Significance
Pancreatic cancer is still one of the major challenges in clinical oncology. Mutant KRAS is a driving oncogene in the majority of human pancreatic cancer cases. We have made an effort to meet this challenge by developing a therapeutic platform for local and prolonged delivery of siRNA. Our results show that the siRNA targeted against KRAS mutations with a local prolonged release system knocks down KRAS expression in vitro and in vivo, leading to an antitumor effect. Our report describes an applicable and efficient delivery method of siRNA that overcomes the major obstacles of toxicity and organ accessibility. Notably our approach enabled the conversion of KRAS from a nondruggable to a potentially druggable cancer target.
Keywords: targeted therapy, gene therapy
Abstract
Pancreatic ductal adenocarcinoma (PDA) represents an unmet therapeutic challenge. PDA is addicted to the activity of the mutated KRAS oncogene which is considered so far an undruggable therapeutic target. We propose an approach to target KRAS effectively in patients using RNA interference. To meet this challenge, we have developed a local prolonged siRNA delivery system (Local Drug EluteR, LODER) shedding siRNA against the mutated KRAS (siG12D LODER). The siG12D LODER was assessed for its structural, release, and delivery properties in vitro and in vivo. The effect of the siG12D LODER on tumor growth was assessed in s.c. and orthotopic mouse models. KRAS silencing effect was further assessed on the KRAS downstream signaling pathway. The LODER-encapsulated siRNA was stable and active in vivo for 155 d. Treatment of PDA cells with siG12D LODER resulted in a significant decrease in KRAS levels, leading to inhibition of proliferation and epithelial–mesenchymal transition. In vivo, siG12D LODER impeded the growth of human pancreatic tumor cells and prolonged mouse survival. We report a reproducible and safe delivery platform based on a miniature biodegradable polymeric matrix, for the controlled and prolonged delivery of siRNA. This technology provides the following advantages: (i) siRNA is protected from degradation; (ii) the siRNA is slowly released locally within the tumor for prolonged periods; and (iii) the siG12D LODER elicits a therapeutic effect, thereby demonstrating that mutated KRAS is indeed a druggable target.
Pancreatic cancer is an aggressive disease that develops in a relatively symptom-free manner and in most cases, is already advanced at the time of diagnosis (1). It has one of the highest fatality rates of all cancers and is one of the leading causes of cancer-related deaths in the Western world (1, 2). Pancreatic ductal adenocarcinoma (PDA) is the most common pancreatic neoplasm, responsible for 95% of pancreatic cancer cases (3). Genetic alterations in the KRAS signaling pathway are involved in over 90% of pancreatic cancer cases (4–6). KRAS mutations were shown to be an early event in the development of pancreatic cancer (5, 7, 8).The most common KRAS mutation of the human pancreas adenocarcinoma is a gain-of-function substitution mutation of glycine at codon 12 to aspartate (G12D) (5, 9–11). Moreover, PDA cancer cell growth was shown to be dependent on the activity of the mutated KRAS (5, 11) and accordingly, silencing KRAS has proven effective in controlling pancreatic cell line proliferation (12). Here, we aimed to harness the advantages of siRNA technology as a therapeutic modality for pancreatic cancer.
Parenteral controlled drug delivery systems are used to improve and advance the therapeutic effects of drug treatments by providing optimized local drug concentrations over prolonged periods of time, reduction of side effects, and cost reduction (13). A prominent method of controlling the release rate of a drug in a pharmaceutical dosage is to embed the active agent within a polymeric matrix (14, 15). The polymer must be biocompatible, and in the case of parenteral administration, preferably biodegradable, to avoid the need to remove empty remnants.
In the present study, we exploited the slow-release characteristics of the biodegradable polymer matrix, which we named local drug eluter (LODER) for the treatment of solid tumors.
Results
LODER Design and Characterization.
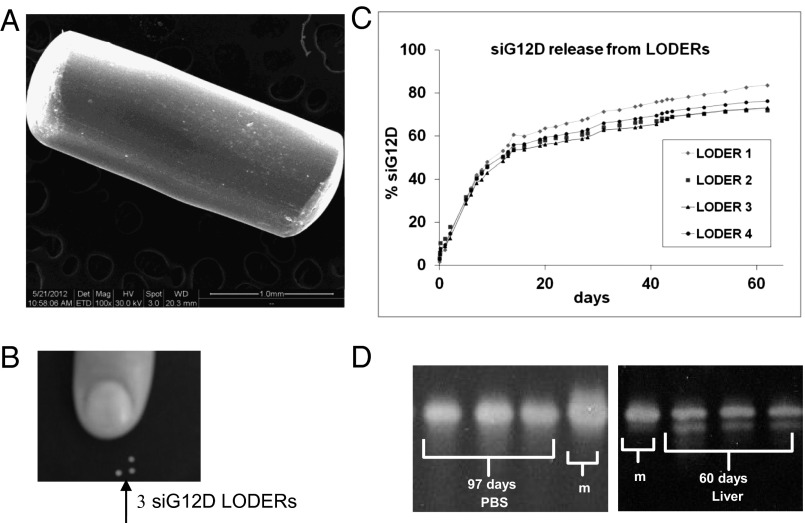
The LODER was developed as a miniature (millimetric) biodegradable polymeric matrix that encompasses siRNA such as anti-KRASG12D siRNA (siG12D LODER; diagram presented in Fig. S1A). It was designed to provide both protection from degradation and a slow and stable local drug release within a tumor over a period of a few months. Fig. 1A shows a SEM image of the siG12D LODER and a picture of the LODER's actual size (Fig. 1B). Fig. S1B exhibits a representative MRI scan, where a number of LODERs implanted into a mouse liver are shown. LODERs could also be detected by microcomputer tomography (Fig. S1C).
