Significance
In the past few years, findings have been controversial in regard to whether human induced pluripotent stem cells (hiPSCs) are distinct from human embryonic stem cells (hESCs) in their molecular signatures and differentiation properties. In this study, hiPSCs and hESCs have overlapping variations in molecular signatures such as RNA expression and DNA methylation. However, some hiPSC clones retained a significant number of undifferentiated cells even after neural differentiation culture and formed teratoma when transplanted into mouse brains. These differentiation-defective hiPSC clones were marked by higher expression levels of several genes, including those expressed from long terminal repeats of specific human endogenous retroviruses. They need to be identified and eliminated prior to applications in regenerative medicine.
Abstract
We examined the gene expression and DNA methylation of 49 human induced pluripotent stem cells (hiPSCs) and 10 human embryonic stem cells and found overlapped variations in gene expression and DNA methylation in the two types of human pluripotent stem cell lines. Comparisons of the in vitro neural differentiation of 40 hiPSCs and 10 human embryonic stem cells showed that seven hiPSC clones retained a significant number of undifferentiated cells even after neural differentiation culture and formed teratoma when transplanted into mouse brains. These differentiation-defective hiPSC clones were marked by higher expression levels of several genes, including those expressed from long terminal repeats of specific human endogenous retroviruses. These data demonstrated a subset of hiPSC lines that have aberrant gene expression and defective potential in neural differentiation, which need to be identified and eliminated before applications in regenerative medicine.
Human pluripotent stem cells possess a robust potential for proliferation and provide useful sources of cells for regenerative medicine and drug discovery. Two types of human pluripotent stem cells have been generated: human embryonic stem cells (hESCs) derived from blastocysts (1) and induced pluripotent stem cells (hiPSCs), which are generated from somatic cells by factor-mediated reprogramming (2, 3).
In the past few years, findings have been controversial in regard to whether hESCs and hiPSCs are distinct cell types. Some reports have argued that they could not be clearly distinguished (4–6), whereas others have reported that they have differences in their gene expression (7–10), DNA methylation (10–13), and capacity for differentiation (14). In the latter papers, relatively small numbers of cell lines were generally compared. In addition, most comparisons used pluripotent cell lines from various laboratories, so the observed differences may be attributable to laboratory-specific variations owing to technical differences (15).
To overcome these problems, we compared the mRNA and microRNA (miRNA) expression and DNA methylation between 10 hESCs and 49 hiPSCs that had been cultured under the same conditions. Furthermore, we compared the in vitro directed neural differentiation of these pluripotent stem cells.
Results
Overlapped Variations of mRNA Expression and DNA Methylation in hiPSCs and hESCs.
We analyzed a total of 49 hiPSCs derived from four types of somatic cells, including human dermal fibroblasts (HDFs), dental-pulp stem cells (DP), cord blood cells (CB), and peripheral blood mononuclear cells (PBMN), generated using three gene delivery methods, including those using retroviruses, nonintegration episomal plasmids, and Sendai viruses (Table 1 and Dataset S1). Most clones were generated in our own laboratory, except for three clones that were established in another laboratory (16). Before the analyses of gene/miRNA expression (Fig. 1 A and B) and DNA methylation (Fig. 1C) we cultured these hiPSCs, as well as 10 hESCs, under the same culture conditions for at least three passages. In addition, we analyzed the original somatic cells, two human embryonic carcinoma cell (hECC) lines (NTera2 cloneD1 and 2102Ep 4D3), and three cancer cell lines (HepG2, MCF7, and Jurkat).
Table 1.
Summary of the iPSC clones used in this study
| Method used to generate clones |
||||
| Origin | Retrovirus | Episomal plasmid | Sendaivirus | Total |
| HDFs | 22 (5, 0) | 3 (0, 0) | 0 (0, 0) | 25 (5, 0) |
| DP | 1 (0, 0) | 2 (1, 1) | 0 (0, 0) | 3 (1, 1) |
| CB | 3 (1, 2) | 4 (0, 0) | 5 (0, 0) | 12 (1, 2) |
| PBMN | 0 (0, 0) | 5 (0, 1) | 4 (0, 0) | 9 (0, 1) |
| Total | 26 (6, 2) | 14 (1, 2) | 9 (0, 0) | 49 (7, 4) |
Total clone numbers with type-1 defective clone numbers and type-2 defective clone numbers in parentheses are shown.
