpin agents.
Results
Design and Characterization of the PAI-1 Inhibitor CDE-096.
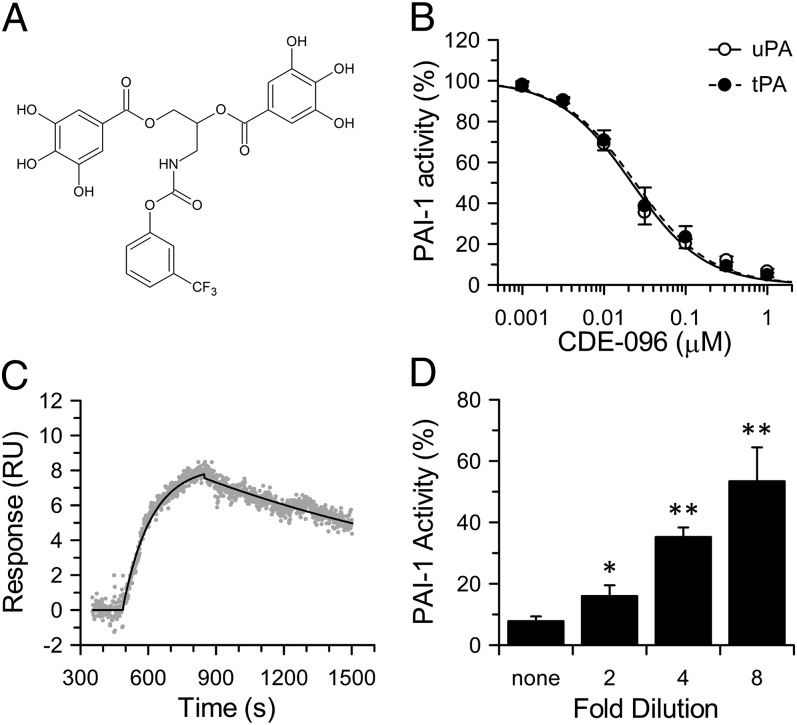
Based on our recent structure-activity relationship study of high-affinity polyphenolic inactivators of PAI-1 (33), CDE-096 was synthesized to explore the mechanism of action of this inhibitor class. CDE-096 contains the minimally required digalloyl moiety plus an additional trifluoromethylphenyl carbamate derivative (Fig. 1A). CDE-096 prevented PAI-1 from inactivating tPA and uPA with similar potency (IC50 = 30 ± 6 and 25 ± 4 nM, respectively; Fig. 1B) and was active against glycosylated PAI-1, as well as PAI-1 derived from several species (Table 1). Showing exquisite specificity for PAI-1, CDE-096 was ineffective against two closely related serpins, antithrombin and α1-protease inhibitor (α1-PI) (Table 1). Using surface plasmon resonance (SPR), we monitored direct binding of CDE-096 to PAI-1 captured on an immobilized monoclonal anti–PAI-1 antibody, measuring a kon of 2.5 ± 0.6 × 104 M−1⋅s−1 and a koff of 5.7 ± 0.9 × 10−4 s−1, (KD of 22 ± 6 nM) (Fig. 1C). Finally, CDE-096 inactivation of PAI-1 was reversible, as its antiproteolytic activity returned on serial dilution of preformed CDE-096/PAI-1 complexes (Fig. 1D).
Fig. 1.
CDE-096 reversibly inactivates PAI-1 with high potency. (A) Structure of CDE-096, a carbamoyl derivative of a synthetic digalloyl polyphenol compound. (B) Inactivation of PAI-1 by CDE-096 as measured with tPA or uPA. The data were fit to an exponential decay equation, yielding IC50 = 30 ± 6 nM and 25 ± 4 nM for tPA and uPA, respectively (n = 6). (C) Real-time sensogram of 200 nM CDE-096 binding to PAI-1 captured on the murine anti-human monoclonal antibody, MA-33H1F7. The data were fit to simultaneous ka/kd using BIAevaluation 4.1.1, yielding a KD of 22 ± 6 nM (n = 4). (D) Reversible inhibition of 5 nM PAI-1 by 1 µM CDE-096, with recovery of PAI-1 activity by serial dilution of preformed PAI-1/CDE-096 complexes into fresh buffer. Activities of diluted samples were compared with that of the nondiluted sample using a one-tailed Student t test (*P < 0.05; **P < 0.01; n = 6).
Table 1.
Target specificity of CDE-096
| Inhibitor | Enzyme | IC50* (nM) |
| PAI-1 | uPA | 25 ± 4 |
| tPA | 30 ± 6 | |
| PAI-114–1B | uPA | 29 ± 3 |
| tPA | 27 ± 4 | |
| PAI-1glyco | uPA | 15 ± 2 |
| Murine PAI-1 | uPA | 19 ± 2 |
| Rat PAI-1 | uPA | 22 ± 2 |
| Porcine PAI-1 | uPA | 18 ± 1 |
| Antithrombin | α-Thrombin | >300,000 |
| α1-PI | Elastase | >300,000 |
Values represent measured IC50 values or the highest CDE-096 concentration tested.
Mechanism of Action of CDE-096 on PAI-1 Antiproteolytic Activity.
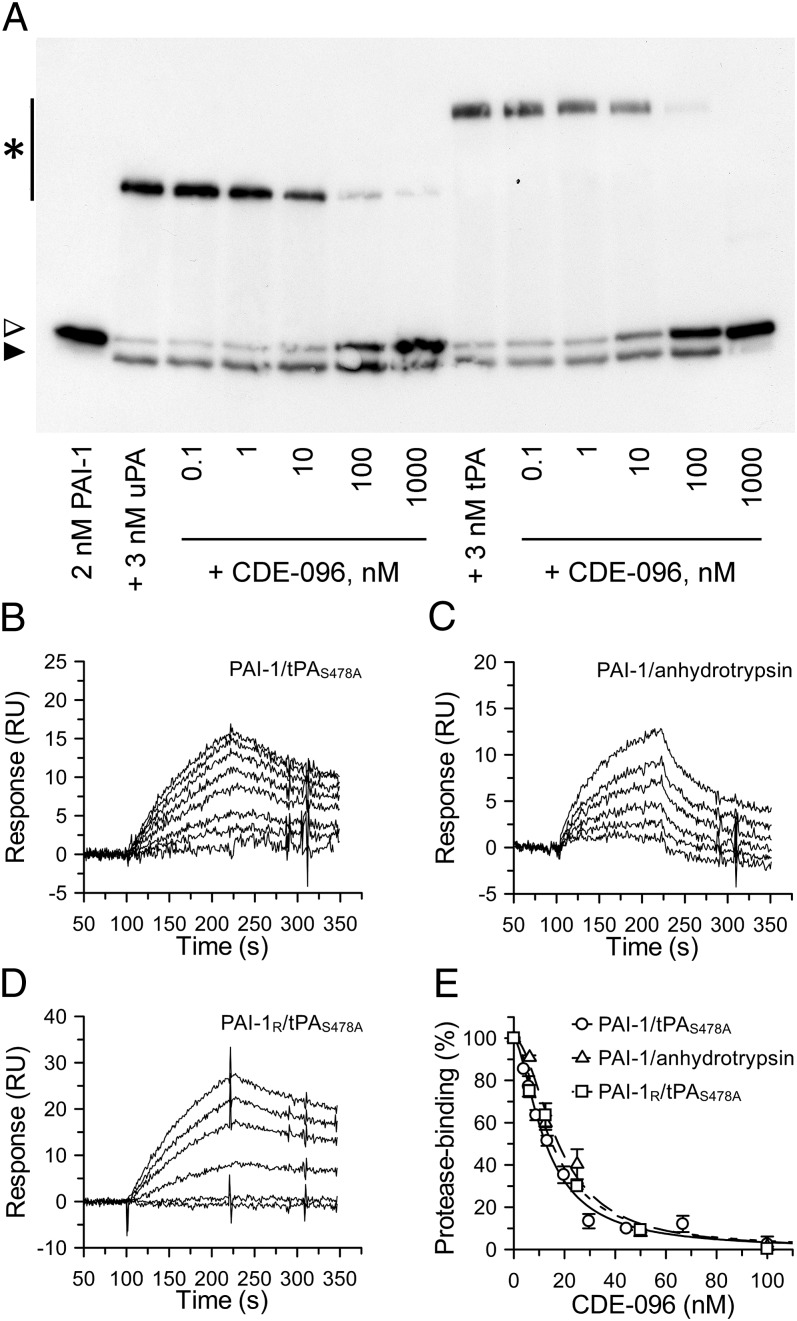
The reversibility of CDE-096 suggests that compound binding does not induce conversion of PAI-1 to the latent conformation or cleavage of PAI-1 as a substrate. Therefore, to determine the mechanism of CDE-096 inactivation of PAI-1, the effect of CDE-096 on PAI-1/protease complex formation was examined by SDS/PAGE. These studies showed that CDE-096 blocked the formation of covalent complexes between PAI-1 and target proteases and led to the accumulation of unreacted PAI-1 (Fig. 2A), suggesting that the compound may prevent Michaelis complex formation. Therefore, to examine whether CDE-096 could disrupt the initial binding between PAI-1 and a target protease, SPR analysis was used. These studies showed that CDE-096 blocked noncovalent complex formation between PAI-1 and an immobilized active site mutant of tPA, tPAS478A, (Fig. 2B) as well as an inactive form of a nonphysiologic target protease, anhydrotrypsin (Fig. 2C). Together these data suggest that CDE-096 prevents the PAI-1 RCL from interacting with the active site of the protease.
