Abstract
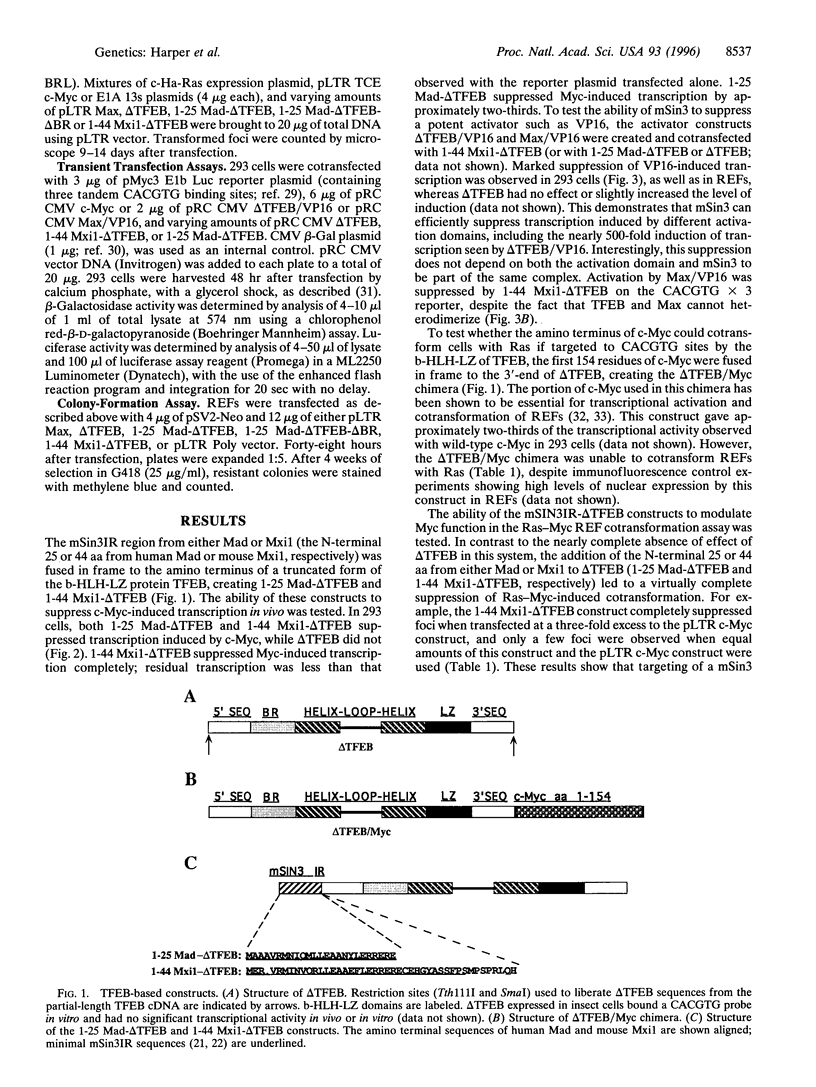
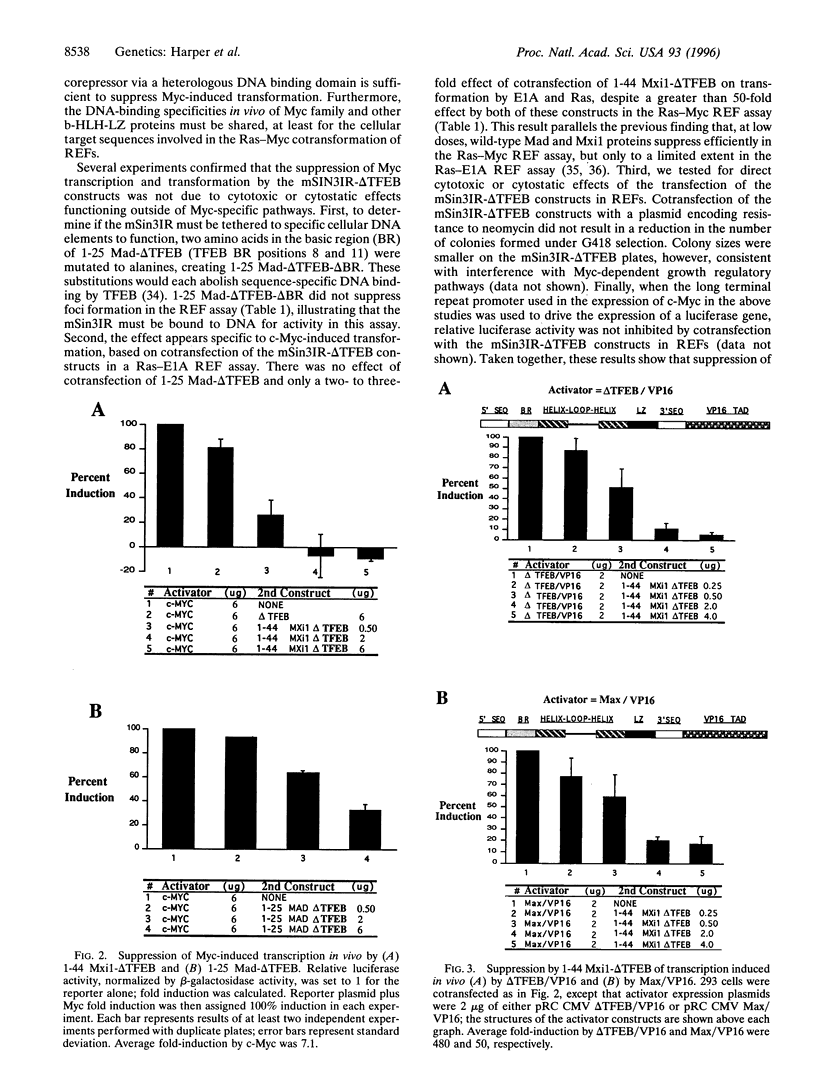
Many basic-helix-loop-helix-leucine zipper (b-HLH-LZ) proteins, including the Myc family and non-Myc family, bind a common DNA sequence CACGTG, yet have quite different biological actions. Myc binds this sequence as a heterodimer with Max in the activation of both transcription and transformation. The Myc family members Mad and Mxi1 are known to suppress Myc-induced transcription and transformation and to dimerize with Max to form ternary complexes with the mammalian Sin3 transcriptional corepressor (mSin3). The b-HLH-LZ domain of TFEB, which cannot heterodimerize within the Myc family, does not suppress Myc-induced transcription or transformation. However, transfer of a 25- to 36-aa region from Mad or Mxi1, which interacts with mSin3, to the b-HLH-LZ of TFEB, mediated profound suppression of Myc-induced transcription and transformation. These results suggest that the DNA binding specificities of the Myc family and non-Myc family b-HLH-LZ proteins, in the context of the cellular genes involved in Myc-induced transformation, are shared. The results also demonstrate that targeting mSin3 to CACGTG sites via a non-Myc family DNA binding domain is sufficient to oppose Myc activity in growth regulation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B., Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994 Feb;4(1):102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Amin C., Wagner A. J., Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993 Jan;13(1):383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Lawrence Q. A., Eisenman R. N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995 Mar 10;80(5):767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Huang J., Ma A., Kretzner L., Alt F. W., Eisenman R. N., Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993 Sep;13(9):5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Brough D. E., Hofmann T. J., Ellwood K. B., Townley R. A., Cole M. D. An essential domain of the c-myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol Cell Biol. 1995 Mar;15(3):1536–1544. doi: 10.1128/mcb.15.3.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C. S., Sharp P. A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990 Aug;10(8):4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerni C., Bousset K., Seelos C., Burkhardt H., Henriksson M., Lüscher B. Differential effects by Mad and Max on transformation by cellular and viral oncoproteins. Oncogene. 1995 Aug 3;11(3):587–596. [PubMed] [Google Scholar]
- Dang C. V. c-myc oncoprotein function. Biochim Biophys Acta. 1991 Dec 10;1072(2-3):103–113. doi: 10.1016/0304-419x(91)90009-a. [DOI] [PubMed] [Google Scholar]