Fig. 1.
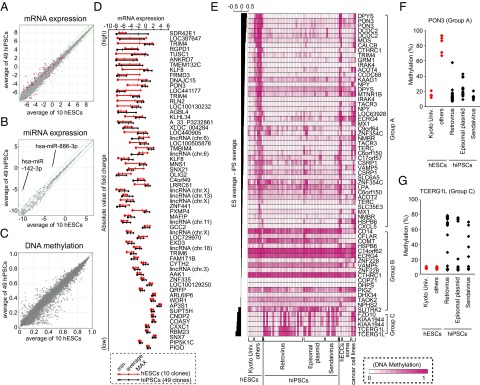
hiPSCs and hESCs have overlapped variations in RNA expression and DNA methylation. Scatter plots of mRNA expression (A), miRNA expression (B), and DNA methylation (C) data comparing the average of 49 hiPSC lines (y axis) to the average of 10 hESC lines (x axis). The RNA expression value is shown on a log 2 scale. Green lines indicate twofold differences in the RNA expression levels between the clones. Differentially expressed probes (t test, FDR < 0.05) are shown in magenta. (D) The variations in the mRNA expression levels of 61 differentially expressed probes in hESCs (red) and hiPSCs (black) are shown. Probes are arranged in order of the absolute value of the FC between hESCs and hiPSCs. (E) The DNA methylation profiles for CpGs contained in reported hES-hiPS DMRs and overlapping with our platform. Probes are arranged in order of the differences between the average DNA methylation level of hESCs and that of hiPSCs. The heat map represents the DNA methylation levels from completely methylated (1, magenta) to unmethylated (0, white) samples. The methylation status of the upstream region of PON3 (F) and TCERG1L (G) was examined by pyrosequencing.
The mRNA microarray analyses (Fig. 1A) identified 61 probes with significant differences in expression between hESCs and hiPSCs [t test, false discovery rate (FDR) <0.05]. Each of the 61 probes showed variable expression among both the hESCs and hiPSCs, and the distributions of the expression levels in the two groups overlapped (Fig. 1D). Of note, hESCs established at Kyoto University (Kyoto hESCs) were more similar to hiPSCs than to the remaining hESCs (other hESCs) in their expression of 15 probes that were differentially expressed between hESCs and hiPSCs [FDR <0.05 and fold change (FC) >3] (Fig. S1A). In contrast, hierarchical clustering using all probes showed no clear-cut separation among Kyoto hESCs, other ESCs, and iPSCs, indicating that the similarities between Kyoto ESCs and iPSCs are confined to a small set of genes (Fig. S1B). In addition, the miRNA array analyses (Fig. 1B) did not find any significant differences between hESCs and hiPSCs (t test, FDR <0.05). The expressions of hsa-miR-886-3p and hsa-miR-142-3p tended to be higher in hiPSCs, but the expression levels of these miRNAs showed overlapped variations among hiPSCs and hESCs (Fig. S1A).
We next compared the global DNA methylation status between hiPSCs and hESCs by the Illumina Infinium Human Methylation27 BeadChip assay. Among 27,445 CpG dinucleotides examined, we did not identify significantly differentially methylated CpG regions (CG-DMR) between the hESCs and hiPSCs (Mann–Whitney U test, FDR <0.05) (Fig. 1C).