Fig. 2.
CDE-096 blocks noncovalent Michaelis complex formation. (A) Immunoblot of the reactions of CDE-096 treated PAI-1 with uPA or tPA, showing SDS-stable PAI-1/protease complexes (asterisk), unreacted PAI-1 (open arrowhead), and cleaved PAI-1 (closed arrowhead). (B–D) Real-time sensograms of the binding of WT PAI-1 or the substrate variant PAI-1R to immobilized inactive tPAS478A or anhydrotrypsin in the presence of CDE-096. In each case, the top-most tracing represents the binding of untreated PAI-1 variant while successively lower tracings represent PAI-1 variant preincubated with increasing amounts of CDE-096 (0, 3.9, 5.8, 8.8, 12.2, 19.8, 29.6, 44.4, 66.7, and 100 nM CDE-096 in B and C; and 0, 6.2, 12.5, 25.0, 50.0, 100.0 nM CDE-096 in D). (E) For the data in B–D, the initial association phase of PAI-1 binding to tPAS478A or anhydrotrypsin or for PAI-1R binding to tPAS478A was linearly fit and the slopes plotted against the CDE-096 concentration with which each PAI-1 was preincubated.
One way in which serpins can be made unavailable to a protease active site is via allosteric changes in the conformation of the RCL. For example, in the native form of the closely related serpin antithrombin, the RCL is partially inserted into the A β-sheet (sA). As a consequence, antithrombin is a relatively poor inhibitor of its target proteases, factors IXa and Xa. However, on interaction of antithrombin with its cofactor heparin, the RCL is fully expelled from sA, resulting in the allosteric activation of antithrombin against target proteases (34). Therefore, to test whether partial loop insertion was required for CDE-096–induced inactivation of PAI-1, the SPR assay was repeated with a variant of PAI-1 (PAI-1R) having substitutions of T333R and A335R within the RCL, which render it unable to insert its RCL into sA (35). These studies showed that CDE-096 blocked the ability of PAI-1R to bind to immobilized inactive tPAS478A (Fig. 2D). Furthermore, the concentration dependence of this effect was indistinguishable from that of CDE-096 inhibition of WT PAI-1 binding to tPAS478A or anhydrotrypsin (Fig. 2E). These data suggest that CDE-096 does not act by inducing partial or full RCL insertion in PAI-1, and taken together with the reversibility of CDE-096 binding and inactivation of PAI-1 (Fig. 1 C and D), further rule out the induction of latency as a mechanism of action.
Effect of CDE-096 on the PAI-1/Vitronectin Interaction.
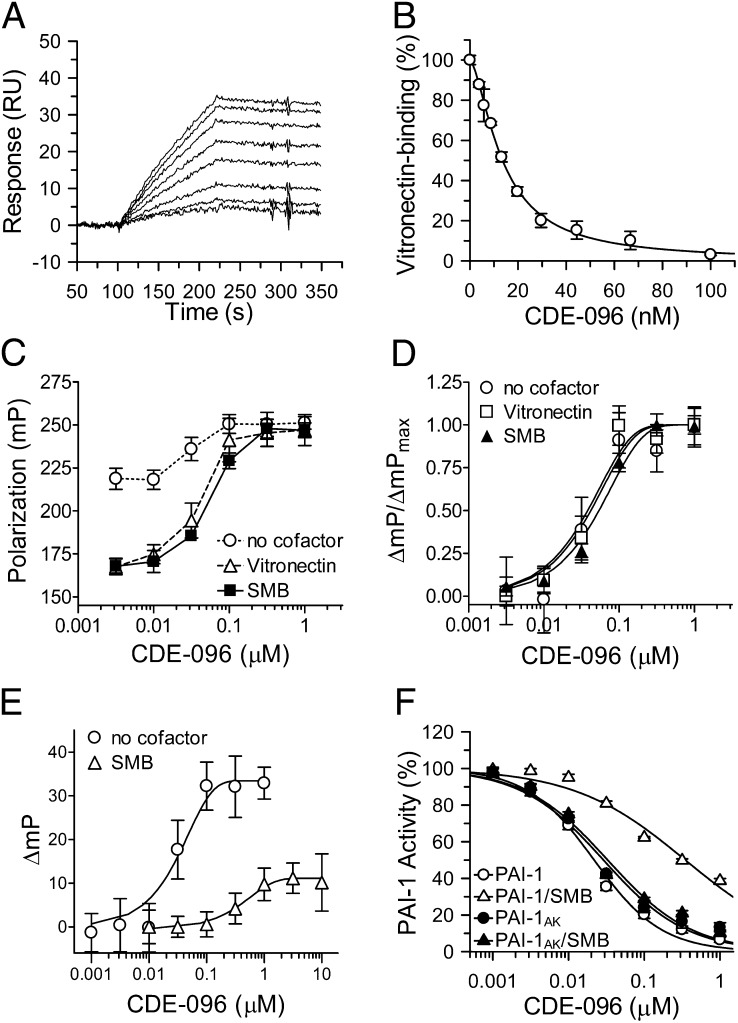
In addition to inhibiting target proteases, PAI-1 also plays an important role in regulating cell adhesion and migration via binding to vitronectin in the extracellular matix (3, 36). PAI-1 also circulates in blood in complex with soluble vitronectin, and this interaction extends the inhibitor's half-life (37). To investigate whether CDE-096 directly affects the PAI-1/vitronectin interaction, SPR and fluorescence polarization (FP) approaches were used. First, SPR was performed to explore the effect of CDE-096 on PAI-1 binding to immobilized vitronectin. These data demonstrated that preincubation of PAI-1 with CDE-096 blocked PAI-1 binding to vitronectin with an apparent IC50 of 20 ± 2 nM (Fig. 3 A and B). This value is essentially identical to the IC50 for the inhibition of the antiproteolytic activity of PAI-1 (Table 1) and to the KD (22 ± 6 nM) determined from the direct binding of CDE-096 to PAI-1 (Fig. 1).
Fig. 3.
CDE-096 and vitronection reciprocally decrease binding for one another to PAI-1. (A) Real-time binding sensograms of PAI-1 to immobilized vitronectin, in the presence of CDE-096 (0, 3.9, 5.8, 8.8, 12.2, 19.8, 29.6, 44.4, 66.7, and 100 nM) with the top-most tracing showing no CDE-096 and the bottom-most tracing showing PAI-1 treated with 100 nM of CDE-096. (B) From the data in A, the initial association phase of PAI-1 binding to vitronectin in the presences of each concentration of CDE-096 was linearly fit and the slopes plotted against the CDE-096 concentration. (C) FP of 5 nM PAI-1S149C-FL in the presence of increasing concentrations of CDE-096 with and without cofactor. Samples of premixed PAI-1S149C-FL and 0–1μM CDE-096 were further incubated with either no cofactor, 100 nM vitronectin, or 100 nM SMB, and the FP signal was measured. (D) Normalization of the FP signal shown in C reveals identical dose dependence of the three samples to CDE-096. (E) Binding of CDE-096 to 5 nM PAI-1S149C-FL with either no cofactor present or after preincubated of PAI-1 with 30 nM SMB, as monitored by FP. (F) Direct binding to SMB is required to protect PAI-1 against inhibition by CDE-096. PAI-1 (open symbols) or a variant of PAI-1 (PAI-1AK) that is unable to bind SMB (closed symbols) was preincubated either alone (○,●) or with 30 nM SMB (△,▲) before the addition of CDE-096 and the determination of PAI-1 inhibitory activity against uPA.