- Desbarats L., Gaubatz S., Eilers M. Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes Dev. 1996 Feb 15;10(4):447–460. doi: 10.1101/gad.10.4.447. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Carr C. S., Parent L. A., Sharp P. A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991 Dec;5(12A):2342–2352. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Parent L. A., Sharp P. A. High affinity DNA-binding Myc analogs: recognition by an alpha helix. Cell. 1993 Feb 12;72(3):467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- Fisher F., Crouch D. H., Jayaraman P. S., Clark W., Gillespie D. A., Goding C. R. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993 Dec 15;12(13):5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Seth A., Davis R. J. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3216–3220. doi: 10.1073/pnas.90.8.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum B. J., Pognonec P., Roeder R. G. Definition of the transcriptional activation domain of recombinant 43-kilodalton USF. Mol Cell Biol. 1992 Nov;12(11):5094–5101. doi: 10.1128/mcb.12.11.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen P. J., Ayer D. E., Eisenman R. N. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995 Jun;6(6):623–629. [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Transcriptional activities of the Myc and Max proteins in mammalian cells. Curr Top Microbiol Immunol. 1992;182:435–443. doi: 10.1007/978-3-642-77633-5_55. [DOI] [PubMed] [Google Scholar]
- Lahoz E. G., Xu L., Schreiber-Agus N., DePinho R. A. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T. D., Amati B., Land H., Evan G. I. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992 Sep;7(9):1783–1792. [PubMed] [Google Scholar]
- Luo X., Sawadogo M. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc Natl Acad Sci U S A. 1996 Feb 6;93(3):1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor G. R., Caskey C. T. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989 Mar 25;17(6):2365–2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Bossone S. A., Patel A. J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Mäkelä T. P., Koskinen P. J., Västrik I., Alitalo K. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992 Apr 17;256(5055):373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Mäkelä T. P., Partanen J., Schwab M., Alitalo K. Plasmid pLTRpoly: a versatile high-efficiency mammalian expression vector. Gene. 1992 Sep 10;118(2):293–294. doi: 10.1016/0378-1119(92)90203-2. [DOI] [PubMed] [Google Scholar]
- Negishi Y., Iguchi-Ariga S. M., Ariga H. Protein complexes bearing myc-like antigenicity recognize two distinct DNA sequences. Oncogene. 1992 Mar;7(3):543–548. [PubMed] [Google Scholar]
- Pellett P. E., McKnight J. L., Jenkins F. J., Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pognonec P., Roeder R. G. Recombinant 43-kDa USF binds to DNA and activates transcription in a manner indistinguishable from that of natural 43/44-kDa USF. Mol Cell Biol. 1991 Oct;11(10):5125–5136. doi: 10.1128/mcb.11.10.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A. I., DePinho R. A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995 Mar 10;80(5):777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Solomon D. L., Amati B., Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993 Nov 25;21(23):5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]