We then validated the CG-DMRs reported in previous studies. Three studies identified a total of 205 regions as CG-DMRs, including 130, 71, and 4 regions identified by comparing five hiPSCs and two hESCs (13), three versus three (12), and nine versus three (10) cell lines, respectively. Of the 205 regions, 46 regions containing 66 CpG dinucleotides were covered by the Infinium platform used in our study (Table S1A). Based on the methylation levels in our hiPSCs and hESCs, these CpGs were clustered into three groups (Fig. 1E). Two-thirds of these CpGs belonged to group A: They tended to be highly methylated in hESCs, ECCs, and cancer cell lines and to be hypomethylated in hiPSCs, as well as somatic cells. However, they were also hypomethylated in Kyoto ESCs (17). The methylation status of the upstream region of the paraoxonase 3 (PON3), a representative example of CpGs in group A, was confirmed by pyrosequencing (Fig. 1F). Thus, the CpG methylation status in group A may distinguish some, but not all, hESCs from hiPSCs. Seventeen CpGs belonged to group B, which showed similar methylation levels in both hESCs and hiPSCs. Five CpGs, representing three genes, belonged to group C and showed higher methylation levels in some, but not all, hiPSCs compared with hESCs. The remaining hiPSCs showed low methylation levels, comparable to those in hESCs. We confirmed the methylation status of a representative example of CpGs in group C, the upstream region of the transcription elongation regulator 1-like (TCERG1L), by pyrosequencing; the methylation levels were low in 21 of the 49 hiPSCs (Fig. 1G). Therefore, the CpGs in group C may distinguish some, but not all, iPSCs from hESCs.
A previous report (12) showed that many CG-DMRs were located in CpG shores, rather than CpG islands. Because only a few (9 out of 71) of their CG-DMRs were covered by our Infinium platform, we analyzed five CG-DMRs in CpG shores by pyrosequencing, including ataxin-2 binding protein 1 (A2BP1), insulin-like growth factor 1 receptor (IGF1R), POU domain, class 3, transcription factor 4 (POU3F4), protein tyrosine phosphatase, receptor type, T (PTPRT), and zinc finger protein 184 (ZNF184) (Fig. S1C). We detected significant differences in the averaged DNA methylation levels between hESCs and hiPSCs in four out of five genes. However, variations of DNA methylation levels in hESCs and hiPSCs are overlapped. They may distinguish some, but not all, hiPSCs from hESCs.
A Subset of hiPSC Clones Retain Undifferentiated Cells After Neural Differentiation.
To examine whether hESCs and hiPSCs have comparable differentiation potential, we performed in vitro directed differentiation into neural stem and progenitor cells using the modified serum-free floating culture of embryoid body-like aggregates (SFEBq) method (Fig. 2A) (18). We initially performed the neural induction of two hESCs and 21 hiPSCs. Fourteen days after induction, the differentiation efficiency was evaluated based on the expression of an early neural marker, polysialylated neural cell adhesion molecule (PSA-NCAM). We found that all hESCs and hiPSCs differentiated into PSA-NCAM+ cells with more than 80% efficiency (Fig. 2B). We also quantified the expression levels of the early neural marker paired box 6 (PAX6) and the late neural marker microtubule-associated protein 2 (MAP2) in neurospheres by quantitative RT-PCR (qRT-PCR) (Fig. S2A). All of the examined hES/iPSCs expressed PAX6 at >100-fold higher levels and MAP2 at >20-fold higher levels in comparison with undifferentiated H9 hESC. However, in some hiPSCs, we noticed slightly lower differentiation efficiency than in the remaining hiPSCs and hESCs (Fig. 2B). This lower efficiency in neural differentiation was inversely correlated with a higher proportion of POU class 5 homeobox 1 (POU5F1, also known as OCT3/4)+ and TRA1-60+ undifferentiated cells (Fig. 2C). We also detected residual undifferentiated cells after a different neural differentiation protocol using adhesion culture (19) (Fig. S2B).
Fig. 2.
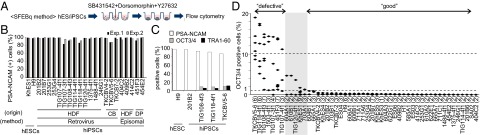
A differentiation-defective phenotype in a subset of hiPSC clones. (A) A schematic diagram of the SFEBq method used for neural differentiation. (B) Neural induction was performed for 2 hESC and 21 hiPSC lines which were established from various origins by retroviral or episomal vector methods. On day 14, we examined the proportion of PSA-NCAM–expressing cells by flow cytometry (n = 2). (C) The proportions of PSA-NCAM- (white), OCT3/4- (gray), and TRA1-60-positive (black) cells 14 d after neural differentiation. (D) The proportions of OCT3/4-positive cells on day 14 after neural differentiation are ranked in order of their maximum value. The numbers in parentheses show the number of trials.