Fig. 1.
LODER characteristics: (A) SEM image of the siG12D LODER removed 45 d after implantation in vivo. (B) LODER actual size picture. (C) Release of siG12D from LODERs containing 10 μg siRNA: siG12D LODERs were incubated in PBS (pH = 7.4), at 37 °C. The siG12D amount was measured in the PBS using NanoDrop (absorption). (D) LODERs protect siG12D from degradation: protection of LODER-embedded siG12D from degradation was assessed by urea-polyacrylamide gel electrophoresis. siG12D was extracted from LODERs that were incubated at 37 °C in PBS or mouse liver tissue ex vivo (liver). m, siG12D size marker.
We measured accumulated siRNA release from the LODER. LODERs containing 10 μg siG12D were incubated in phosphate buffer solution (PBS), at 37 °C, in a humidified incubator. Fig. 1C shows the cumulative release of siG12D from LODERs incubated in PBS for over 2 mo. The release curve reveals that after 60 d in PBS, ∼75% of the encapsulated siG12D was released. Notably, variations in release curves measured for different LODERs did not exceed 12%. In parallel, we measured the amount of siRNA retained in the LODERs (Fig. S1D). To assess their ability to protect the siRNA from degradation, LODERs containing 10 μg siG12D were incubated in PBS or placed in a mouse liver tissue ex vivo; siG12D was later extracted from the LODERs and its quantity and integrity were measured using HPLC, gel electrophoresis, and light absorption. Both gel electrophoresis and HPLC show that siG12D remained in its intact form for at least 97 d in PBS and at least 60 d in a liver tissue (Fig. 1D and Fig. S1E). These data substantiate the potential of the LODER to function as an efficient and stable delivery device for siRNA in vivo.
LODER-Derived siG12D Significantly Inhibits Growth of Pancreatic Cancer Cells in Vitro.
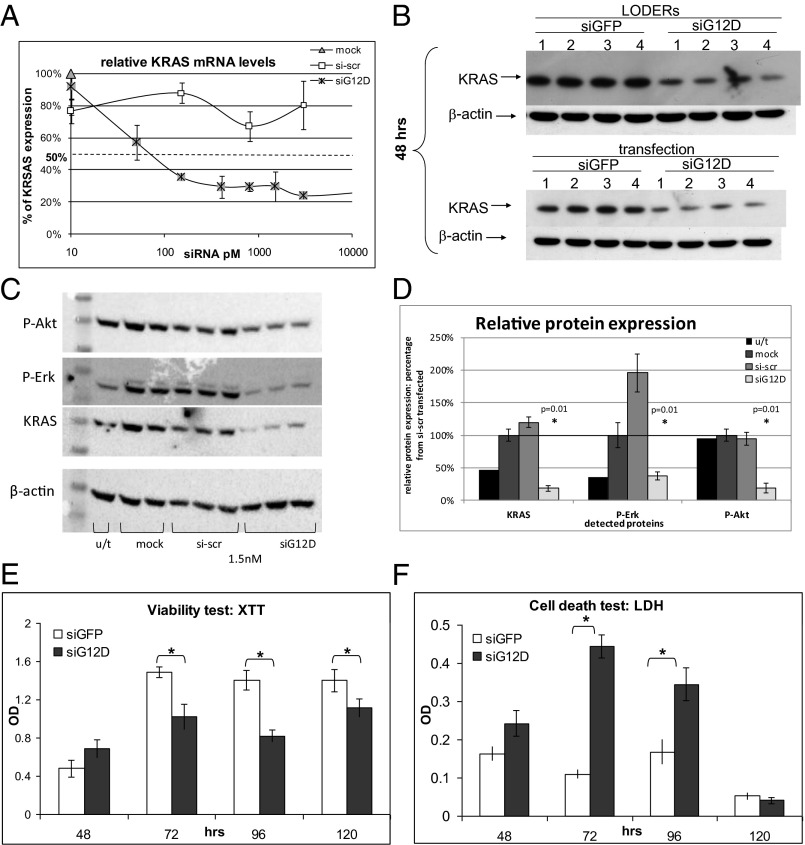
To determine the silencing potential of siG12D, a specific siRNA directed against the mutant KRASG12D, we used the Panc1 human pancreatic carcinoma cells that harbor this specific mutation in the KRAS gene. Cultured Panc1 cells were transfected with siG12D, control scrambled siRNA (si-scr) in decreasing concentrations or mock transfected. Following 24 h, mRNA levels of KRAS were assessed by quantitative PCR (qPCR) (Fig. 2A). This analysis revealed that siG12D led to reduced KRAS mRNA levels, with a half maximal inhibitory concentration (IC50) of 67 pM. We confirmed the site-specific, siG12D-directed cleavage of the KRAS message, by rapid amplification of cDNA ends (RACE) (Fig. S2).
Fig. 2.
