Significance
Arbuscular mycorrhizal (AM) symbiosis is a mutualistic interaction formed between most land plants and soil fungi. During symbiosis the fungus develops branched hyphae, known as arbuscules, inside the root cortical cells. Arbuscules are critical to the symbiosis and function in phosphate delivery to the plant. Here we show that arbuscule formation is regulated by DELLA proteins. DELLA proteins are negative regulators of gibberellic acid (GA) signaling and repress plant growth and development. Our data provide insights into regulation of arbuscule formation and identify a potential mechanism by which the plant can coordinate the symbiosis with its growth and nutrient status.
Keywords: endosymbiosis, phytohormone, biotrophic, cyclops, Lotus japonicus
Abstract
Most flowering plants are able to form endosymbioses with arbuscular mycorrhizal fungi. In this mutualistic association, the fungus colonizes the root cortex and establishes elaborately branched hyphae, called arbuscules, within the cortical cells. Arbuscule development requires the cellular reorganization of both symbionts, and the resulting symbiotic interface functions in nutrient exchange. A plant symbiosis signaling pathway controls the development of the symbiosis. Several components of the pathway have been identified, but transcriptional regulators that control downstream pathways for arbuscule formation are still unknown. Here we show that DELLA proteins, which are repressors of gibberellic acid (GA) signaling and function at the nexus of several signaling pathways, are required for arbuscule formation. Arbuscule formation is severely impaired in a Medicago truncatula Mtdella1/Mtdella2 double mutant; GA treatment of wild-type roots phenocopies the della double mutant, and a dominant DELLA protein (della1-Δ18) enables arbuscule formation in the presence of GA. Ectopic expression of della1-Δ18 suggests that DELLA activity in the vascular tissue and endodermis is sufficient to enable arbuscule formation in the inner cortical cells. In addition, expression of della1-Δ18 restores arbuscule formation in the symbiosis signaling pathway mutant cyclops/ipd3, indicating an intersection between DELLA and symbiosis signaling for arbuscule formation. GA signaling also influences arbuscule formation in monocots, and a Green Revolution wheat variety carrying dominant DELLA alleles shows enhanced colonization but a limited growth response to arbuscular mycorrhizal symbiosis.
Arbuscular mycorrhizal (AM) symbiosis, formed by most angiosperms and fungi of the order Glomeromycota (1, 2), occurs widely in terrestrial ecosystems, where it plays a significant role in plant phosphorus nutrition and the carbon cycle and consequently impacts ecosystem productivity (1, 3). Given the limitations of global phosphate rock reserves, sustainable use of Pi fertilizer in agriculture will become increasingly important, and AM symbiosis may contribute to this sustainability (4).
To initiate AM symbiosis, root cells, primed by signals from the fungus, activate a symbiosis signaling pathway (SSP), which triggers cellular rearrangements that permit growth of the fungal hyphae through the epidermal cells and into the cortex. A second phase of differentiation results in hyphal growth into cortical cells and development of branched hyphae called arbuscules (reviewed in refs. 5 and 6). Phosphorus and nitrogen are delivered to the root through arbuscules, and in return the fungus gains access to carbon (7–10). The SSP was first identified in legumes, where it also functions in symbiosis with nitrogen-fixing rhizobia (11). Several components of the pathway have been identified, and in SSP mutants AM symbiosis is blocked at different stages of development. In three SSP mutants, including those with a mutation in a calcium calmodulin-dependent protein kinase (CCAMK/DMI3), hyphal growth is arrested in the epidermis (12–15). CYCLOPS/IPD3, a protein of unknown function that interacts with CCAMK, influences cortical colonization, and in Lotus japonicus and rice cyclops mutants, AM fungal hyphae grow into the cortex, but arbuscules are not formed (14, 16). The phenotypes of some Medicago truncatula ipd3 alleles are similar to those of cyclops, but arbuscules are formed in others (17–19). VAPYRIN/PAM2, also of unknown function, likewise is required for arbuscule formation, although currently it is unclear whether this protein is a signaling protein (20–22). Our current understanding of the SSP and how it functions to regulate AM symbiosis is incomplete. In rhizobium legume symbiosis (RLS), signaling through the SSP ultimately activates transcription factors that control infection and nodule development (23). A similar model is proposed for AM symbiosis, and GRAS factors that regulate hyphopodia development (24) and cortical colonization levels have been identified (25), but regulators specific for arbuscule formation are unknown. Transcript profiling and promoter–reporter gene analyses indicate complex changes in plant gene expression in the root cortex during arbuscule development, suggesting that multiple signaling pathways may be involved in arbuscule formation (14, 26–30). Complex regulation of arbuscule formation might be expected, because the symbiosis is modulated in response to the plant’s phosphate status, nitrogen status, and photosynthetic capacity (31–34). It is not known if regulation in response to the plant’s physiological status involves the SSP.