We then increased the number of clones and examined the proportions of OCT3/4+ undifferentiated cells after neural induction from 10 hESCs and 40 hiPSCs. The 50 clones were ranked according to their proportions of OCT3/4+ cells on day 14 (Dataset S1 and Fig. 2D). The proportions of OCT3/4+ cells varied from 0 to ∼20%. Thirty-eight clones, including nine hESCs and 29 hiPSCs, showed less than 1% OCT3/4+ cells in all experiments. We designated these clones as “good” clones. However, seven hiPSCs contained more than 10% OCT3/4+ cells after neural differentiation in at least one experiment. We designated these clones as “differentiation-defective” clones. Clones that were not good or defective were categorized as “intermediate.”
Activation of Specific LTR7 Elements in Differentiation-Defective Clones.
To identify molecular signatures that can predict differentiation-defective clones, we compared the global gene expression patterns of 38 good clones and seven differentiation-defective clones under the culture conditions used for the undifferentiated state. We identified 19 probes (13 putative transcripts) that showed greater than fivefold differences in expression, with an FDR <0.05, shown by magenta dots in Fig. 3A and listed in Table S1B.
Fig. 3.
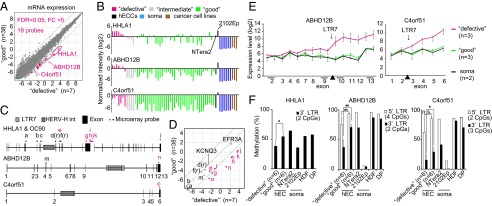
Activation of specific endogenous retroviral LTR7s in defective clones. (A) A scatter plot of the mRNA expression data comparing the average of 38 good clones (y axis) to the average of seven defective clones (x axis). Green lines indicate fivefold differences in expression. A total of 19 differentially expressed probes are colored magenta. (B) The expression levels of LTR7-related genes (HHLA1, ABHD12B, and C4orf51) were examined by microarray. (C) A schematic diagram of three LTR7-related genes. HHLA1 and OC90 are neighboring genes. Dots indicate microarray probes. Magenta dots show probes that are located after LTR7 regions, which were up-regulated in defective clones. (D) A scatter plot of array probes that recognized LTR7-related genes and two other genes [EFR3 homolog A (S. cerevisiae, EFR3A) and potassium voltage-gated channel, KQT-like subfamily, member 3 (KCNQ3)], which are genes neighboring HHLA1 and OC90, respectively. (E) The exon array of the ABHD12B and C4orf51. The average levels of the normalized exon expression are shown. (F) The DNA methylation status of LTR7 and its neighboring regions of HHLA1, ABHD12B, and C4orf51 was examined by pyrosequencing. (n.s., not significant; *P < 0.05, **P < 0.01, Mann–Whitney U test).
Of the 19 probes identified, five probes recognized HHLA1 (human endogenous retrovirus-H LTR-associating 1). Previous reports have shown that HHLA1 is regulated by a long terminal repeat (LTR) of a human endogenous retrovirus-H (HERV-H) (20). The LTR in HHLA1 is classified as LTR7. Moreover, among the genes recognized by the 19 probes, we found that at least two others, abhydrolase domain containing 12B (ABHD12B) and chromosome 4 open reading frame 51 (C4orf51), also contained LTR7 sequences in their gene bodies. According to a microarray analysis, we confirmed that these three LTR7-containing genes were up-regulated in the differentiation-defective hiPSCs, as well as the nullipotent hECC line 2102Ep 4D3 (21), but they were expressed at lower levels in the good hiPSCs, hESCs, and pluripotent hECC line NTera2 cloneD1. They were almost not expressed in the original somatic cells (Fig. 3B).
The Agilent Technologies microarray platform has 12 probes, including two reverse probes [d(r), f(r)], for HHLA1 and its neighboring gene, otoconin 90 (OC90), which is reported to make a fusion transcript with HHLA1 (20) (Fig. 3C). Among them, seven probes located downstream of LTR7 showed higher expression levels in differentiation-defective clones than in good clones (Fig. 3 C and D). Similarly, there are two probes for ABHD12B, designed for exons 4 and 13 (Fig. 3C). Only the exon 13 probe, located downstream of LTR7, showed a higher expression in differentiation-defective clones than in good clones (Fig. 3D). We also performed an exon array (Affymetrix) of ABHD12B and C4orf51 and found that exons downstream of LTR7 were preferentially up-regulated in differentiation-defective hiPSC clones (Fig. 3E). We also found that the methylation status of LTR7s in these three genes were lower in differentiation-defective hiPSC clones than in good clones (Fig. 3F). These results indicate that the three genes are transcribed from activated LTR7.