The silencing effect of the siG12D LODER in human pancreatic cancer cells. (A) Panc1 cells were transiently transfected with siG12D, nontargeting scrambled siRNA (si-scr) or mock transfected using Lipofectamine 2000. A relative level of KRAS mRNA was assessed by real-time PCR. HPRT and UBC were used as endogenous controls. The result is shown as an average of three different samples ± SEM. (B) Panc1-Luc cells were incubated with LODERs containing either siGFP or siG12D in the presence of Lipofectamine 2000 or transiently transfected with these siRNAs using Lipofectamine 2000. After 48 h, cells were lysed and the total level of KRAS protein was assessed by Western blot analysis. Shown are representative blots of 48 h of four independent experiments. (C and D) Panc1 cells were left untreated (u/t) or transiently transfected with siG12D, nontargeting scrambled siRNA (si-scr), or mock transfected using Lipofectamine 2000. At 36 h after the transfection, cells were lysed and relative levels of KRAS, P-Erk, and P-Akt proteins were assessed by Western blot analysis. The results were normalized to the level of β-actin. (C) Representative blots of KRAS, P-Erk, P-Akt, and β-actin. (D) The graph shows the average relative protein level as percentage of mock transfected cells ± SEM. Student t test (p) was calculated compared with mock transfected cells. (E and F) Panc1-Luc cells were incubated with LODERs containing either siGFP or siG12D in the presence of Lipofectamine 2000 for 48, 72, 96, or 120 h. Cell viability (E) and cell death (F) were assessed using XTT and LDH tests, respectively. Representative results of six different samples are shown ± SEM, *P < 0.05; **P < 0.01 according to the Student t test.
Next, we compared the effects of LODER-derived siG12D with directly applied siG12D on KRAS protein levels in vitro. Panc1 cells were incubated with siGFP, siG12D, or with LODERs containing these siRNAs, in the presence of a transfection reagent. At 48 h posttreatment, the protein levels of KRAS were found to be lower in both siG12D groups compared with the siGFP. This decrease was met whether siG12D was applied directly or through a LODER (Fig. 2B and Fig. S3A). These results show that the LODER-driven siRNA can efficiently silence target gene expression, comparable to nonbound siRNA. To verify the specific effect of siG12D on KRAS signaling pathway, we measured the levels of downstream signaling proteins that are activated by KRAS—including Erk (P-Erk) and Akt (P-Akt). Western blot results revealed that inhibition of KRAS caused a decrease in the levels of P-Erk and P-Akt (Fig. 2 C and D).
Previous studies reported that inhibition of KRAS expression led to the inhibition of pancreatic cancer cell growth (12, 16). To examine the potential growth-inhibiting effect of siG12D, we measured both cell viability and cell death of Panc1 cells treated with the siG12D LODER. A time-dependent decrease in cell viability correlating with an increase in cell death was met in the presence of siG12D LODERs compared with the siGFP LODERs (Fig. 2 E and F). To confirm the effects of LODER-derived siG12D on cell growth, Panc1-Luc cells, a subclone of Panc1 cells stably expressing the luciferase reporter gene under the constitutive promoter [CMV early enhancer/chicken β actin (CAG) promoter], were used for vitality assays. These cells were incubated with siG12D LODER, siLuc LODER (as a positive technical control), or the siGFP LODER. Fig. S3B shows that in the presence of either siLuc or siG12D LODERs, a significant inhibition of luciferase expression is met compared with the siGFP LODER. These results both emphasize the ability of LODERs to efficiently release functional siRNA and confirm the feasibility of Panc1-Luc cells to serve as indicators of effects on growth for further in vivo assessment.
The Effect of the siG12D LODER on Epithelial–Mesenchymal Transition.
In the course of tumor development cancer cells may undergo an epithelial–mesenchymal transition (EMT). In this process, the cells acquire mesenchymal characteristics and lose epithelial ones. It was previously reported that KRAS inhibition reduces EMT, as manifested by a decrease in cell migration and reduction in contact inhibition (16). To study the effects of siG12D on EMT, we conducted a series of experiments in which we examined different aspects of this phenomenon. We analyzed the migration characteristics of Panc1 cells following transfection with siG12D or scrambled siRNA, using the scratch and Transwell-migration assays (17, 18). TGFβ was used as a positive control. Migration ability was assessed using a modified Boyden chamber assay, which revealed that siG12D inhibited Panc1 migration by more than 30% (Fig. S4A). The scratch assay revealed that after 24 h, the nontreated and the scrambled siRNA transfected cells migrated and narrowed the gap created by the scratch, whereas the siG12D transfected cells barely migrated (Fig. S4B).
LODER-Derived siRNA Decreases Gene Expression in Vivo.
To confirm the functionality of LODER-driven siRNA in vivo, we tested the ability of a LODER containing siRNA targeting the luciferase gene, siLuc LODER, to reduce luciferase expression in normal and tumor tissues constitutively expressing the luciferase gene. We therefore implanted empty or siLuc LODERs into the livers of transgenic mice expressing the luciferase gene (MUP-Luc) in liver cells (19). In vivo imaging results showed that siLuc LODERs led to a significant decrease in luciferase levels compared with empty LODERs (Fig. S5A). Next, LODERs carrying siLuc or siGFP were implanted into CT26 cell-derived s.c. synograft tumors. Three days after implantation, measurements of luciferase activity in vivo revealed that siLuc released from the LODERs inhibited luciferase expression (Fig. S5B). This decrease in luciferase activity was not correlated to nonspecific effects on tumor growth, as tumor weights were similar within both groups (Fig. S5C). Together these results show that siRNA is released in vivo from the LODER and silences the target gene in vivo, thus demonstrating that the LODER can serve as a delivery platform for active siRNA in both normal and tumor tissues in vivo.