Transcriptome analyses reveal substantial alterations in the expression of genes encoding enzymes of gibberellic acid (GA) biosynthesis, degradation, and signaling during AM symbiosis (28, 35–39). Consistent with these alterations, GA levels increase significantly in mycorrhizal roots (40). Furthermore, researchers studying polyamine biosynthesis in mycorrhizal roots observed that GA treatment inhibited the development of AM symbiosis and in particular arbuscule number (41).
GA is a phytohormone that controls many aspects of plant growth and development and also influences responses to abiotic and biotic stress (42–47). GAs are synthesized from carotenoid precursors, and bioactive GA levels are modulated by a combination of GA synthesis and GA degradation (48, 49). Through studies in Arabidopsis and rice, a mechanistic understanding of GA perception and signaling has been obtained (reviewed in refs. 50–52). DELLA proteins, a unique group of GRAS transcriptional regulators, are central players in GA signaling and repress GA responses and restrain growth (53, 54). There are five DELLA proteins in Arabidopsis, two in pea, and one in rice (55–61). DELLA proteins contain domains typical of other GRAS transcription factors (62), but in addition they contain a unique DELLA domain at the N terminus. In the presence of GA, the DELLA domain mediates interaction with the GID1 receptor, and DELLA proteins subsequently are degraded via the 26S proteasome (51–54). Removal of DELLA proteins thus relieves repression and enables growth and other GA-activated responses (53, 54, 63). DELLA proteins interact physically with many transcriptional regulators in diverse signaling pathways and interfere with or modulate transcription factor function (64–70). As a consequence, GA has wide-reaching effects on gene expression, and DELLA proteins mediate cross-talk between many signaling pathways. In Arabidopsis, a role for DELLA proteins and GA signaling in Pi-starvation signaling has been established, and DELLA proteins regulate a subset of the adaptive responses to Pi starvation, including alterations in root architecture (71).
Here, we demonstrate that in M. truncatula DELLA1 and DELLA2 are required for arbuscule formation, as is consistent with a recent report that arbuscule formation is impaired in a pea cry, la mutant (72). Surprisingly, in M. truncatula della1/della2 mutants, hyphal growth in the cortex is not impaired by the lack of arbuscules, suggesting that intercellular hyphae have the capacity to access carbon. A dominant DELLA mutant restores arbuscule formation in cyclops, indicating an intersection between DELLA signaling and the SSP. Thus, DELLA proteins provide a mechanism for connecting symbiosis signaling with plant growth and development.
Results
M. truncatula DELLA1 and DELLA2 Are Required for Arbuscule Formation.
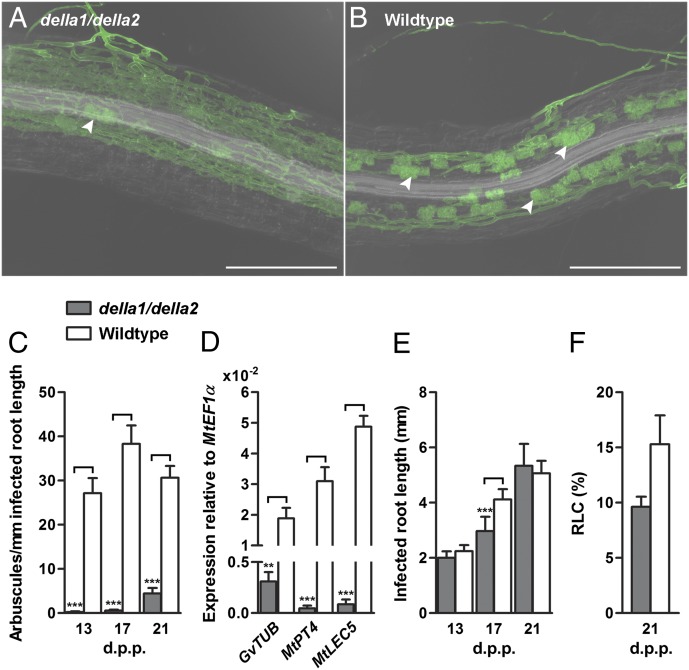
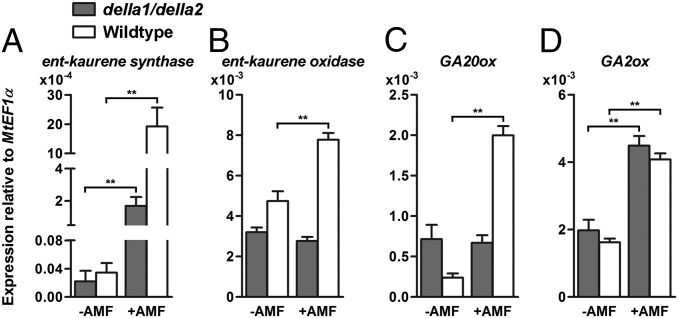
A reverse genetic screen that aimed to identify M. truncatula genes involved in AM symbiosis revealed that RNAi knockdown of a DELLA gene resulted in aberrant AM symbiosis. In DELLA RNAi roots inoculated with Glomus versiforme, hyphal growth into the roots occurred as in wild-type roots, but development in the cortex was altered, and arbuscule formation was markedly reduced (Fig. S1). There are three DELLA genes in the M. truncatula genome database Mt3.5: MtDELLA1 (contig_170694 and contig_69957), MtDELLA2 (contig_52215), and MtDELLA3 (contig_55897). The encoded proteins share 56–68% identity with DELLA proteins of Arabidopsis (Fig. S2), and MtDELLA1 and MtDELLA2 are orthologs of Pea LA and CRY (60), which recently were shown to influence arbuscule formation (72). To confirm and extend the RNAi results, M. truncatula lines containing Tnt1 insertions in