DNA Hypomethylation Exists in Some, but Not All, LTR7s in Differentiation-Defective Clones.
According to the Repeatmasker software program, there are 3,523 LTR7 elements in the human genome. To extract microarray probes that are potentially affected by LTR7s, we first selected genes containing LTR7s in their gene bodies or regions 2 kb upstream from their transcription start sites. We then retrieved the microarray probes located between each LTR7 and the 3′ end of the corresponding gene body. As a result, we selected 763 probes as LTR7-related probes (Fig. S3A and Table S1C). We found that most of these probes showed comparable expression levels in good and defective lines (Fig. S3B), with the exception of some probes, such as those corresponding to arrestin, beta 1 (ARRB1), fatty acid amide hydrolase 2 (FAAH2), and TBC1 domain family, member 23 (TBC1D23), that were differentially expressed between good and defective clones (FDR <0.05 and FC >2) and showed slightly higher expression in defective lines.
We then checked the DNA methylation status of the LTR7 regions in these three genes and three other genes DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B), ATP-binding cassette, sub-family A (ABC1), member 1 (ABCA1), and amyloid beta (A4) precursor protein (APP)] whose expression levels were not significantly different between the good and defective clones. By pyrosequencing and clonal bisulfite sequencing, we found that the LTR7 regions in four genes (ARRB1, FAAH2, TBC1D23, and APP) were hypomethylated in defective clones compared with good clones. In contrast, the LTR7 regions in two genes (DNMT3B and ABCA1) did not show such hypomethylation (Fig. S3C). Therefore, the activation of LTR7 is not confined to HHLA1, ABHD12B, and C4orf51; DNA hypomethylation exists in some, but not all, LTR7s in defective hiPSCs.
Differentiation-Defective hiPSC Clones Form Teratomas in Mouse Brains.
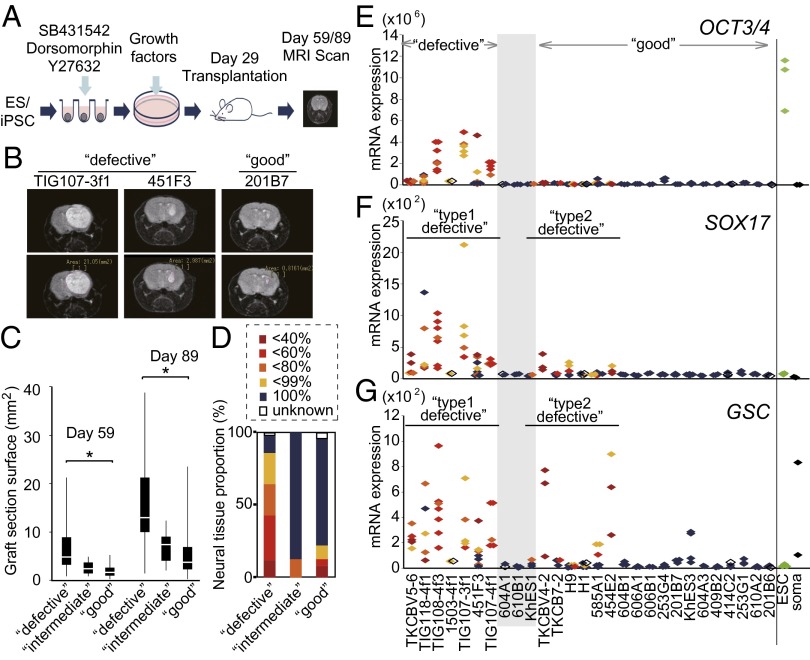
To further evaluate the defective hiPSCs, we induced their differentiation into dopaminergic neurons, which were then transplanted into the striata of nonobese diabetic/severe combined immune-deficient (NOD/SCID) mouse brains (Fig. 4A). Thirty and 60 d after transplantation, we obtained T2-weighted images of the mouse brains with an MRI scanner to observe the graft sizes at the transplanted sites (Fig. 4B). The quantification of the MRI images showed that defective hiPSC clones resulted in significantly larger graft sizes than good clones (Fig. 4C). Notably, some mice that had received defective clones died or developed symptoms that required euthanasia before day 60 (Table S2). Therefore, we could not obtain the graft size data on day 60 in these mice.