To study the toxicity of the LODER-driven siRNA, we compared the effect of the siG12D LODER (16, 32, and 64 μg siG12D/kg body weight) to that of the empty LODER, sham-operated, or untreated animals. LODERs were implanted into the pancreas of normal mice and rats. Animals of both species, males and females, were tested. In parallel, the same and higher doses of siG12D were intraperitoneally injected into normal mice and rats (16, 160, and 320 μg siG12D/kg body weight). The duration of this study was 2 wk in mice and 8 wk in rats. The results demonstrated that the siG12D LODER had no effect on animal mortality, behavior, or body and liver weight. Hematological and biochemical tests failed to reveal any statistically significant differences between the tested groups. Gross and histopathology analyses revealed that all changes were of minimal severity and typical in untreated mice or rats of the same age and strain. Based on the lack of adverse reaction following siRNA treatment, the maximum dose of 0.32 mg siRNA/kg body weight was considered as not representing an acute toxic risk.
LODER-Driven siG12D Inhibits Tumor Growth in Vivo.
We next assessed the ability of LODERs to inhibit the growth of tumors derived from human pancreatic tumor cell lines in vivo. We used human pancreatic cancer cell lines, Panc1 and Capan1, constitutively expressing the luciferase gene. Both these cell lines bear mutations in the KRAS gene, G12D in Panc1 cells and G12V in Capan1 cells. Immune-deficient mice bearing Capan1-Luc–derived s.c. tumors at an average size of 1 cm3 were divided into four groups: intratumorally implanted with siG12D, siG12V, or empty LODERs, and untreated. In Fig. S6A, the immunohistochemistry (IH) staining results are shown for the proliferation marker CDC47 at 1 mo after LODER implantation. Measurement of the percentage of necrotic area reveals that in both siG12D and siG12V LODER-treated groups, necrotic areas were significantly larger than in the untreated and empty LODER-treated control groups (Fig. S6B). Thus, the siG12D and siG12V LODERs severely hampered the growth of Capan1-Luc–derived tumors. In mice bearing s.c. Panc1-Luc tumors, results revealed that tumors treated with the siG12D LODER had significantly smaller volumes than those treated with the siLuc LODERs (Fig. S6C). Furthermore, histological analysis of LODER-treated Panc1 tumor tissues showed in the siG12D group a case of tumor tissue that was almost totally replaced by an adipose tissue (Fig. S6D). Correlated to these, the survival of mice bearing s.c. Panc1 tumors was significantly increased (P < 0.05) compared with each of the other three groups: untreated, empty LODER, and intraperitoneally (i.p.) injected siG12D (Fig. S6E).
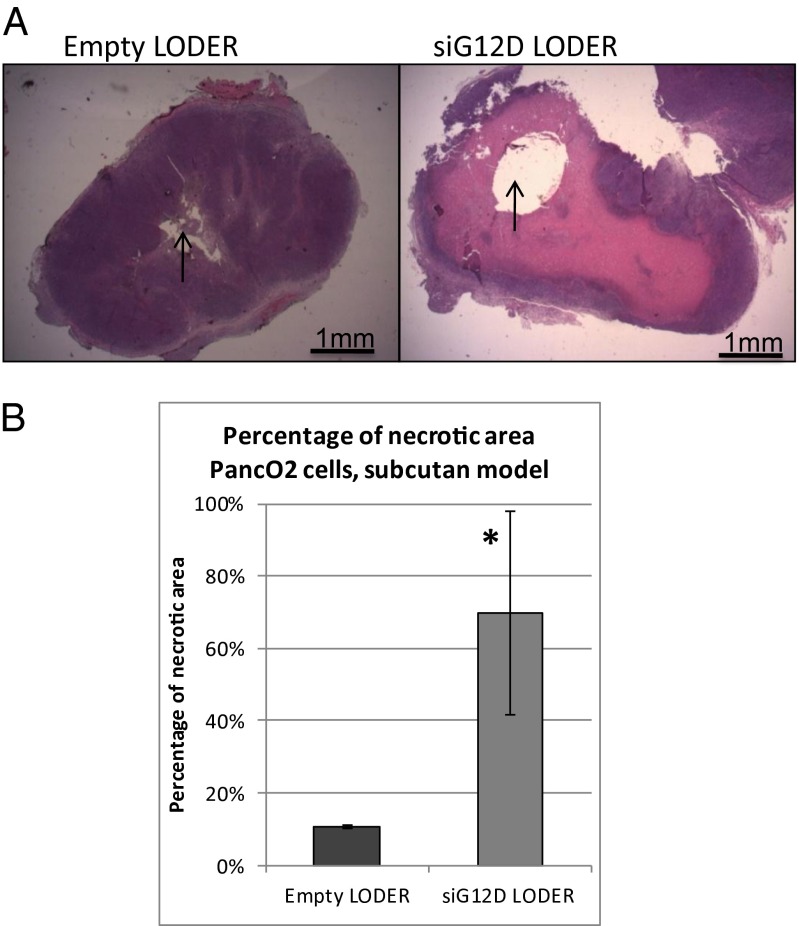
Next, we tested the effects of the siG12D LODER treatment on tumor growth in immune proficient settings. Toward this end, we used in a syngeneic mouse model, the Panc02 mouse pancreatic cell line bearing a G12D mutation. Notably, in vitro, siG12D inhibited Panc02 cell growth in a similar mode as in Panc1 cells (Fig. S7). In vivo, siG12D LODERs implanted in the s.c. Panc02-derived tumors resulted in extensive necrosis 1 wk following implantation (Fig. 3A). Quantification of the necrotic area showed that it exceeds 60% of the total tumor tissue area (Fig. 3B).
Fig. 3.
Silencing of KRAS inhibited s.c. tumor growth in immune-proficient settings. (A and B) Syngeneic s.c. PancO2 tumors were implanted with empty LODERs or LODERs containing siG12D. (A) Representative pictures of H&E immunostaining are shown 1 wk after the implantation. (B) Calculated percentage of necrotic area. The calculation was done using the ImageJ program. *P < 0.05 according to the Student t test.