The effect of CDE-096 on PAI-1 binding to vitronectin in solution was examined by monitoring changes in FP of a fluorescein-labeled PAI-1 variant (PAI-1S149C-FL). These studies were performed with both full-length vitronectin and with the isolated high-affinity PAI-1–binding domain of vitronectin, the somatomedin B domain (SMB) (38). Binding of both vitronectin and SMB induced a dose-dependent decrease in the polarization of PAI-1S149C-FL, yielding nearly identical solution phase affinities of PAI-1 for each ligand (15 ± 5 and 13 ± 4 nM, respectively; Fig. S1). In contrast, preincubation of PAI-1S149C-FL with increasing concentrations of CDE-096 before vitronectin or SMB addition blocked the reduction in polarization and instead increased the polarization signal to one consistent with compound binding exclusively (Fig. 3C). In addition, the effect of CDE-096 on both vitronectin and SMB binding was similar, yielding nearly identical IC50s of 43 ± 8 and 54 ± 7 nM, respectively (Fig. 3D). These data indicate that pretreatment of PAI-1 with CDE-096 blocks the binding of PAI-1 to the SMB domain of vitronectin.
FP was next used to examine the reciprocal interaction, where PAI-1S149C-FL was first preincubated with SMB followed by the addition of increasing concentrations of CDE-096. This study showed that CDE-096 retained the ability to induce changes in polarization of PAI-1S149C-FL when bound to SMB (Fig. 3E, open triangles). However, the magnitude of the change in polarization was markedly reduced compared with that seen with free PAI-1S149C-FL (Fig. 3E, open circles). In addition, the change in polarization induced by CDE-096 binding to the binary complex of PAI-1S149C-FL/SMB was shifted 7.3-fold compared with free PAI-1S149C-FL (EC50 = 397 ± 22 and 54 ± 7 nM, respectively). These data indicate that CDE-096 not only binds to SMB-bound PAI-1, but also that compound binding does not displace SMB from PAI-1, because if CDE-096 binding were able to disrupt the PAI-1/SMB interaction, then the magnitude of the change in polarization would have matched that of free PAI-1S149C-FL (compare the maximum signal of the open triangles to that of the open circles in Fig. 3E). Thus, the binding of CDE-096 and SMB to PAI-1 are not strictly mutually exclusive events, although each ligand reduces PAI-1's affinity for the other.
To determine whether CDE-096 could also inhibit the anti-protease activity of cofactor-bound PAI-1, the inhibitory activity of PAI-1 against uPA was analyzed in the presence or absence of SMB. CDE-096 effectively inhibited the antiproteolytic activity of PAI-1 in the presence of SMB with an IC50 of 360 ± 16 nM (Fig. 3F, open triangles). Compared with an IC50 of 25 ± 4 nM for free PAI-1 (Fig. 3F, open circles), this indicates that there is 14-fold reduction in the efficacy of the compound when PAI-1 is bound to SMB. Furthermore, this shift in IC50 is in agreement with the effect of cofactor on CDE-096 binding seen in Fig. 3E, suggesting that the shift in efficacy is due to a decrease in the affinity of CDE-096 for the PAI-1/SMB complex. To confirm that the shift in IC50 was not due to a direct effect of SMB on CDE-096, the experiment was repeated with mutant PAI-1AK (R101A and Q123K), this PAI-1 variant inhibits uPA normally but does not interact with SMB (39). These data showed that there was no difference in inactivation of PAI-1AK by CDE-096 in the presence (Fig. 3F, closed triangles) or absence of SMB (Fig. 3F, closed circles), suggesting that direct interaction between PAI-1 and SMB was required to reduce PAI-1's affinity for the compound and thereby its efficacy. Taken together with the studies above, these data suggest that the effect of CDE-096 on the PAI-1/SMB interaction is allosteric and not a direct competition for the same binding site. Likewise, the effect of SMB on CDE-096 binding to PAI-1 suggests that the CDE-096 binding site on PAI-1 may also be allosterically affected by SMB binding.
Effect of CDE-096 on the PAI-1 Structure.
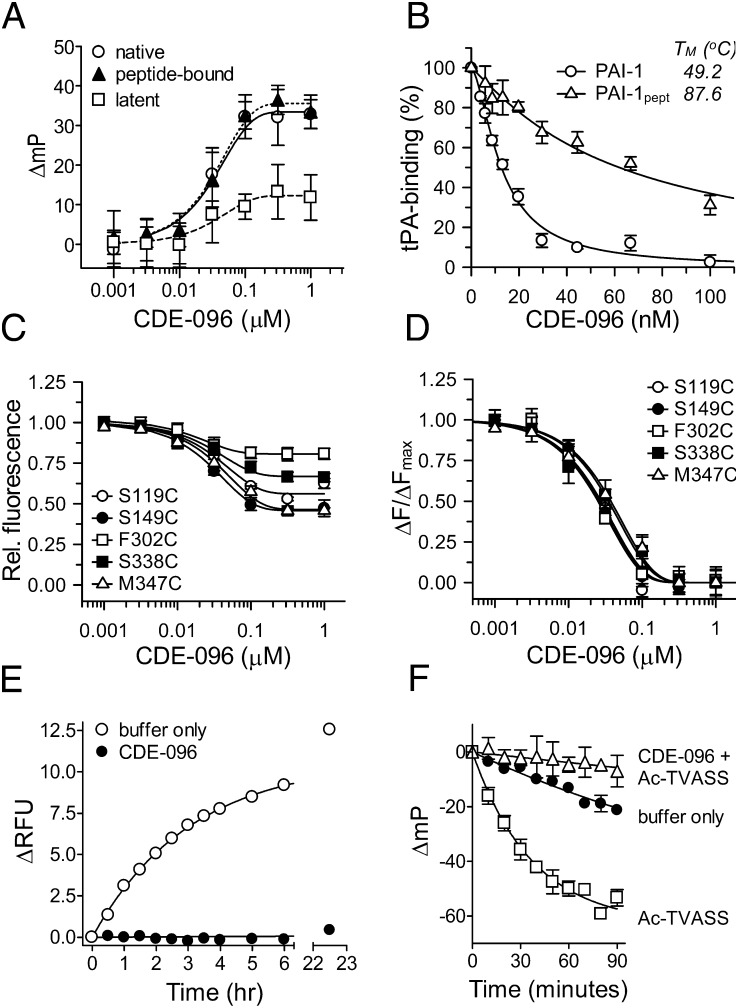
The most extensively studied PAI-1 inactivating compound to date, tiplaxtinin, binds to active but not latent PAI-1 (11). This selectivity is a consequence of structural differences in the binding site between the two serpin conformations. To test whether CDE-096 has a similar conformational bias, three conformationally distinct forms of PAI-1S149C-FL were assayed for compound-induced changes in FP, including active PAI-1, latent PAI-1, and peptide-annealed PAI-1 (PAI-1pept). In active PAI-1, the RCL is not inserted into β-sheet A, which exists as a five-stranded antiparallel β-sheet. This conformation is metastable and is often referred to as the stressed serpin conformation (40), which has a relatively low thermostability with a TM = 49.2 °C. In contrast, the RCL of latent PAI-1 is fully inserted into β-sheet A (24), stabilizing β-sheet A in a state known as the relaxed conformation (40) with a reported TM of 67.5 °C (41). PAI-1pept is a complex of PAI-1 and an RCL mimicking octapeptide that is bound to β-sheet A in place of an inserted RCL (42), thereby exhibiting structural features of both active and latent PAI-1 (43). Specifically, PAI-1pept adopts a structurally stable relaxed serpin conformation with a TM of 87.6 °C, but contains an exposed RCL. The KD values determined from the CDE-096–dependent changes in polarization of PAI-1S149C-FL in the active, latent, and peptide-bound forms were 30 ± 6, 36 ± 6, and 31 ± 5 nM, respectively (Fig. 4A). These data indicate that CDE-096 binds both stressed and relaxed forms of PAI-1 with essentially identical affinities and that an exposed RCL is not required for compound binding.
Fig. 4.
The mechanism of action of CDE-096 involves conformational changes in PAI-1. (A) Binding of CDE-096 5 nM of the stressed or relaxed PAI-1 conformers, native, peptide-bound, or latent PAI-1S149C-FL. Changes in FP were plotted against the CDE-096 concentration, and the data were fit to an exponential association equation. (B) Incomplete inactivation of a thermostable PAI-1 variant by CDE-096. Relative residual binding of PAI-1 or PAI-1pept to immobilized tPAS478A was plotted against the CDE-096 concentration. (C and D) Effect of CDE-096 on the fluorescence intensity of 5 nM NBD-conjugated PAI-1, singly labeled at different sites as shown. For each PAI-1 variant, relative (C) and normalized (D) fluorescence signal were plotted against the CDE-096 concentration, and the data fit to an exponential decay equation. (E) CDE-096 blocks the latency transition. PAI-1S338C-NBD at 5 nM was incubated with buffer alone or with 1 µM CDE-096 at 37 °C, and the fluorescence intensity measured at the times shown. Arbitrary fluorescence intensity units were plotted against time, and the data fit to an exponential association equation. (F) CDE-096 blocks binding of the RCL-peptide, Ac-TVASS. PAI-1S149C-FL at 5 nM was incubated in buffer alone at 37 °C, in buffer, and then with 250 µM of the Ac-TVASS peptide, or with 1 µM CDE-096 followed by 250 µM of Ac-TVASS and changes in FP determined at the times shown.