Fig. 4.
Transplantation of neural cells derived from hiPSCs and hESCs into mouse brains. (A) A schematic diagram of the SFEBq method used for dopaminergic neural differentiation. On day 29, the cells were transplanted into NOD/SCID mouse brains. (B) Magnetic resonance images of coronal sections of the grafted brains. The section surface of grafted cells indicated as the white shadow in right brain was measured as described in the lower panels. (C) A box-and-whisker plot of the surface sizes of graft sections 30 and 60 d after transplantation. The median, quartile, and range are shown. *P < 0.05 (t test). (D) The proportion of each kind of graft. Grafts were categorized according to their components as determined in H&E sections and were classified by the proportion of neural tissues by a microscopic observation. The expression levels of the undifferentiated cell marker, OCT3/4 (E), the endoderm marker, SOX17 (F), and the mesoderm and endoderm marker, GSC (G), in pretransplantation cultures, undifferentiated hESC lines, and somatic cells (HDF and DP) were examined by qRT-PCR. The colors of the dots were identical to the proportion of neural tissues in D.
To identify the composition of the surviving grafts, we performed a histological analysis of the brains of animals that died or that became moribund after transplantation. The remaining healthy mice were euthanized 14–41 wk after transplantation. Sections were stained with H&E. Thirty-six of the 42 grafts (85.7%) from defective clones contained nonneural lineage tissues, such as intestine-like epithelial cells, cartilage, or mesenchymal cells (Fig. 4D and Fig. S4A). In contrast, grafts from good clones largely consisted of neural tissues. Immunostaining confirmed grafts were positive for human neural cell adhesion molecule (NCAM) (Fig. S4B). A qRT-PCR analysis of pretransplanted cells from defective clones revealed higher expression levels of OCT3/4, suggesting that some undifferentiated cells still remained even after 29 d of neural induction (Fig. 4E). We then depleted the TRA-1-60+ cells on day 22 during neural induction and transplanted cells on day 29. The TRA-1-60–depleted cells from defective clones resulted in significantly smaller grafts that did not contain nonneural tissues (Fig. S4C).
We also observed that 14 out of the 63 (22.2%) grafts from good clones, including those from hESCs, contained a nonneural component in the graft tissue after transplantation (Fig. 4D and Table S2), although these clones did not show high expression levels of OCT3/4 in the pretransplantation samples (Fig. 4E). We referred to these clones as “type-2 defective” clones, which were distinct from “type-1 defective” clones that contained OCT3/4+ undifferentiated cells in the pretransplantation samples. We observed higher expression levels of SRY (sex determining region Y)-box 17 (SOX17, an endoderm marker) and goosecoid homeobox (GSC, an endoderm and mesoderm marker) in the pretransplantation samples of type-2 defective clones (Fig. 4 F and G), demonstrating the presence of other lineages in these pretransplantation samples.