To determine the long-term potential effect of the siRNA LODERs, we analyzed the remaining siRNA content of LODERs isolated from tumors after implantation. The results revealed that the LODER structure protected the siG12D from degradation for over a 70-d period. Collectively, these results demonstrate that the siG12D LODER effectively inhibited tumor growth and can lead to necrotic destruction of tumor tissue in vivo.
siG12D LODERs Impede Orthotopic Pancreatic Tumor Growth.
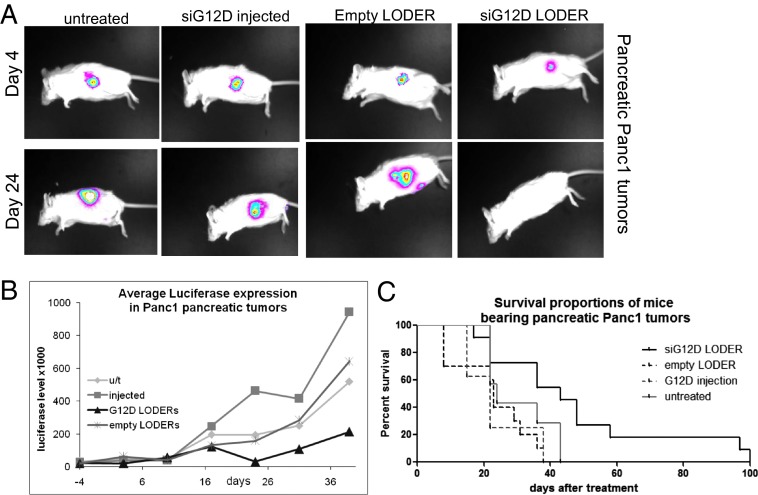
We established an orthotopic pancreatic cancer model in which mice were operated on and Panc1-Luc or Capan1-Luc cells were injected into the tail of the mouse pancreas. Tumor growth was monitored by in vivo luciferase imaging. When tumors were detected, mice were divided into the different treatment groups, keeping a similar average level of luciferase intensity in each group. Mice then underwent laparotomy and two LODERs were stitched to the pancreatic tumor mass. In Panc1-Luc–derived tumors, in vivo luciferase measurements revealed that tumor growth was retarded in the siG12D LODER group throughout the study in comparison with the control groups (Fig. 4 A and B). Importantly, the overall survival of siG12D LODER-treated mice was considerably prolonged compared with all of the different control groups (Fig. 4C). Similar results were obtained with Capan1-Luc cells (Fig. S8).
Fig. 4.
siG12D LODERs inhibit growth of orthotopic pancreatic tumors of human origin. (A and B) Representative mouse pictures (A) and luciferase activity graph (B) in mice bearing Panc1-Luc pancreatic tumors from the different treatment groups: untreated, i.p. injected with siG12D (injected) or implanted with empty or siG12D LODERs. (C) Survival curve in mice bearing Panc1 pancreatic tumors from the different treatment groups: untreated, i.p. injected with siG12D, or implanted with empty or siG12D LODERs.
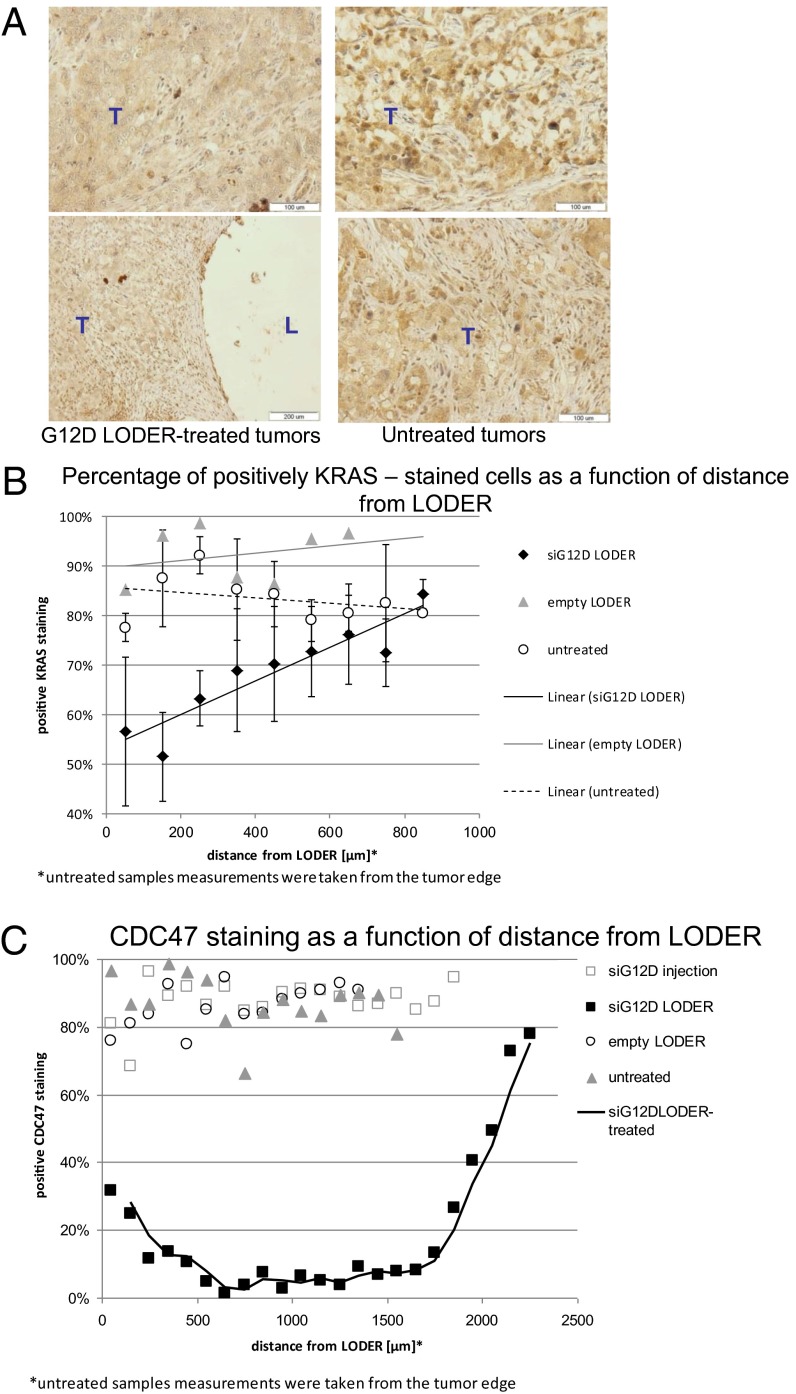
To show the direct effects of siG12D LODER on tumor tissue, we assessed the effect of the siG12D LODER on KRAS protein expression in xenograft tumor tissues. IH with a KRAS-specific antibody was performed on tumor tissue sections obtained from xenograft tumors, 24 d after treatment with: siG12D LODERs, empty LODERs, or untreated. This analysis shows that KRAS staining in siG12D LODER-treated tumors was weaker compared with the empty LODER or untreated tumors (representative tissue sections in Fig. 5A). Image analysis software was used to calculate the percentage of KRAS positive cells vs. nonstained cells, in 0.1 × 0.1 mm2 squares, in various consecutive distances from the LODER implantation sites (representative micrographs shown in Fig. S9A). The percent of KRAS positive cells was found to be in an anticorrelation with the distance from the LODER, whereas in other groups, untreated and empty LODER implanted, the graph remained linear (Fig. 5B and Fig. S9B). Notably, the ratio of KRAS positive stained versus nonstained cells (positive/negative ratio) in siG12D LODER-treated tumors was ∼10-fold lower compared with the two other groups. These data attest to the specific silencing effect of siG12D LODER treatment on KRAS expression.
Fig. 5.