The data above suggest that the degree of PAI-1 structural stability does not affect CDE-096 binding to PAI-1. However, the effect of CDE-096 on vitronectin binding suggests that CDE-096’s effects on PAI-1 may involve an allosteric mechanism, which could be influenced by changes in the structural stability of PAI-1. To examine this possibility, the two PAI-1 variants with exposed RCLs, but with differing degrees of thermostability (active PAI-1 and PAI-1pept), were analyzed by SPR for their susceptibility to CDE-096 inhibition of Michaelis complex formation with tPAS478A. In contrast to direct compound binding (Fig. 4A), the structurally stable variant, PAI-1pept, exhibited markedly reduced CDE-096–dependent inhibition of binding to immobilized tPAS478A (Fig. 4B). These findings suggest that even though CDE-096 binds both active PAI-1 and PAI-1pept with similar affinity, the increased structural stability of PAI-1pept is associated with reduced efficacy of CDE-096 for inhibition of PAI-1 Michaelis complex formation with tPA. Taken together with the similar affinity of CDE-096 for active and latent PAI-1, these data suggest that CDE-096 is not blocking Michaelis complex formation by direct interaction with the RCL. Instead these data are consistent with an allosteric mechanism for CDE-096 inactivation of PAI-1’s anti-protease activity, because increased structural stability of PAI-1 would likely affect the efficiency of CDE-096 induced allosteric changes in the serpin conformation.
The ability of CDE-096 to affect binding sites at opposite ends of PAI-1, namely the RCL and the vitronectin binding site, suggests that its mechanism of action against both proteases and vitronectin involves allosteric changes to PAI-1. Therefore, to examine whether CDE-096 binding could affect the structure of PAI-1 at multiple sites, a series of PAI-1 mutants with single-site cysteine substitutions conjugated to the environmentally sensitive fluorescent probe nitrobenzoxadiazole (NBD) were used. The probes were localized to the RCL (S338C or M347C), the vitronectin-binding site (S119C), the s3A/hF loop (S149C), or a loop at the base of sA (F302C). Each NBD-containing variant can report local changes in the probe environment. If CDE-096 binding to a single site can induce global changes in the PAI-1 structure, then each NBD-containing mutant should show a change in NBD fluorescence. Analysis of these data with each PAI-1 variant demonstrated that CDE-096–induced fluorescence changes of different magnitudes in each mutant but with nearly identical EC50 values of 25 ± 2 to 37 ± 4 nM (Fig. 4 C and D). These values are similar to the independently determined KD for CDE-096 binding to PAI-1 (22 ± 6 nM), suggesting that a single CDE-096 binding interaction with PAI-1 can affect the PAI-1 structure at distant sites and that CDE-096-binding is inducing conformational changes throughout the PAI-1 structure.
The reversibility of CDE-096's inactivation of PAI-1 suggests that it does not accelerate the latency transition as part of its mechanism of action. To determine if CDE-096 might instead reduce the rate of this transition, studies were performed using a PAI-1 variant, PAI-1S338C-NBD, previously shown to report the conformational change associated with RCL insertion into sA (44). PAI-1S338C-NBD is a variant with a NBD probe conjugated to a cysteine substitution at the P9 position of the RCL, and it shows a significant enhancement in NBD fluorescence on conversion to the latent structure. Incubation of PAI-1S338C-NBD at 37 °C in the absence of CDE-096 demonstrated an expected time-dependent increase in fluorescence intensity with a half-life of 2.1 h (Fig. 4E), consistent with the previously reported rate of the latency transition for PAI-1 (45). In contrast, CDE-096 markedly delayed the latency transition, with no significant change in fluorescence intensity measured over 20 h (Fig. 4E). Because RCL insertion into sA during the latency transition requires opening between β-strands s3A and s5A (24), the effects of CDE-096 on the accessibility of the s3A-s5A groove was probed using the intercalating RCL-mimicking peptide, Ac-TVASS (43), coupled with the FP of PAI-1S149C-FL. These studies monitored the insertion of Ac-TVASS into β-sheet A in the presence and absence of CDE-096. PAI-1 alone showed a slow decrease in FP with a half-life of ∼2.6 h, again consistent with the latency transition of PAI-1. In contrast, in the absence of CDE-096, the addition of Ac-TVASS induced a rapid decrease in FP with a half-life of 23 ± 1 min, consistent with incorporation of the peptide into β-sheet A (Fig. 4F). However, the addition of CDE-096 protected PAI-1 from both the latency transition and peptide insertion (Fig. 4F), suggesting that the compound may stabilize sA in PAI-1 in a closed conformation.
Identification of the CDE-096 Binding Site.
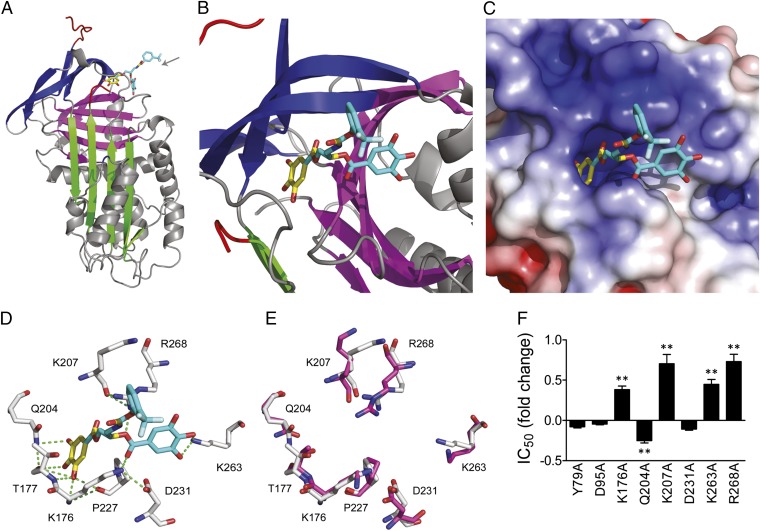
X-ray crystallography was used to identify the high-affinity CDE-096 binding site on PAI-1. Most crystal structures of PAI-1 have been obtained using the PAI-114–1B variant, which contains four point mutations that prevent the conversion to the latent form and maintain PAI-1 in an active conformation throughout the timeframe necessary for crystallization (46, 47). Inactivation of PAI-114–1B by CDE-096 was indistinguishable from that of WT PAI-1 (Table 1), suggesting that CDE-096 interacts with the same binding site on both WT PAI-1 and PAI-114–1B. Both cocrystallization and crystal soaking approaches were used. In the cocrystallization study, a site composed of residues from the s3A/s4C loop, β-sheets B and C, and α-helix H (the sB/sC pocket) was identified (Fig. 5 A and B). In addition, a second candidate binding site was observed only in the crystal soaking studies, where high concentrations (>0.5 mM) of CDE-096 was incubated with preformed PAI-114–1B crystals. The second site was consistent with a previously described organochemical compound binding site, bordered by β-strand 1A, strand 2A, and α-helix D (sA/hD pocket; Fig. S2) (9). However, as discussed below, biochemical and mutational analyses of these two regions revealed that the sB/sC pocket site most likely represented the high-affinity functional binding site for CDE-096 and suggested that the sA/hD pocket interaction was likely a result of the high concentrations of CDE-096 used for crystal soaking studies. Accordingly, the second site is only presented in the supplementary data section.
Fig. 5.