Discussion
We identified two types of defective pluripotent stem cell lines in this study. The first type consisted of hiPSCs that retained a substantial number of undifferentiated cells after in vitro directed neural differentiation. Seven out of the 40 iPSCs (17.5%) examined in this study fell into this category. In contrast, we did not observe such defects in any of the 10 hESCs. More clones should be analyzed to confirm that hESCs are free from this deficiency. Nevertheless, it is likely that type-1 defectiveness is more common in hiPSCs than in hESCs. The type-1 defective hiPSCs are accompanied by an aberrant epigenetic status. Among the 13 putative transcripts that were highly expressed in these defective clones, at least three were expressed from the LTR of endogenous retroviruses. Normally, these LTRs are silenced by various epigenetic modifications, including DNA methylation (22–24). In type-1 defective iPSC clones, the LTR locus in the three genes showed lower DNA methylation levels than in good clones and original somatic cells. Notably, the same regions were hypomethylated in the nullipotent hECC line, 2102Ep 4D3, suggesting that the loss of DNA methylation in these LTR loci is correlated with the lower ability to differentiate. At present, the biological significance and relationship between activation of specific LTRs and the defective phenotype is unclear. Recent reports showed that endogenous retrovirus may play roles in the establishment and maintenance of transcription network in pluripotent stem cells (25, 26). Furthermore, updated annotations revealed that one of the differentially expressed probes in type-1 defective hiPSCs (A_19_P00325604) encoded large intergenic noncoding RNA regulator of reprogramming (Linc-ROR), which contained LTR7 in its 5′ region. Linc-ROR is reported to have multiple roles in the induction and maintenance of pluripotency (27, 28). Future studies should be undertaken to clarify why these epigenetic abnormalities occur and how they are related to the defective differentiation.
Kim et al. (29) showed that there is an inverse correlation between the hsa-mir-371–373 expression and the efficiency of neural differentiation. They also showed that Kruppel-like factor 4 (gut) (KLF4) may induce the expression of hsa-mir-371–373. In our analyses, the hsa-mir-371–373 cluster was highly expressed in all of the seven type-1 defective hiPSC clones (Fig. S5A). However, the cluster was also highly expressed in many good clones. KLF4 was highly expressed in some defective clones (Fig. S5B), and four out of six retroviral defective clones failed to silence KLF4 retroviral transgenes (Fig. S5C). There was no correlation between the OCT3/4 transgene expression and type-1 defectiveness (Fig. S5D). Taken together, these findings indicate that high expression levels of the hsa-mir-371–373 cluster, KLF4, and transgenes cannot function as absolute markers for type-1 defectiveness.
We previously reported that the origin of mouse iPSCs was a major determinant of defectiveness in directed neural differentiation; mouse iPSCs from adult tail tip fibroblasts showed the highest incidence of resistance to differentiation (30). In the present study using human iPSCs, five out of seven type-1 defective clones were derived from fibroblasts of donors of various ages, and six out of the seven clones were generated using retroviruses (Table 1). This may suggest that type-1 defectiveness is associated with fibroblast origin and retroviral induction. However, in this study, most of the fibroblast-derived iPSCs were generated by retroviruses, and most of the nonfibroblast iPSCs were generated by nonretroviral methods. Future studies will need to be undertaken to determine whether the origin or the generation method (or both) has a significant impact on the frequency of type-1 differentiation-defective iPSCs.
The second type of defective group includes hiPSCs and hESCs that contained differentiated cells of nonneural lineages after in vitro directed differentiation into dopaminergic neurons. We have previously shown that the optimal conditions for hepatic differentiation are different for each clone (16). By optimizing the protocols, it may be possible to induce complete neural differentiation to avoid type-2 defective clones. Alternatively, purification of neural cells using a cell sorter may work to avert type-2 defective clones.
Several studies have reported sets of genes whose DNA methylation status is different between hiPSCs and hESCs. We validated these CG-DMRs and found that many of them can distinguish some hESCs from hiPSCs (group A in Fig. 1E). They are highly methylated in some hESCs, but not in hiPSCs or original somatic cells. Thus, these CG-DMRs may represent epigenetic memories of somatic cells in iPSCs. However, we found a set of hESCs that showed low methylation status of these CG-DMRs, which were comparable to hiPSCs. We also found another set of the reported CG-DMRs that showed high methylation status in some hiPSCs but not in original somatic cells or hESCs (group C in Fig. 1E). These likely represent aberrant methylation associated with reprogramming. However, we also found many hiPSCs showed normal methylation patterns of these CG-DMRs. A more recent study identified nine genes that can segregate hiPSCs from hESCs in DNA methylation and gene expression (31). However, we did not observe such a clear distinction in gene expression of these genes between our hiPSCs and hESCs (Fig. S6). Two of these genes, TCERG1L and FAM19A5, may distinguish some, but not all, hiPSCs from hESCs.