siG12D LODER inhibits KRAS expression and growth of orthotopic pancreatic tumors of human origin. (A) Representative pictures of KRAS immunostaining of Panc1 pancreatic tumor tissues 24 d posttreatment: implanted with siG12D LODERs or untreated. (B) Graph represents percentage of KRAS positively stained cells in squares 0.1 × 0.1 mm2 as a function of distance from LODER or tumor edge, measured in three groups: untreated, empty LODER, and siG12D LODER treated. (C) Graphs represent percentage of CDC47 positively stained versus unstained cells as a function of distance from LODER or tumor edge, measured in four groups: untreated, siG12D injected, empty LODER, and siG12D LODER treated.
Having shown the effects of KRAS silencing on pancreatic tumor cell proliferation, we performed IH staining for CDC47, a marker for dividing cells. Quantification of this staining (Fig. 5C and Fig. S9 C and D) reveals that the siG12D LODER inhibited the growth of tumor cells with increased efficiency in the immediate periphery of the LODER. These results confirm that the observed tumor growth inhibition was due to the silencing of KRAS by the siG12D LODER.
Altogether, our results demonstrate that the LODER-driven siG12D hampered KRAS expression and significantly inhibited the in vivo growth of pancreatic tumors in both s.c. and orthotopic, xenograft, and synograft mouse models.
Discussion
In this study, we present a unique platform technology for siRNA delivery. We show that the mutated KRAS, previously considered as an undruggable oncogene, can be targeted by siRNA in vivo in mouse models of pancreatic cancer. Our strategy of siRNA delivery to cancer cells was through embedding of the siRNA within a miniature biodegradable polymeric matrix that both protects and enables the release of the siRNA drug for an extended duration of time regionally within tumor tissue. We show the efficacy of this drug and this delivery system as being efficient, specific, and safe.
The notion of oncogene addiction lies at the base of the bulk of cancer research performed today (20). KRAS has long been underlined as a beneficial target for cancer therapeutics, because its activation drives many of the tumor-cell–related traits, especially growth and proliferation (21). Specific KRAS targeting in tumors—especially in pancreatic cancer—offers hope for cancer patients by attenuating tumor growth through the deprivation of this oncogene (22).
In conclusion, the siG12D LODER overcomes the current siRNA delivery obstacles, including enzymatic degradation of the drug, renal clearance, and the need for active targeting. Moreover, it improves efficacy by eliminating the need for distal vectors and/or chemical modifications and reduces the required siRNA dose. Importantly, this approach is universal and can be easily applied to target other genes that are pivotal for progression and growth of various solid tumors, using different sets of siRNAs. In addition to being safe and effective, the strategy we propose is applicable, reproducible, cost-effective, and withstands a much lower dose than siRNAs administered systemically.
Materials and Methods
The LODER.
LODER is a biopolymeric cylindrical implant comprising an siRNA drug that is released throughout a period of months into a tumor. Dimensions are optimized for insertion into the pancreatic tumor by a 19-gauge endoscopic ultrasound needle (D ∼ 0.8 mm). The used drug load was from submicrogram levels according to in vivo requirements, up to 375 µg for the optimal dose in clinical use.
For siRNA integrity tests, siRNA was separated on 10% (wt/vol) urea-polyacrylamide gels followed by ethidium bromide staining. Band intensity was quantified using TINA software.