CDE-096 binds to a unique inhibitory site in PAI-1. (A and B) Diagrammatic representation of the 3D structure of the high-affinity PAI-1/CDE-096 complex. The protein is shown in ribbon style, with sheet A in green, sheet B in purple, sheet C in blue, and the RCL in red. The resolved portion of the compound is represented by the yellow sticks, whereas the remainder of the compound is approximated using mutational and biochemical data and is shown in cyan. The gray arrow designates the direction from which the binding site is viewed in B. (C) Electrostatic surface rendering of PAI-1 at the CDE-096 binding site (more positively charged regions in blue, and more negatively charged regions in red). (D) Residues predicted to hydrogen bond with CDE-096. PAI-1 residues are shown in white, CDE-096 in yellow, and cyan with oxygen atoms in red and nitrogen atoms in blue, and predicted hydrogen bonds as green dashes. (E) Overlay of the putative high-affinity binding site as predicted in D with the same residues in latent PAI-1, with residues from native PAI-1 shown in white, and those from latent PAI-1 shown in purple. (F) Inhibitory activity analysis of the alanine substitutions of residues in WT PAI-1 from two candidate binding sites. The IC50 of inhibition of each mutant by CDE-096 is shown relative to that of WT PAI-1. Mutants that showed significant differences in their IC50s compared with WT PAI-1 as determined by a two-tailed Student t test are indicated (**P < 0.01).
Examination of the binding of CDE-096 to the sB/sC site shows a single gallate group of CDE-096 inserted snugly into a deep pocket that is rimmed by basic residues, with the remainder of the CDE-096 ligand unresolved (cyan). Several hydrogen bond interactions were predicted between the gallate and residues Lys176, Thr177, Gln204, Lys207, and Pro227 (Fig. 5 C and D). We previously reported that the minimal structural requirements for high-affinity binding of polyphenols to PAI-1 include two gallates connected by a nonlinear linker of optimal length (33). Manual inspection of the area surrounding the sB/sC pocket for additional elements that would be compatible with these requirements revealed a surface groove between the sB/sC pocket and hH, and modeling the second gallate into this groove predicted possible additional interactions with residues Lys263 and Arg268 (Fig. 5 C and D). A major functional finding was that CDE-096 could bind to both native and latent PAI-1 with the same affinity, implying significant structural similarities between the compound-binding site in the two PAI-1 conformations. Comparison of the sB/sC pocket in these two structures showed that the putative hydrogen bonding residues at the sB/sC pocket are positioned almost identically in the two conformations (Fig. 5E). In contrast, the structure of the sA/hD pocket shows a dramatic reorganization in latent PAI-1 compared with the native conformation (Fig. S2 E and F).
The two candidate binding sites were also evaluated via site-directed mutagenesis on WT PAI-1. Residues for mutagenesis were selected based on potential hydrogen bonding or hydrophobic interactions with CDE-096 as predicted by manual inspection of the X-ray crystal structures. Five mutations around the sB/sC pocket were shown to significantly affect the IC50 of CDE-096, consistent with altered compound-binding relative to WT PAI-1 (Fig. 5F). In contrast, two residues predicted to interact with CDE-096 in the sA/hD pocket seen in the soak-in structure (Y79 and D95; Fig. S2 C and D) showed no effect on CDE-096 activity against PAI-1 (Fig. 5F). Replacement of Lys176, Lys207, Lys263, or Arg268 with alanine increased the IC50, suggesting these basic residues interact with the surface-exposed polar electronegative galloyl group in CDE-096. Interestingly, alanyl substitution of Gln204 in the wall of the hydrophobic pocket resulted in a reduction of IC50, implying that expansion of the hydrophobic pocket by removal of a larger Gln side-chain modestly improved CDE-096 binding. The detrimental effects of alanyl substitution of Lys263 and Arg268 on the IC50 strongly indicate that the CDE-096/PAI-1 interaction extends beyond the sB/sC pocket and toward hH. Together, the data suggest that the previously uncharacterized sB/sC pocket mediates PAI-1 inactivation by providing a high-affinity CDE-096 binding site that includes residues Lys176, Lys207, Lys263, and Arg268.
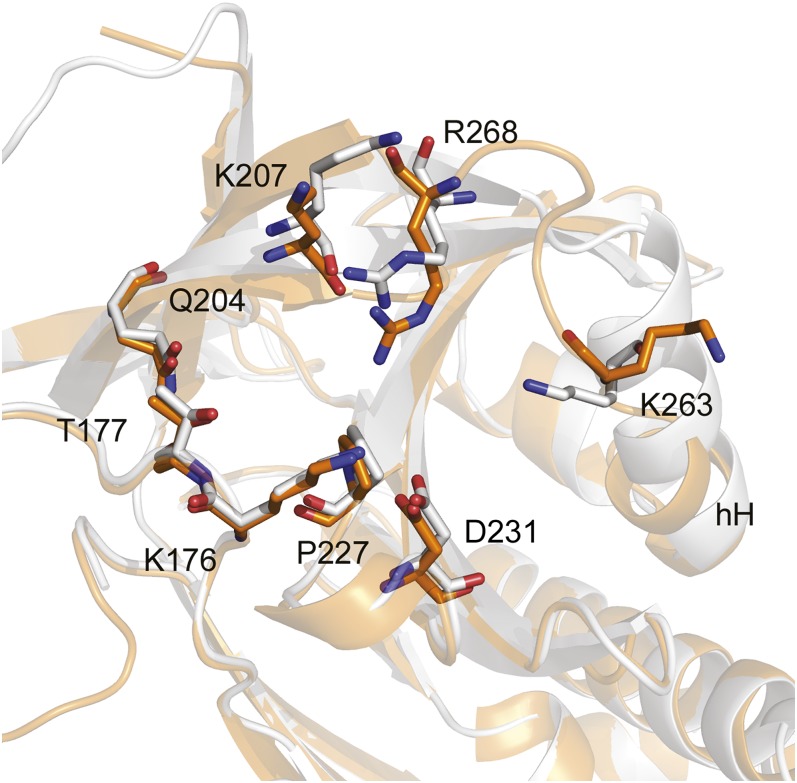
Further examination of the sB/sC pocket suggested underlying structural relationships that may explain many of the biochemical findings, including target specificity and the effects on vitronectin binding. Although structures of native murine, rattus, and porcine PAI-1 are unavailable for comparison with that of human PAI-1, Lys176, Lys207, Lys263, and Arg268 are completely conserved across these species, consistent with their common susceptibility to CDE-096. Furthermore, inspection of the sB/sC pockets of PAI-1, antithrombin (PDB ID code 1T1F), and α1-PI (PDB ID code 1QLP) reveals marked differences in their composition. Rather than the deep, well-formed pocket with a positively charged rim found in PAI-1, the same region in antithrombin contains a poorly defined, negatively charged pocket, whereas α1-PI contains a sB/sC pocket that is smaller than the pocket on PAI-1 (Fig. S3). The latter two structural arrangements appear incompatible with the binding of an acidic, relatively bulky polyphenolic compound, and correlate with the insensitivity of antithrombin and α1-PI to inactivation by CDE-096 (Table 1). Finally, the crystal structure of SMB-bound PAI-1 (PDB ID code 1OC0) shows a rearrangement of the C-terminal region of α-helix H into an ordered loop with the ε-amino group of Lys263 facing away from the sB/sC pocket and minor associated changes in Lys207 and Arg268 (Fig. 6). This rearrangement is consistent with SMB-binding to PAI-1 decreasing the affinity and IC50 of CDE-096 ∼10-fold (Fig. 3 E and F). However, it should be noted that this region in the PAI-1/SMB structure may be also be influenced by crystal contacts between different molecules in the crystal lattice, and therefore, it is uncertain whether these differences are responsible for the effects of SMB binding on CDE-096’s activity against PAI-1.
Fig. 6.
Binding of SMB to PAI-1 is associated with changes in the CDE-096 binding site. Overlay of the high-affinity binding site as predicted in Fig. 5D shown in white and those from PAI-1/SMB shown in orange. The PAI-1/CDE-096 cocrystal structure presented here was fit to the isolated PAI-1 structure from the PAI-1/SMB cocrystal (PDB ID code 1OC0) (RMS = 0.531, 303–303 atoms). Notably the H helix (hH) is differentially oriented in the two structures, affecting side chains of Lys263 and Arg268 in the CDE-096 binding site. Local secondary structures are shown in ribbon style.
Discussion
Both Galloyl Groups Interact with PAI-1.
Gallate-containing polyphenolic compounds are the highest affinity class of small molecule inhibitors of PAI-1 known to date (33). Based on a structure-activity relationship study of this scaffold, the compound CDE-096 was synthesized to provide an optimal reference compound for mechanistic and structural studies of this class of PAI-1 inactivators. Cocrystallization of PAI-1 with CDE-096 supported an earlier finding that a minimum of two galloyl moieties was required to inhibit PAI-1 at low nanomolar compound concentrations (33). In this structure, one gallate group is anchored into the relatively static sB/sC pocket, and mutational analysis indicates that the other extends through a basic surface groove that is contiguous with the pocket to contact residues from the more mobile hH. Thus, the digalloyl structure may provide a means of stabilizing one region of PAI-1 against another.