MtDELLA1 and MtDELLA2 were obtained from a mutant population generated at the Samuel Roberts Noble Foundation. We focused on these two DELLA genes because they are coexpressed and because their transcript levels are high in M. truncatula Pi-deprived roots and remain high during AM symbiosis (Fig. S3 A and B). Expression of both genes is reduced after the addition of phosphate (Fig. S3 C and D). Two lines with insertions in MtDELLA1 and one line with an insertion in MtDELLA2 were obtained (Fig. S3E) and crossed to create a della1/della2 double mutant. The della1/della2 mutant showed developmental phenotypes characteristic of Arabidopsis and rice della mutants, namely, a slender shoot (61), early flowering, and reduced root fresh weight (Fig. S4). After inoculation with G. versiforme, della1 and della2 single mutants each showed an AM phenotype that did not differ from that of the wild-type segregant controls; however, the AM phenotype of the della1/della2 double mutant largely recapitulated the phenotype of the RNAi roots, and arbuscule formation was drastically reduced relative to the controls (Fig. 1 A–C and Fig. S5). On average, the arbuscule density in della1/della2 mycorrhizal roots was 85% lower than in control roots. The AM symbiosis phenotype of the della1/della2 mutant was examined in a range of substrates and nutritional conditions, and arbuscules were entirely absent in some experiments (Fig. S5). MtPT4 and MtLEC5 transcripts, which are markers of cells containing arbuscules (73–75), were extremely low in della1/della2 roots, and G. versiforme α-tubulin transcripts, which provide an indication of fungal biomass, likewise are lower in della1/della2 mutant than in wild-type roots (Fig. 1D). In Arabidopsis, several GA biosynthesis genes, including GA20ox2, are direct targets of DELLA proteins (76). In M. truncatula, genes encoding enzymes of GA biosynthesis, which are highly induced in wild-type roots during symbiosis, either are not induced or are induced only marginally in della1/della2 roots (Fig. 2 A–C). Expression of GA-2-oxidase, an enzyme that degrades bioactive GA, is similar in wild type and della1/della2 mutants (Fig. 2D). Despite the lack of arbuscules in della1/della2 mutants, intercellular hyphal growth in the cortex was not impaired, and infected root lengths in the double mutant did not differ significantly from those of wild-type roots (Fig. 1 E and F and Fig. S5C). The extensive growth of intercellular hyphae in della1/della2 roots suggests that the fungus is able to obtain carbon in the absence of arbuscules. However, the fungus fails to produce new spores and thus cannot complete its life cycle in della1/della2 roots (Table 1).
Fig. 1.
AM phenotype of della1/della2 roots colonized by G. versiforme. (A and B) Laser-scanning confocal microscope images of G. versiforme in della1/della2 (A) and wild-type (B) roots. Arrowheads mark arbuscules. (Scale bars: 250 µm.) (C) Arbuscule density in della1/della2 and wild-type roots at 13, 17, and 21 d post planting (d.p.p.). Data are averages ± SEM (n ≥ 24, where N denotes the number of infected root sections). (D) Expression of G. versiforme α-tubulin (GvTUB), MtPT4, and MtLEC5 in roots 21 d post inoculation (d.p.i.) assayed by quantitative RT-PCR. Data are averages ± SEM (n ≥ 5, where N denotes the number of independent root samples). (E) Infected root length in della1/della2 roots does not differ dramatically from that in wild-type roots, although significant changes are seen at 17 d.p.p. Data are averages ± SEM (n ≥ 24, where N denotes the number of infected root sections). (F) Level of colonization of della1/della2 and wild-type roots. Data are averages ± SEM (n = 3, where N denotes the number of independent root samples). RLC, root length colonized. **P ≤ 0.01; ***P ≤ 0.001.