Cell Lines and Cell Culture Conditions.
Panc1 and Capan1 cell lines were obtained from American Type Culture Collection (ATCC). The Panc02 cell line is a gift from Dr. M. Elkin (Sharett Institute, Hadassah-Hebrew University Medical Center, Jerusalem). The three cell lines were cultured in RPMI-1640 medium, supplemented with 10% (vol/vol) heat-inactivated FCS, 2 mM glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin. The cell cultures were maintained in a humidified atmosphere of 5% (vol/vol) CO2 at 37 °C.
Transfections, Stable Transfections, and Gene Silencing Assays.
For establishing stably transfected Panc1-Luc, Panc02-Luc, and Capan1-Luc cell lines, pCDNA3.1 (Invitrogen) plasmid bearing the neomycin gene was cotransfected with the pCAG-Luc plasmid (obtained from E. Zeira, Hebrew University, Jerusalem) in a ratio of 1:5, using TransIT-LT1Mirus transfection reagent, according to the manufacturer's instructions. The selection was performed using G418. Luciferase level was assessed using the Steady-Glo luciferase assay (Promega). siRNA transfections were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cell viability was assessed using 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) (Biological Industries) or the lactate dehydrogenase (LDH) (Roche) kit, according to the manufacturer's instructions.
Synthetic siRNAs.
KRAS-targeting siRNA sequences were described previously (12, 16). Control siRNA sequences were 5-CUUACGCUGAGUACUUCGAdTdT and
5′-UCGAAGUACUCAGCGUAAGdTdT for si-Luc;
5′-GCUGACCCUGAAGUUCAUCdTdT and
5-'GAUGAACUUCAGGGUCAGCdTdT for siGFP;
and 5′-GUUGGAGGCGGUAUGUGAGdTdT and
5′-CUCACAUACCGCCUCCAACdTdT for scrambled control. All siRNAs were synthesized as ready-to-use duplexes by Sigma-Aldrich or by Biospring.
Real-Time PCR.
RNA was isolated using TRIzol (Invitrogen; 15596). cDNA was synthesized using qScript cDNA Synthesis kit (Quanta BioSciences; 95047). The primers used were as follows: for KRAS, forward 5′-GAGGCCTGCTGAAAATGACTG-3′ and reverse 5-ATTACTACTTGCTTCCTGTAGG-3′; for hypoxanthine-guanine phosphoribosyltransferase (HPRT), forward
5′-GGTCCTTTTCACCAGCAAGCT-3′ and reverse
5′-TGACACTGGCAAAACAATGCA-3′; and for human ubiquitin C promoter (hUBC), forward
5′-ATTTGGGTCGCGGTTCTTG-3′ and reverse 5′-TGCCTTGACATTCTCGATGGT-3′.
RACE.
Panc1 cells were transfected with siG12D using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were harvested at 1, 4, and 8 h posttransfection. RNA was extracted from cells using TRIzol Reagent (Ambion) according to the manufacturer's instructions. A total of 2 µg RNA was reverse transcribed using KRAS-specific primer (5′-CTGTTCTAGAAGGCAAATCAC-3′) and M-MLV reverse transcriptase (Promega). The 5′ poly-A tails were added using terminal transferase (NEB) and dATP. PCR amplification of the RACE-specific target was done using the 3′ KRAS-specific primer (5′-CTGTTCTAGAAGGCAAATCAC-3′) and an oligo-dT primer containing 4 bp at the 3′ complementary to the 5′ nick site (5′-TTTTTTTTTTTTTTTTTTGATG-3′).
Western Blot Analysis.
Western blot assays were carried out using routine procedures. Briefly, cells were homogenized in lysis buffer A [0.25 M sucrose, 20 mM Tris pH 7.6, 1.5 mM Mg Cl2, 10% glycerol, 1 mM EDTA, and ‘‘Complete mini’’ protein inhibitor mixture (Roche Diagnostics)], incubated on ice for 10 min and centrifuged at 15,000 × g for 15 min at 4 °C for supernatant collection. Primary Abs are as follows: anti-KRAS (Abcam; ab84573), anti-P-Erk (Sigma; M 8159), anti-P-Akt (Cell Signaling; 4060), and anti–β-actin (ICN/MP Biomedicals; 691001). Secondary Abs are as follows: Dako EnVision system labeled polymer-HRP anti-mouse (Dako; K4001) and anti-rabbit (Dako; K4003). Proteins were visualized by the EZ-ECL chemiluminescence detection kit for HRP (Biological Industries; 20-500-120). Results are expressed as a ratio protein of interest/β-actin to correct for loading for each sample.
Animals.
Female C57B/6 5-wk old mice, nude and SCID/bg female 6-wk-old mice were purchased from Harlan Laboratories. All mice were kept in a specific pathogen-free facility. Mice were handled according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals (23) prepared by the National Academy of Sciences and published by the National Institutes of Health. All experiments were approved by the Animal Care Committee of Hebrew University.
Tumor Models.
The mice were allowed to acclimate to the facility for at least 1 wk before manipulation. Mice had free access to water and chow at all times. All animal procedures were performed under general anesthesia with i.p. administered xylazine 10 mg/g body weight (Chanelle Pharmaceuticals Manufacturing) and ketamine, 450 mg/g body weight (Fort Dodge Animal Health). After surgery, mice were allowed food and water ad libitum.
S.c. tumors.
Tumor xenografts were established by s.c. injection of log-phase growth viable cells, 107 (in 150 µL PBS) in the case of Panc1 cells or 106 (in 100 µL PBS) in the case of PancO2 cells; the cells were injected into the flanks of the mice. When tumors reached an average volume of 80 mm3, mice were divided into equal groups. LODERs were implanted into tumors under anesthesia. The tumor volume was calculated according to the following formula: V = largest diameter × small diameter2/2.