Inhibitory Mechanism Against Protease Binding.
The localization of CDE-096 in the sB/sC pocket is strongly supported by both structural and mutagenesis data. Structural analyses reveal that the CDE-096 pocket is in relatively close proximity to the RCL and provide a potential explanation of how CDE-096 can prevent target proteases from binding to PAI-1. Identification of the sB/sC pocket in the X-ray crystal structure of the noncovalent complex between PAI-1 and low molecular weight uPA illustrates how the CDE-096 binding could sterically preclude the RCL from adopting a conformation favorable for protease-binding (Fig. S4). Consistent with this possibility, the effect of CDE-096 on the fluorescence intensity of environmentally sensitive probes conjugated to the RCL demonstrated that the compound does influence the loop conformation. However, a mechanism of action based solely on steric clashes between the compound and the RCL does not explain the resistance of the thermostable variant, PAI-1pept, to CDE-096 inhibition of Michaelis complex formation, even though CDE-096 binding to PAI-1pept was indistinguishable from its binding to active WT PAI-1. These data imply that CDE-096 allosterically sequesters the RCL in a limited conformational space that is incompatible with protease-docking and in a manner that requires a certain degree of structural flexibility in PAI-1 (Fig. 4B).
Many studies reveal that the interaction of serpins with target proteases requires RCL rearrangement into a conformation suitable for initial binding to a protease active site (34, 48, 49), and such movements would be expected to involve structural elements at or near the top of sA. These movements of the central sheet are similarly required for the insertion of RCL mimicking peptides into sA and for conversion to the latent conformation (24, 43, 50), and CDE-096 strongly inhibits both peptide insertion and the latency transition. This effect is consistent with a mechanism where CDE-096 binding to the sB/sC pocket stabilizes PAI-1 in a conformation with sA closed at the top.
Inhibitory Mechanism Against Vitronectin Binding.
CDE-096 also blocks vitronectin binding through an allosteric mechanism. Previous studies have shown that vitronectin can allosterically influence both the top of sA and the RCL (51, 52). Unexpectedly, our data also suggest that there are reciprocal effects and that in order for a high affinity PAI-1/vitronectin interaction to be achieved, mobility around the top of the A-sheet is likely required. The binding of CDE-096 to PAI-1 likely stabilizes PAI-1 in a conformation that prevents this mobility and thereby allosterically inhibits vitronectin binding. This reciprocity in allostery is also suggested by a comparison of the structures of CDE-096 bound PAI-1 and SMB-bound PAI-1, because in the PAI-1/SMB complex, the C-terminal end of hH appears as an ordered loop with the ε-amino group of Lys263 facing away from the sB/sC pocket, whereas in the PAI-1/CDE-096 complex, Lys263 is likely to interact with the surface gallate group.
The serpin inhibitory mechanism is a multistep process of coordinated conformational changes, and as with any complex mechanism there are many moving parts within the serpin structure that have to shift position in an integrated fashion. Taken together, our data suggest that CDE-096 can be thought to act as a grain of sand in a finely tuned watch mechanism. We propose that CDE-096, which sits directly behind the point of initial RCL insertion, reduces flexibility in this region such that the RCL is no longer able to adopt key conformations necessary for protease binding or latency, and unexpectedly, for a high affinity PAI-1/vitronectin interaction to be achieved.
Conserved Target to Modulate the Serpin Mechanism.
The high degree of structural and functional homology among serpins (53) suggests that the sB/sC pocket is potentially a site that could be targeted to modify the activity of multiple serpin family members. For example, the binding of citrate to the sB/sC pocket of α1-PI is known to stabilize this serpin against misfolding and aggregation (Fig. S5), highlighting the role of the sB/sC pocket as a crucial “gatekeeper” that can profoundly influence serpin conformation and function. In addition, since serpins evolved to exquisitely regulate their affinities for other molecules via conformational control, the sB/sC pocket could also be targeted to modulate their binding to other molecules, such as hormones, cell-surface receptors, or extracellular matrix components. Thus, given the ability to develop serpin-specific compounds, the sB/sC pocket might be exploited to pharmacologically regulate a variety of serpin functions and interactions.
Methods
Synthesis of CDE-096.
Synthetic procedures and spectroscopic data for CDE-096 are provided in SI Methods, along with a schematic of the synthesis reaction (Scheme 1).
Assay Conditions.
Unless otherwise specified, all assays were performed in assay buffer (40 mM Hepes, pH 7.8, 100 mM NaCl, 0.005% Tween 20, and 0.1% DMSO) at 25 °C.
Inactivation of PAI-1.
PAI-1 inactivation by CDE-096 and SDS/PAGE were performed as described (33). In some experiments, 30 nM SMB was preincubated with 2 nM PAI-1 at 25 °C (15 min) before adding CDE-096, protease, and fluorogenic substrate. The reversibility of PAI-1 inactivation by CDE-096 was studied as described previously (33), with the exception that the concentrations of PAI-1, CDE-096, and uPA used were 5 nM, 1 μM, and 6 nM, respectively.
Surface Plasmon Resonance.
Direct binding of CDE-096 to PAI-1 was studied using a BIAcore 2000 optical sensor. The anti-human PAI-1 murine monoclonal antibody, MA-33H1F7 (54, 55), was immobilized to a CM5 SPR chip at ∼3,000 response units (RUs) under previously described conditions (33). The monoclonal antibody was saturated by injecting 100 nM PAI-1 in assay buffer (2.5 min, 30 µL/min). After washing with buffer (4 min), 200 nM CDE-096 was flowed over the captured PAI-1 (5 min) for compound association, followed by assay buffer (7 min) for compound dissociation. The data were fit to simultaneous ka/kd using BIAevaluation 4.0.
Binding of PAI-1 variants treated with vehicle or CDE-096 to vitronectin, tPAS478A, or anhydrotrypin was monitored as described (33). Vitronectin, tPAS478A, or anhydrotrypsin was immobilized to CM5 SPR chips at ∼1,000, ∼1,000, or ∼2,000 RUs, respectively (33).
Thermal Denaturation.
Thermostability studies were performed as previously described using assay buffer (41).
FP.
FP measurements were performed on a SpectraMax M5 spectrofluorometric microplate reader (Molecular Devices) using the FP template [excitation 485 nm, emission 525 nm, cutoff 515n m, high photomultiplier tube (PMT) setting]. The FP of a FITC standard was 27 ± 2 mP up to 1 µM of free fluorophore. The equilibrium binding of PAI-1 to CDE-096, vitronectin, or SMB was monitored by preincubating 5 nM PAI-1S149C-FL with 0−1 µM CDE-096 or 0−300 nM vitronectin or SMB in assay buffer (15 min), followed by FP measurement. To study the effect of prebinding CDE-096 on the subsequent binding of vitronectin or SMB, 2 nM PAI-1S149C-FL was first incubated with 0−1 µM CDE-096 (15 min), followed by 30 nM cofactor (30 min), before FP measurement. The effect of cofactor on CDE-096 binding to PAI-1 was studied by preincubating 2 nM PAI-1S149C-FL with 30 nM SMB (15 min), followed by incubation with 0−1 µM CDE-096 (30 min). For each reaction, tandem samples were assembled for background subtraction and contained all reaction components except PAI-1S149C-FL.
The effect of CDE-096 on the binding between PAI-1 and the pentapeptide, Ac-TVASS, could not be assessed by the standard measurement of peptide-induced loss of PAI-1 activity due to the inhibitory effect of the compound. Thus, 5 nM PAI-1S149C-FL was preincubated with or without 1 µM CDE-096 at 25 °C in assay buffer (15 min) and incubated at 37 °C in the presence or absence of 250 µM Ac-TVASS. The time-dependent change in PAI-1S149C-FL FP was monitored.
Environmentally Sensitive Fluorescent Probes.
Conformational changes in PAI-1 were probed using the environmentally sensitive fluorescent label NBD. The effect of CDE-096 on the PAI-1 latency transition was measured by preincubating 10 nM PAI-1S338C-NBD with buffer or 1 µM CDE-096 at 25 °C in assay buffer (15 min) followed by incubation at 37 °C. The time-dependent change in NBD fluoresence was monitored on a spectrofluorometric microplate reader (excitation 480 nm, emission 520 nm, 515-nm auto-cutoff, high PMT, 20 reads per well), and the data were fit to an exponential association equation.