Fig. 2.
Transcript levels of genes encoding enzymes of GA biosynthesis in noncolonized (−AMF) della1/della2 and wild-type roots and in della1/della2 and wild-type roots colonized with G. versiforme (+AMF) at 21 d.p.i. as assayed by quantitative RT-PCR. GAs are synthesized from geranylgeranyl diphosphate (GGDD) (116). (A and B) ent-kaurene synthase is involved in the conversion of GGDD to ent-kaurene, which is further converted to ent-kaurenoic acid, catalyzed by ent-kaurene oxidase. Ent-kaurenoic acid is subsequently converted to GA12. (C) GA 20-oxidases (GA20ox) catalyze the sequential oxidation of C-20 to convert GA12 to bioactive GAs. (D) GA 2-oxidases (GA2ox) catalyze GA inactivation reactions. Data are averages ± SEM (n ≥ 6, where N denotes the number of independent roots samples). **P ≤ 0.01.
Table 1.
G. versiforme spore production during association with della1/della2
| Genotype | Spores per 1-cm2 area |
| della1/della2 | 0 (n = 65 sections) |
| Wild type | 10 ± 2 (n = 56 sections) |
Spores were counted in 1-cm2 areas across the root system.
A Dominant DELLA Mutant Protein Promotes Arbuscule Formation.
DELLA proteins are negative regulators of GA signaling, and their repressive function is relieved by GA-induced proteolytic degradation (52, 54, 77, 78). Thus application of GA to mycorrhizal roots would be expected to phenocopy the della1/della2 mutant, and a dominant della mutant that is insensitive to GA but maintains function (55) would be expected to enable arbuscule formation even in the presence of GA. It has been shown previously that GA treatment suppresses arbuscule development in pea (41, 72), although in these experiments the effects on the fungus and plant could not be easily distinguished. The application of GA3 to M. truncatula roots inoculated with G. versiforme resulted in the expected alterations in plant growth and had a significant impact on development of symbiosis. G. versiforme was able to develop intercellular hyphae in the cortex, but arbuscule development was abolished completely (Fig. 3 A and B). As observed in della1/della2 mycorrhizal roots, intercellular hyphal growth in the cortex was unimpaired, and infection unit lengths in the GA-treated roots did not differ from those in control roots (Fig. 3C).
Fig. 3.
AM phenotype of M. truncatula roots and transgenic roots expressing a dominant DELLA1 gene (della1-Δ18) after GA3 treatment. (A and B) Fluorescence/differential interference contrast overlay images of G. versiforme in roots of M. truncatula after treatment with solvent (A) or GA3 (B). The arrow in A marks an arbuscule. Arbuscules are absent in GA3-treated roots. (C) Infected root length in GA3-treated roots does not differ from control. Data are averages ± SEM (n ≥ 31). (D) Morphology of G. versiforme in roots expressing della1-Δ18 after treatment with GA3. Arbuscule formation is restored. (E) Percentage of infected root sections containing only intercellular hyphae or arbuscules after GA3 treatment in roots expressing a vector control or della1-Δ18. Data are averages ± SEM from two independent experiments (n ≥ 10). (Scale bars: 50 µm.)
In Arabidopsis, the semidominant gai-1 allele encodes a mutant DELLA protein that lacks the DELLA domain; the repressor function is unimpaired, but the protein is insensitive to GA (55). Guided by this information, we created a dominant version of MtDELLA1 (della1-Δ18) by deleting the DELLA domain, and this version was introduced into wild-type roots under the control of the 35S promoter. After inoculation and GA3 treatment, the vector control roots showed a typical GA-treated AM phenotype with intercellular hyphae and no arbuscules. In contrast, arbuscule formation was restored in roots expressing della1-Δ18, with 40–50% of the infections showing arbuscules (Fig. 3 D and E). Likewise, arbuscule formation was restored in della1/della2 mutants expressing MtDELLA1 promoter:della1-Δ18 (Fig. S6 A and B). In both cases, arbuscules resulting from della1-Δ18 expression showed a normal morphology (Fig. S6C). Taken together, the phenotypes of the della1/della2 mutant and the effects of the della1-Δ18 constructs indicate that GA signaling through DELLA proteins regulates arbuscule formation.
Expression of della1-Δ18 in the Vascular Tissue and Endodermis Enables Arbuscule Formation in the Inner Cortical Cells.