Intrapancreatic orthotopic tumors.
The mice were anesthetized, their abdomens were sterilized with alcohol (70%), and they were positioned laterally. A small, left abdominal flank incision was made, and the pancreas tail with the spleen was carefully exposed under aseptic conditions. The tumor cells (106 cells/30 µL PBS) were injected into the tail of pancreas using a 27-gauge tuberculin syringe. After replacement of the pancreas into the abdominal cavity, the incision was closed in two layers using an absorbable surgical 6-0 vicryl suture for the peritoneum and a 4-0 vicryl suture for the skin. After surgery, mice were inspected daily. Tumor growth was followed by measurement of luciferase level. When the tumors were detected, mice were stratified and divided into treatment groups according to the luciferase levels and treated as noted. For LODER implantation, mice were anesthetized, pancreas was exposed as described, and LODERs were attached to the tumor using a 7-0 vicryl suture. The abdominal cavities were closed as described. Pancreatic tumor growth was followed by luciferase measurement twice a week.
Immunohistochemical Staining.
Immunohistochemistry was done on 4-μm-thick formalin-fixed paraffin-embedded tissue sections by standard procedure. Deparaffinization and rehydration were followed by antigen retrieval using a pressure cooker with citrate buffer (pH 6) for P-ERK and glycine buffer (pH 9) for KRAS and CDC47. Primary antibodies were diluted 1:100 for KRAS (Abnova; PAB14811) and ERK (M8159; Sigma); 1:50 for CDC47 (Biocare Medical CM137b; Pharmatrade). Secondary antibodies were from Dako. Staining was developed with diamonobenzine using a kit from Zymed.
Statistical Analysis.
All data were subjected to statistical analysis using the Excel software package (Microsoft). A two-tailed Student t test was used to determine the difference between the groups. Differences were considered significant at P < 0.05. Data are given as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Ms. Evelyne Zeira for her contribution to establishing the in vivo tumor models, Dr. Daniel Goldenberg for MUP-Luc mice, Prof. Nahum Goldberg for numerous useful comments, and Dr. Eliel Ben-David for CT interpretation.
Footnotes
Conflict of interest statement: E.Z.K., R.G., I.-H.R., E.H., Z.B., A.O., Adva Shemi, and Amotz Shemi are employed by Silenseed. E. Galun, T.G., A.J.D., E.Y., and A.K. are advisers to Silenseed.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314307110/-/DCSupplemental.
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Karamitopoulou E. Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front Oncol. 2012;2:209. doi: 10.3389/fonc.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delpu Y, et al. Genetic and epigenetic alterations in pancreatic carcinogenesis. Curr Genomics. 2011;12(1):15–24. doi: 10.2174/138920211794520132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belda-Iniesta C, et al. Molecular biology of pancreatic cancer. Clin Transl Oncol. 2008;10(9):530–537. doi: 10.1007/s12094-008-0247-6. [DOI] [PubMed] [Google Scholar]
- 6.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann G, Beaty R, Hruban RH, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary Pancreat Surg. 2007;14(3):224–232. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008;28(8):2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C, et al. Characterization of the mutations of the K-ras, p53, p16, and SMAD4 genes in 15 human pancreatic cancer cell lines. Oncol Rep. 2001;8(1):89–92. doi: 10.3892/or.8.1.89. [DOI] [PubMed] [Google Scholar]
- 10.Villanueva A, et al. Diagnostic utility of K-ras mutations in fine-needle aspirates of pancreatic masses. Gastroenterology. 1996;110(5):1587–1594. doi: 10.1053/gast.1996.v110.pm8613066. [DOI] [PubMed] [Google Scholar]
- 11.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Réjiba S, Wack S, Aprahamian M, Hajri A. K-ras oncogene silencing strategy reduces tumor growth and enhances gemcitabine chemotherapy efficacy for pancreatic cancer treatment. Cancer Sci. 2007;98(7):1128–1136. doi: 10.1111/j.1349-7006.2007.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leucuta SE. Drug delivery systems with modified release for systemic and biophase bioavailability. Curr Clin Pharmacol. 2012;7(4):282–317. doi: 10.2174/157488412803305786. [DOI] [PubMed] [Google Scholar]
- 14.Mohtaram NK, Montgomery A, Willerth SM. Biomaterial-based drug delivery systems for the controlled release of neurotrophic factors. Biomed Mater. 2013;8(2):022001. doi: 10.1088/1748-6041/8/2/022001. [DOI] [PubMed] [Google Scholar]
- 15.Hudson D, Margaritis A. Biopolymer nanoparticle production for controlled release of biopharmaceuticals. Crit Rev Biotechnol. 2013 doi: 10.3109/07388551.2012.743503. [DOI] [PubMed] [Google Scholar]
- 16.Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: Justification for K-ras-directed therapy. Mol Cancer Res. 2005;3(7):413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 17.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 18.Kramer N, et al. In vitro cell migration and invasion assays. Mutat Res. 2013;752(1):10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Klopstock N, Levy C, Olam D, Galun E, Goldenberg D. Testing transgenic regulatory elements through live mouse imaging. FEBS Lett. 2007;581(21):3986–3990. doi: 10.1016/j.febslet.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Sharma SV, Settleman J. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21(24):3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 21.Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochim Biophys Acta. 2005;1756(2):127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Zimmermann G, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497(7451):638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 23. Committee on Care and Use of Laboratory Animals (1985) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.