In our analyses, 35 hiPSCs had records of the donor’s genetic background; 14 were derived from Caucasians and 21 were from Japanese subjects (Dataset S1). Thus, the similarity of some signatures between the Kyoto hESCs and our hiPSCs cannot be attributed to the racial or ethnic backgrounds of the donors. Another possible cause of the differences is the method used to establish the hESCs and the subsequent culture conditions. The Kyoto hESCs were generated on feeders consisting of a 1:1 mixture of mouse embryonic fibroblasts and SL10 cells (17, 32), which were subcloned from STO cells. Most of our hiPSCs were established on SNL feeders, which were also derived from STO cells. A recent report showed that the feeders have profound effects on established hiPSCs (33). To confirm the importance of the culture conditions, more studies comparing hESC/hiPSCs established under different conditions will be needed.
In conclusion, our results revealed that a subset of hiPSCs is defective in neural differentiation and marked with activation of endogenous retroviruses. We also confirmed that some hiPSCs are different from hESCs in molecular signatures, including CG-DMRs, which has been previously reported. It remains to be determined whether these molecular signatures specific for some hiPSCs have functional consequences.
Materials and Methods
Gene expression profiling was carried out using the SurePrint G3 human GE microarray (Agilent Technologies). Most of the data were analyzed using the GeneSpring GX 11.5.1 software program (Agilent Technologies). Neural induction was performed as described previously (18). Detailed descriptions of methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Takashi Aoi, Yoshinori Yoshida, Masato Nakagawa, and other members of the S.Y. research group for their valuable scientific discussions; Drs. Takuya Yamamoto, Toshiki Taya, and Tomoaki Tsubota for advice about bioinformatics; Dr. Hirofumi Suemori for hES cell lines; Drs. Koji Eto and Naoya Takayama for TKCB hiPS cell lines; Akiko Otsuka for RNA and genomic DNA samples; Dr. Takafumi Kimura and Takaomi Futakami for HLA analysis; and Aki Sasaki, Tomomi Ito, Midori Yokura, and Ayumi Ichikawa for technical assistance. We also thank Naoki Nagata, Yoko Miyake, Rie Kato, Eri Minamitani, Sayaka Takeshima, and Ryoko Fujiwara for their valuable administrative support. This work was supported in part by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science; from the Ministry of Education, Culture, Sports, Science, and Technology; by a grant from the Leading Project of the Ministry of Education, Culture, Sports, Science, and Technology; and by a grant from the Funding Program for World-Leading Innovative Research and Development on Science and Technology (First Program) of the Japanese Society for the Promotion of Science. M.K.-A. and M.O. were Research Fellows of the Japanese Society for the Promotion of Science.
Footnotes
Conflict of interest statement: S.Y. is a member without salary of the scientific advisory boards of iPierian, iPS Academia Japan, Megakaryon Corporation, and HEALIOS K. K. Japan.
Data deposition: Gene expression, miRNA expression, DNA methylation, and exon array data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE49053).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319061110/-/DCSupplemental.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Bock C, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7(2):258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7(2):249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetto MC, et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4(9):e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh Z, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE. 2010;5(2):e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohi Y, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13(5):541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27(4):353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamanaka S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell. 2012;10(6):678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Kajiwara M, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(31):12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suemori H, et al. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem Biophys Res Commun. 2006;345(3):926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 18.Morizane A, Doi D, Kikuchi T, Nishimura K, Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J Neurosci Res. 2011;89(2):117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- 19.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalski PE, Freeman JD, Mager DL. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics. 1999;57(3):371–379. doi: 10.1006/geno.1999.5787. [DOI] [PubMed] [Google Scholar]
- 21.Bahrami AR, Matin MM, Andrews PW. The CDK inhibitor p27 enhances neural differentiation in pluripotent NTERA2 human EC cells, but does not permit differentiation of 2102Ep nullipotent human EC cells. Mech Dev. 2005;122(9):1034–1042. doi: 10.1016/j.mod.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411(2):273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285(27):21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, et al. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell. 2011;8(6):695–706. doi: 10.1016/j.stem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27(8):743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz S, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(40):16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawase E, et al. Strain difference in establishment of mouse embryonic stem (ES) cell lines. Int J Dev Biol. 1994;38(2):385–390. [PubMed] [Google Scholar]
- 33.Tomoda K, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11(1):91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.