In the AM symbiosis formed between M. truncatula and G. versiforme, arbuscules develop in the inner cortical cell layers. However, based on analysis of an MtDELLA1 promoter:GUS construct, MtDELLA1 is expressed strongly in the vascular tissue and endodermis but is detected in inner cortical cells only in rare instances and then only at extremely low levels (Fig. 4A). The vascular/endodermal expression pattern of MtDELLA1 promoter:GUS is maintained after colonization, and GUS expression was not observed in cells with arbuscules (Fig. 4B). These data are consistent with MtDELLA1 transcript levels, which do not change significantly after colonization (Fig. S3B). To monitor the DELLA protein, we created an MtDELLA1 promoter:GFP-MtDELLA1 fusion but were unable to detect its expression in M. truncatula roots. To increase the sensitivity, a GFP fusion to the GA-insensitive della1-Δ18 mutant protein was created; this construct complemented the arbuscule defect of the della1/della2 mutant, indicating that the fusion protein was functional (Fig. S6 A and B). When expressed from the MtDELLA1 promoter, GFP- della1-Δ18 was visible in the nuclei of cells in the vascular tissue and endodermis (Fig. S6D) and also, sporadically, in some of the inner cortical cells (Fig. 4 C and D and Fig. S6 E and F).
Fig. 4.
Expression and localization of MtDELLA1 in M. truncatula roots and AM phenotype of della1/della2 expressing della1-Δ18 driven by a vascular tissue-specific promoter. (A and B) Localization of MtDELLA1 promoter activity in noncolonized roots (−AMF) (A) and in roots colonized by G. versiforme (+AMF) (B). GUS-stained roots expressing pMtDELLA1:UidA indicate promoter activity in the vascular tissue and endodermis. Arrowheads mark cells containing arbuscules. Stars mark fungal hyphae. (C and D) Roots expressing pMtDELLA1:GFP-della1-Δ18 show consistent GFP signals in the nuclei of cells of the vascular tissue and endodermis; weaker signals are seen in some, but not all, cortical cells. Note that in this image the vascular tissue is not entirely in the focal plane (see also Fig. S6D). (E and F) Laser-scanning confocal microscope images of G. versiforme in roots of the della1/della2 mutant transformed with a control construct (E) or pMtPT9:della1-Δ18 (F). Expression of pMtPT9:della1-Δ18 restores arbuscule formation. (Scale bars: 100 µm in A and B; 250 µm in C–F.)
Because MtDELLA1 promoter activity was detected consistently in the vascular tissue/endodermis and was sporadic or almost absent in the cortex, we questioned whether DELLA activity in the vascular tissue/endodermis was sufficient to enable arbuscule formation. In Arabidopsis, DELLA activity in the endodermal layer plays a major role in the control of root elongation, and the endodermis is particularly sensitive to expression of gai, a dominant DELLA protein (79). To investigate further the potential site of DELLA action, the della1-Δ18 gene was placed under the control of the MtPT9 promoter. MtPT9 (Medtr4g083960) encodes a phosphate transporter of the Pht1 family and is expressed in roots and shoots and during AM symbiosis in both wild type and della1/della2 mutants (Fig. S7 A, B, and E). The MtPT9 promoter is highly active in the vascular tissue and endodermis (Fig. S7 C, D, and F). Expression of pMtPT9:della1-Δ18 restored arbuscule formation in the inner cortical cells of della1/della2 roots (Fig. 4 E and F) and also in wild-type roots treated with GA. These data indicate that expression of DELLA in the vascular tissue and endodermis is sufficient to drive arbuscule formation in the inner cortex cells. Consequently, we suggest that, when expressed from the native promoter, DELLA activity in these cell types may control arbuscule formation, although an additional contribution from the cortex cannot be ruled out.
DELLA Proteins and the Symbiosis Signaling Pathway.
Development of AM symbiosis is controlled by the SSP; however, current data indicate that activation of the SSP pathway alone is not sufficient to induce the complete cortical transcriptional program associated with arbuscule formation (80) and that other pathways may be involved. To test the hypothesis that DELLA signaling for arbuscule formation might intersect with the SSP, the della1-Δ18 gene was expressed from the 35S promoter in a L. japonicus cyclops mutant, a M. truncatula ipd3 mutant, and in two additional symbiosis mutants, M. truncatula dmi3 and M. truncatula vapyrin, that are positioned upstream and downstream of cyclops/ipd3 in the SSP, respectively. In dmi3 mutants hyphal growth is arrested at the epidermis (12), whereas in cyclops mutants and some ipd3 mutants the hyphae grow into the outer cortex, but arbuscules are absent (16, 18). In vapyrin mutants, hyphae reach the inner cortex, but arbuscules are not formed (20, 21).