The effect of CDE-096 binding on local environmental changes in PAI-1 was monitored using NBD conjugated to individual preparations of PAI-1 at S119C, S149C, F302C, S338C, or M347C. Each labeled PAI-1 mutant (5 nM) was incubated with 0−1 µM CDE-096 in assay buffer (15 min) before measuring the NBD fluorescence intensities. The data were expressed as relative fluorescence intensity (Frelative; Eq. 1) and normalized intensity (Fnormalized; Eq. 2)
wherein F is the raw fluorescence intensity, and Fmax and Fmin are the maximum and minimum fluorescence intensities, respectively. The relative and normalized intensities were fit to an exponential decay equation.
Binding Site Mutagenesis.
PAI-1 mutagenesis and expression was performed as previously described (29). Cell extract was prepared from bacterial pellets by washing in ice cold 0.85% NaCl followed by centrifugation at 6,000 × g at 4 °C (20 min). Washed cells were resuspended in lysis buffer consisting of 0.05 M sodium phosphate, pH 7.0, 0.1 M NaCl, and 4 mM EDTA, and incubated with 1 mg/mL T4 lysozyme at 25 °C (30 min). Cells were lysed by three cycles of liquid nitrogen (1 min), followed by 37 °C incubation (10 min). Lysates were diluted fivefold into lysis buffer, passed through a 25-gauge needle 10 times, and centrifuged at 16,000 × g at 4 °C (20 min). The supernatant was collected as cell extract (CE). The concentrations of active mutant PAI-1 in CE were titrated as described (29), and the total protein concentration was measured using the BCA protein assay kit (Pierce Biotechnology). PAI-1 mutant inactivation by CDE-096 in CE was performed as described previously (33), with the total protein concentrations normalized using CE from nontransformed bacteria.
X-Ray Crystallography.
Crystallographic techniques are provided in SI Methods, and Omit maps for the CDE-096 binding sites are shown in Figs. S6 and S7. The PAI-1/CDE-096 cocrystal and soaked forms were deposited into the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB ID codes 4G8O and 4G8R, respectively). Structural renderings and fits were performed using the PyMOL Molecular Graphics System, Version 1.4.1 (Schrödinger, LLC), and electrostatic potentials for Fig. 5 and Figs. S2 and S3 were calculated with the Adaptive Poisson-Boltzmann Solver plug-in PyMOL, with high and low e-levels of 5 and −5, respectively (56). Data collection and refinement statistics are shown in Table S1.
Statistical Analyses.
Unless otherwise specified, all assays were performed in at least triplicate except for SPR binding assays, which were performed in duplicate. Results are reported as means ± SEs. Reversibility studies and inhibitory activity analysis of the binding site mutants were analyzed for significance by a Student t test, with *P ≤ 0.05 and **P ≤ 0.01.
Supplementary Material
Acknowledgments
We thank Mark Warnock for excellent technical assistance. We thank K. Chinnaswamy for help with crystallization experiments. This work was supported, in whole or in part, by National Institutes of Health Grants HL55374, NS079639, and HL089407 (to D.A.L.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Footnotes
Conflict of interest statement: S.-H.L., A.A.R., C.D.E., J.A.S., and D.A.L. are inventors on patent applications related to CDE-096.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4G8O and 4G8R).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216499110/-/DCSupplemental.
References
- 1.Hamsten A, Wiman B, de Faire U, Blombäck M. Increased plasma levels of a rapid inhibitor of tissue plasminogen activator in young survivors of myocardial infarction. N Engl J Med. 1985;313(25):1557–1563. doi: 10.1056/NEJM198512193132501. [DOI] [PubMed] [Google Scholar]
- 2.McMahon GA, et al. Plasminogen activator inhibitor-1 regulates tumor growth and angiogenesis. J Biol Chem. 2001;276(36):33964–33968. doi: 10.1074/jbc.M105980200. [DOI] [PubMed] [Google Scholar]
- 3.Stefansson S, Lawrence DA. The serpin PAI-1 inhibits cell migration by blocking integrin α V β 3 binding to vitronectin. Nature. 1996;383(6599):441–443. doi: 10.1038/383441a0. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, et al. A mutant, noninhibitory plasminogen activator inhibitor type 1 decreases matrix accumulation in experimental glomerulonephritis. J Clin Invest. 2003;112(3):379–388. doi: 10.1172/JCI18038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitzman DT, et al. Bleomycin-induced pulmonary fibrosis in transgenic mice that either lack or overexpress the murine plasminogen activator inhibitor-1 gene. J Clin Invest. 1996;97(1):232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinsky DJ, et al. Coordinated induction of plasminogen activator inhibitor-1 (PAI-1) and inhibition of plasminogen activator gene expression by hypoxia promotes pulmonary vascular fibrin deposition. J Clin Invest. 1998;102(5):919–928. doi: 10.1172/JCI307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorquist P, et al. Identification of the binding site for a low-molecular-weight inhibitor of plasminogen activator inhibitor type 1 by site-directed mutagenesis. Biochemistry. 1998;37(5):1227–1234. doi: 10.1021/bi971554q. [DOI] [PubMed] [Google Scholar]
- 8.Neve J, et al. Sideroxylonal C, a new inhibitor of human plasminogen activator inhibitor type-1, from the flowers of Eucalyptus albens. J Nat Prod. 1999;62(2):324–326. doi: 10.1021/np980286+. [DOI] [PubMed] [Google Scholar]
- 9.Egelund R, et al. A regulatory hydrophobic area in the flexible joint region of plasminogen activator inhibitor-1, defined with fluorescent activity-neutralizing ligands. Ligand-induced serpin polymerization. J Biol Chem. 2001;276(16):13077–13086. doi: 10.1074/jbc.M009024200. [DOI] [PubMed] [Google Scholar]
- 10.Crandall DL, et al. Characterization and comparative evaluation of a structurally unique PAI-1 inhibitor exhibiting oral in-vivo efficacy. J Thromb Haemost. 2004;2(8):1422–1428. doi: 10.1111/j.1538-7836.2004.00829.x. [DOI] [PubMed] [Google Scholar]
- 11.Gorlatova NV, et al. Mechanism of inactivation of plasminogen activator inhibitor-1 by a small molecule inhibitor. J Biol Chem. 2007;282(12):9288–9296. doi: 10.1074/jbc.M611642200. [DOI] [PubMed] [Google Scholar]
- 12.Liang A, et al. Characterization of a small molecule PAI-1 inhibitor, ZK4044. Thromb Res. 2005;115(4):341–350. doi: 10.1016/j.thromres.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Gardell SJ, et al. Neutralization of plasminogen activator inhibitor I (PAI-1) by the synthetic antagonist PAI-749 via a dual mechanism of action. Mol Pharmacol. 2007;72(4):897–906. doi: 10.1124/mol.107.037010. [DOI] [PubMed] [Google Scholar]
- 14.Rupin A, et al. S35225 is a direct inhibitor of Plasminogen Activator Inhibitor type-1 activity in the blood. Thromb Res. 2008;122(2):265–270. doi: 10.1016/j.thromres.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Izuhara Y, et al. Inhibition of plasminogen activator inhibitor-1: Its mechanism and effectiveness on coagulation and fibrosis. Arterioscler Thromb Vasc Biol. 2008;28(4):672–677. doi: 10.1161/ATVBAHA.107.157479. [DOI] [PubMed] [Google Scholar]
- 16.Einholm AP, et al. Biochemical mechanism of action of a diketopiperazine inactivator of plasminogen activator inhibitor-1. Biochem J. 2003;373(Pt 3):723–732. doi: 10.1042/BJ20021880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yepes M, Loskutoff DJ, Lawrence DA (2006) Hemostasis and Thrombosis: Basic Principles and Clinical Practice, eds Colman RW, Marder VJ, Clowes AW, George JN, Goldhaber SZ (Lippincott Williams & Wilkins, Baltimore), pp 365–380.