Expression of p35S:della1-Δ18 did not affect the AM symbiosis phenotype of dmi3 or vapyrin; however it altered the L. japonicus cyclops and M. truncatula ipd3 phenotypes. After inoculation with G. versiforme, L. japonicus cyclops roots expressing p35S:della1-Δ18 showed arbuscules in 14% of the infections, but arbuscules were absent in cyclops roots expressing a control construct (Fig. 5 A–C). Expression of p35S:della1-Δ18 also increased intraradical hyphal growth in the cortex of cyclops roots by an average of 3.3-fold (Fig. 5C). The effect was not as obvious in M. truncatula ipd3 roots because, although ipd3-2 is reported to lack arbuscules (18), arbuscules were present in both ipd3-1 and ipd3-2 in our experimental conditions, although overall colonization levels were lower than in the wild-type controls (Fig. S8 A and B). However, in ipd3-1 roots expressing p35S:della1-Δ18 there was a small, but statistically significant increase in arbuscule numbers (Fig. S8C) and in the overall level of colonization (Fig. S8D). Thus, repression of GA signaling via the expression of della1-Δ18 promotes arbuscule formation and increases hyphal development in the absence of CYCLOPS/IPD3.
Fig. 5.
AM phenotype of cyclops expressing della1-Δ18 and gene expression in cyclops. (A and B) Laser-scanning confocal microscope images of G. versiforme in L. japonicus cyclops-3 roots expressing a control construct (A) or p35S:della1-Δ18 (B) at 26 d.p.i. Arrowheads mark arbuscules. Arbuscule formation is restored in cyclops expressing p35S:della1-Δ18. (Scale bars: 250 µm in A; 100 µm in B). (C) Percentage of infected root sections containing arrested infections, intercellular hyphae, or arbuscules. The growth of intercellular hyphae is increased significantly in roots expressing p35S:della1-Δ18 relative to roots expressing a control construct. Data are averages ± SEM (n ≥ 7, where N denotes the number of independent root samples). (D–I) Gene expression in roots of L. japonicus cyclops and wild-type plants either mock-inoculated (−AMF) or colonized by G. versiforme (+AMF) at 8 wk post planting (w.p.p.), assayed by quantitative RT-PCR. Data are averages ± SEM (n ≥ 4, where N denotes the number of independent root samples). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
The L. japonicus cyclops AM symbiosis phenotype is similar to that of the della1/della2 mutant in that it lacks arbuscules (17) (Fig. 5 D and E); however DELLA and GA-20-oxidase transcript levels in cyclops roots do not differ significantly from those in wild-type roots, suggesting that the cyclops phenotype does not arise from a reduction in DELLA transcripts or from an increase in GA biosynthetic gene expression (Fig. 5 F and G). NSP1 and NSP2 transcript levels likewise are similar in cyclops and wild-type roots, although cyclops roots do not show increased NSP1 transcripts following colonization (Fig. 5 H and I).
To evaluate DELLA and the SSP further, we analyzed transcript levels of the symbiosis-signaling genes DMI3 and IPD3 and two GRAS transcription factors, NSP1 and NSP2, in della1/della2 mutants. There were no changes in DMI3 and IPD3 transcript levels in della1/della2 roots relative to the wild-type segregant controls (Fig. 6 A and B). However, NSP1 and NSP2 transcript levels were significantly lower in the della1/della2 mutant than in the corresponding wild-type segregant, particularly during AM symbiosis (Fig. 6 C and D), suggesting that DELLA regulates NSP1 and NSP2 transcript levels either directly or indirectly. A link between GA and NSP2 has been reported during nodulation when Nod factor-induced expression of NSP2 was suppressed by GA treatment (81). Consistent with these data, we found that nodulation in the della1/della2 mutant was reduced significantly (Fig. S9). NSP1 and NSP2 are GRAS factors that are positioned downstream of DMI3 and CYCLOPS/IPD3 and are required for nodulation (11). In addition, recent data suggest that both transcription factors influence AM symbiosis in a quantitative manner but do not influence the morphology of the symbiosis (25, 82, 83). As reported previously (84), we found that an nsp1/nsp2 double mutant showed lower overall colonization levels relative to wild type (Fig. S10A). Expression of della1-Δ18 in nsp1/nsp2 roots resulted in increased overall colonization levels and arbuscule formation, suggesting that these GRAS factors are not essential for the della1-Δ18–driven changes in arbuscule formation or intercellular hyphal development (Fig. S10 B and C). Consequently, we predict that DELLA proteins influence other, as yet unknown, transcription factors to control arbuscule formation.
Fig. 6.
Expression of SPP genes in roots of della1/della2 and wild-type plants. (A–D) Transcript levels of SSP genes in roots either mock-inoculated (−AMF) or colonized by G. versiforme (+AMF) at 21 d.p.i., assayed by quantitative RT-PCR. Data are averages ± SEM (n ≥ 5, where N denotes the number of independent root samples). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
AM Symbiosis in Green Revolution Crop Varieties.
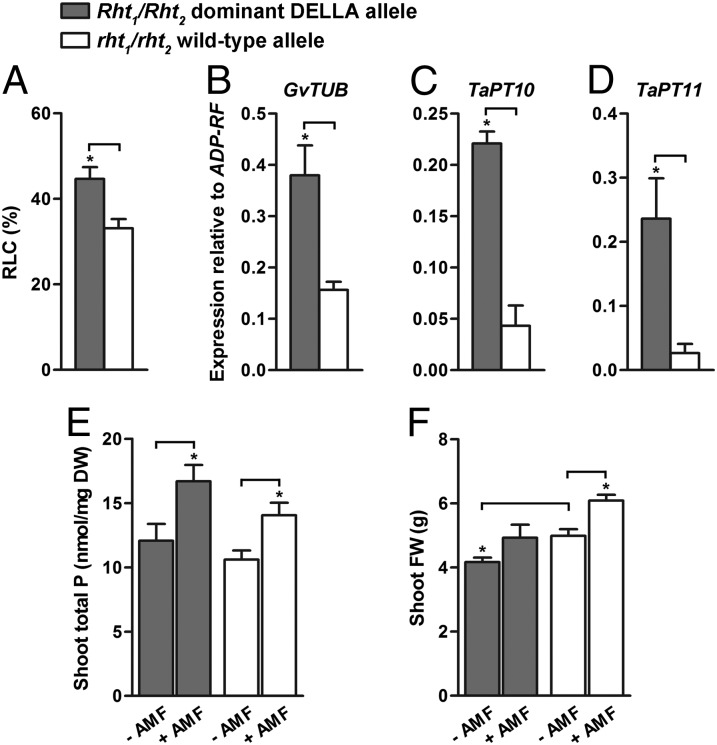
Wheat, maize, and rice are major food sources for the world’s population, and several of the Green Revolution cereal crop varieties are dominant DELLA mutants (85). For example, most wheat varieties grown worldwide are high-yielding dwarf varieties that express Reduced height (Rht) alleles which encode dominant DELLA proteins (86). To determine whether breeding for short stature and high yield has inadvertently resulted in plants with altered responses to AM symbiosis, wheat lines carrying the Rht1/Rht2 dominant DELLA alleles or the rht1/rht2 wild-type alleles (87–89) were inoculated with G. versiforme, and colonization and growth were monitored. The wheat line expressing Rht1/Rht2 alleles showed a small but significant increase in colonization relative to the line expressing rht1/rht2 wild-type alleles (Fig. 7A), and G. versiforme a-tubulin transcript levels were significantly higher in the Rht1/Rht2 line (Fig. 7B). Furthermore, TaPT10 and TaPT11 transcripts, which encode AM-induced phosphate transporters belonging to the MtPT4 subfamily (90), also were significantly higher in the Rht1/Rht2 line than in the wild type (Fig. 7 C and D), suggesting an increase in arbuscules numbers. These data are consistent with the effects observed in M. truncatula expressing della1-Δ18, and we observed a similar effect in Maize D8 (85, 91), a dominant della mutant. DXR transcripts, which serve as markers of arbuscule-containing cells (92), were higher in D8 than in the wild-type control (Fig. S11). In the wheat Rht1/Rht2 line, the arbuscules appear to be functional, because this line shows a significant increase in shoot phosphorus content during symbiosis (Fig. 7E). Despite the high colonization levels and the increase in shoot phosphorus content, the Rht1/Rht2 line did not show a symbiosis-associated increase in shoot biomass (Fig. 7F), likely because the dominant DELLA proteins restrain shoot growth. In conclusion, under the growth conditions tested, expression of the dominant DELLA Rht1/Rht2 proteins does not greatly impact the functionality of AM symbiosis in wheat.
Fig. 7.
AM phenotype of a Rht1/Rht2 dominant DELLA dwarf wheat line colonized by G. versiforme. (A) Level of colonization of the Rht1/Rht2 dominant DELLA dwarf and rht1/rht2 wild type at 31 d.p.i. The Rht1/Rht2 line differs significantly from the rht1/rht2 wild-type line. Data are averages ± SEM (n = 3, where N denotes the number of independent root samples). (B–D) Gene expression of G. versiforme α-tubulin (GvTUB) (B) and the phosphate transporters TaPT10 (C) and TaPT11 (D) in dwarf and wild-type roots at 31 d.p.i. relative to the Triticum aestivum ADP ribosylation factor (ADP-RF) assayed by quantitative RT-PCR. TaPT10 and TaPT11 transcript levels are significantly higher in the dwarf than in the wild-type roots. Data are averages ± SEM (n = 3, where N denotes the number of independent root samples). (E) Phosphorus content in shoots of noncolonized (−AMF) and colonized (+AMF) dwarf and wild-type plants at 13 wk post inoculation (w.p.i.). Data are averages ± SEM (n = 4, where N denotes the number of independent plants). DW, dry weight. (F) Shoot fresh weight (FW) of noncolonized and colonized dwarf and wild-type plants at 13 w.p.i. Data are averages ± SEM (n = 4, where N denotes the number of independent plants). *P ≤ 0.05.