- 18.Lawrence DA, et al. Partitioning of serpin-proteinase reactions between stable inhibition and substrate cleavage is regulated by the rate of serpin reactive center loop insertion into beta-sheet A. J Biol Chem. 2000;275(8):5839–5844. doi: 10.1074/jbc.275.8.5839. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence DA, et al. Serpin-protease complexes are trapped as stable acyl-enzyme intermediates. J Biol Chem. 1995;270(43):25309–25312. doi: 10.1074/jbc.270.43.25309. [DOI] [PubMed] [Google Scholar]
- 20.Stratikos E, Gettins PG. Formation of the covalent serpin-proteinase complex involves translocation of the proteinase by more than 70 A and full insertion of the reactive center loop into beta-sheet A. Proc Natl Acad Sci USA. 1999;96(9):4808–4813. doi: 10.1073/pnas.96.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407(6806):923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 22.Dementiev A, Dobó J, Gettins PG. Active site distortion is sufficient for proteinase inhibition by serpins: Structure of the covalent complex of alpha1-proteinase inhibitor with porcine pancreatic elastase. J Biol Chem. 2006;281(6):3452–3457. doi: 10.1074/jbc.M510564200. [DOI] [PubMed] [Google Scholar]
- 23.Smith KF, Harrison RA, Perkins SJ. Structural comparisons of the native and reactive-centre-cleaved forms of alpha 1-antitrypsin by neutron- and X-ray-scattering in solution. Biochem J. 1990;267(1):203–212. doi: 10.1042/bj2670203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mottonen J, et al. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355(6357):270–273. doi: 10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence DA, et al. Characterization of the binding of different conformational forms of plasminogen activator inhibitor-1 to vitronectin. Implications for the regulation of pericellular proteolysis. J Biol Chem. 1997;272(12):7676–7680. doi: 10.1074/jbc.272.12.7676. [DOI] [PubMed] [Google Scholar]
- 26.Hekman CM, Loskutoff DJ. Endothelial cells produce a latent inhibitor of plasminogen activators that can be activated by denaturants. J Biol Chem. 1985;260(21):11581–11587. [PubMed] [Google Scholar]
- 27.Andreasen PA, Egelund R, Jensen S, Rodenburg KW. Solvent effects on activity and conformation of plasminogen activator inhibitor-1. Thromb Haemost. 1999;81(3):407–414. [PubMed] [Google Scholar]
- 28.Gils A, Declerck PJ. Modulation of plasminogen activator inhibitor 1 by Triton X-100—identification of two consecutive conformational transitions. Thromb Haemost. 1998;80(2):286–291. [PubMed] [Google Scholar]
- 29.Gorlatova NV, Elokdah H, Fan K, Crandall DL, Lawrence DA. Mapping of a conformational epitope on plasminogen activator inhibitor-1 by random mutagenesis. Implications for serpin function. J Biol Chem. 2003;278(18):16329–16335. doi: 10.1074/jbc.M208420200. [DOI] [PubMed] [Google Scholar]
- 30.Dupont DM, et al. Evidence for a pre-latent form of the serpin plasminogen activator inhibitor-1 with a detached beta-strand 1C. J Biol Chem. 2006;281(47):36071–36081. doi: 10.1074/jbc.M606851200. [DOI] [PubMed] [Google Scholar]
- 31.Mathiasen L, et al. A peptide accelerating the conversion of plasminogen activator inhibitor-1 to an inactive latent state. Mol Pharmacol. 2008;74(3):641–653. doi: 10.1124/mol.108.046417. [DOI] [PubMed] [Google Scholar]
- 32.Li SH, Lawrence DA. Development of inhibitors of plasminogen activator inhibitor-1. Methods Enzymol. 2011;501:177–207. doi: 10.1016/B978-0-12-385950-1.00009-2. [DOI] [PubMed] [Google Scholar]
- 33.Cale JM, et al. Characterization of a novel class of polyphenolic inhibitors of plasminogen activator inhibitor-1. J Biol Chem. 2010;285(11):7892–7902. doi: 10.1074/jbc.M109.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DJ, Li W, Adams TE, Huntington JA. Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation. EMBO J. 2006;25(9):2029–2037. doi: 10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefansson S, et al. Inhibition of angiogenesis in vivo by plasminogen activator inhibitor-1. J Biol Chem. 2001;276(11):8135–8141. doi: 10.1074/jbc.M007609200. [DOI] [PubMed] [Google Scholar]
- 36.Deng G, Curriden SA, Wang S, Rosenberg S, Loskutoff DJ. Is plasminogen activator inhibitor-1 the molecular switch that governs urokinase receptor-mediated cell adhesion and release? J Cell Biol. 1996;134(6):1563–1571. doi: 10.1083/jcb.134.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer EJ, Fujita T, Gardell SJ, Shebuski RJ, Reilly CF. The pharmacokinetics of plasminogen activator inhibitor-1 in the rabbit. Blood. 1990;76(8):1514–1520. [PubMed] [Google Scholar]
- 38.Seiffert D, Loskutoff DJ. Evidence that type 1 plasminogen activator inhibitor binds to the somatomedin B domain of vitronectin. J Biol Chem. 1991;266(5):2824–2830. [PubMed] [Google Scholar]
- 39.Xu Z, et al. Conservation of critical functional domains in murine plasminogen activator inhibitor-1. J Biol Chem. 2004;279(17):17914–17920. doi: 10.1074/jbc.M314197200. [DOI] [PubMed] [Google Scholar]
- 40.Carrell RW, Owen MC. Plakalbumin, alpha 1-antitrypsin, antithrombin and the mechanism of inflammatory thrombosis. Nature. 1985;317(6039):730–732. doi: 10.1038/317730a0. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence DA, Olson ST, Palaniappan S, Ginsburg D. Engineering plasminogen activator inhibitor-1 (PAI-1) mutants with increased functional stability. Biochemistry. 1994;33(12):3643–3648. doi: 10.1021/bi00178a022. [DOI] [PubMed] [Google Scholar]
- 42.Komissarov AA, Declerck PJ, Shore JD. Protonation state of a single histidine residue contributes significantly to the kinetics of the reaction of plasminogen activator inhibitor-1 with tissue-type plasminogen activator. J Biol Chem. 2004;279(22):23007–23013. doi: 10.1074/jbc.M401383200. [DOI] [PubMed] [Google Scholar]
- 43.Xue Y, et al. Interfering with the inhibitory mechanism of serpins: Crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactive-centre loop peptide. Structure. 1998;6(5):627–636. doi: 10.1016/s0969-2126(98)00064-1. [DOI] [PubMed] [Google Scholar]
- 44.Verhamme I, et al. Accelerated conversion of human plasminogen activator inhibitor-1 to its latent form by antibody binding. J Biol Chem. 1999;274(25):17511–17517. doi: 10.1074/jbc.274.25.17511. [DOI] [PubMed] [Google Scholar]
- 45.Lindahl TL, Sigurdardóttir O, Wiman B. Stability of plasminogen activator inhibitor 1 (PAI-1) Thromb Haemost. 1989;62(2):748–751. [PubMed] [Google Scholar]
- 46.Berkenpas MB, Lawrence DA, Ginsburg D. Molecular evolution of plasminogen activator inhibitor-1 functional stability. EMBO J. 1995;14(13):2969–2977. doi: 10.1002/j.1460-2075.1995.tb07299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharp AM, et al. The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion. Structure. 1999;7(2):111–118. doi: 10.1016/S0969-2126(99)80018-5. [DOI] [PubMed] [Google Scholar]
- 48.Gils A, Declerck PJ. Proteinase specificity and functional diversity in point mutants of plasminogen activator inhibitor 1. J Biol Chem. 1997;272(19):12662–12666. doi: 10.1074/jbc.272.19.12662. [DOI] [PubMed] [Google Scholar]
- 49.Gooptu B, et al. Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: Implications for inhibitory function and conformational disease. Proc Natl Acad Sci USA. 2000;97(1):67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner R, et al. Implications for function and therapy of a 2.9 A structure of binary-complexed antithrombin. J Mol Biol. 1998;283(1):9–14. doi: 10.1006/jmbi.1998.2083. [DOI] [PubMed] [Google Scholar]
- 51.Li SH, Gorlatova NV, Lawrence DA, Schwartz BS. Structural differences between active forms of plasminogen activator inhibitor type 1 revealed by conformationally sensitive ligands. J Biol Chem. 2008;283(26):18147–18157. doi: 10.1074/jbc.M709455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fa M, et al. Time-resolved polarized fluorescence spectroscapy studies of plasminogen activator inhibitor type 1: Conformational changes of the reactive center upon interations with target protease. vitronectin and heparin. Biochem. 1995;34(42):13833–13840. doi: 10.1021/bi00042a015. [DOI] [PubMed] [Google Scholar]
- 53.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10(12):1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- 54.Debrock S, Declerck PJ. Neutralization of plasminogen activator inhibitor-1 inhibitory properties: identification of two different mechanisms. Biochim Biophys Acta. 1997;1337(2):257–266. doi: 10.1016/s0167-4838(96)00173-2. [DOI] [PubMed] [Google Scholar]
- 55.Debrock S, Declerck PJ. Identification of a functional epitope in plasminogen activator inhibitor-1, not localized in the reactive center loop. Thromb Haemost. 1998;79(3):597–601. [PubMed] [Google Scholar]
- 56. Lerner MG, Carlson HA (2006) APBS plug-in for PyMOL. University of Michigan, Ann Arbor.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.