Discussion
The AM symbiosis is of central importance to plant mineral nutrition, and arbuscules are critical for nutrient exchange between the fungal and plant symbionts. Arbuscule development is controlled in part by the plant; however, the regulatory mechanisms are largely unknown (reviewed in ref. 5). Here we show that two DELLA proteins, MtDELLA1 and MtDELLA2, are required for arbuscule formation. Treatment of M. truncatula roots with GA results in a phenocopy of della1/della2, and arbuscules can be restored in GA-treated roots by the expression of a GA-insensitive della1-Δ18 mutant protein. Taken together, these data confirm a role for GA as a negative regulator of arbuscule formation. In mycorrhizal roots, it is possible that the site of DELLA action may not be directly in the cortical cells where arbuscules develop, because ectopic expression of della1-Δ18 specifically in the vascular tissue and endodermis enables arbuscule formation in GA-treated roots and also in the della1/della2 mutant. These experiments establish that DELLA can act from the vascular tissue and endodermis, but in the native situation a contribution from DELLA in the cortex cannot be ruled out. If the site of action is the vascular tissue and endodermis, DELLA proteins may interact with transcription factors that subsequently move to the cortex; alternatively, DELLA regulation of arbuscule formation may be indirect. DELLA proteins influence several other phytohormone-signaling pathways, so responses in the cortex could result from changes in other mobile signaling molecules (66, 69); alternatively, a temporary restraint of root growth (79) may be necessary to enable arbuscule formation.
The SSP controls the development of both AM symbiosis and RLS, and the effects of GA on RLS have been described previously (81). GA treatment of L. japonicus roots altered several aspects of RLS and resulted in a partial phenocopy of nsp2, leading to the suggestion that GA inhibits nodulation signaling between DMI3 and NSP2 (81). By expressing della1-Δ18 in several SSP mutants, we were able to show an intersection between DELLA and symbiosis signaling for arbuscule formation. Constitutive expression of della1-Δ18 partially restores arbuscule formation in a cyclops mutant but not in a dmi3 mutant, thus indicating a requirement for CCAMK. One interpretation of these data is that della1-Δ18–driven arbuscule development is successful only when some aspects of the cortical program have been activated by CCaMK. In dmi3 (ccamk) mutants the fungus fails to enter the cortex, but in cyclops mutants hyphal growth in the cortex occurs, suggesting partial activation of the cortical program. In L. japonicus, a gain-of-function ccamk mutant showed cellular changes in cortex which also suggest that CCAMK influences this phase of the symbiosis (80). An alternative interpretation is that arbuscule formation driven by della1-Δ18 has a less stringent requirement for CYCLOPS.
Both NSP1 and NSP2 transcript levels are reduced significantly in della1/della2 relative to wild type. This finding is consistent with nodulation studies in which GA application was shown to reduce NSP2 transcripts and suggests that DELLA proteins regulate NSP1 and NSP2 either directly or indirectly. Despite effects on their transcripts, these two GRAS factors are not essential for the increases in arbuscule formation or hyphal development driven by della1-Δ18. Consequently, it seems likely that DELLA proteins regulate other, as yet unknown, transcription factors to influence arbuscule formation and hyphal development. Because della1-Δ18 restores arbuscules in cyclops mutants, we propose that at least some of these factors are regulated by cyclops. Alternatively, because DELLA proteins are known to influence many signaling pathways (reviewed in ref. 52), overexpression of the dominant DELLA1 could promote signaling through other pathways, for example root growth- or phosphate-signaling pathways, and could sensitize the root to enable some arbuscule formation even in the absence of complete SSP signaling. An analogous situation occurs in Arabidopsis, in which a dominant DELLA increases sensitivity to jasmonic acid and alters interactions with plant pathogens (46, 47, 93).
In Arum-type symbioses, arbuscules are the site of Pi delivery to the plant, and in vapyrin mutants, where arbuscules are absent, or mtpt4 mutants, where arbuscules are not functional, intercellular hyphal growth is reduced substantially, and the hyphae show septa, which are a sign of death (20–22, 94). These data suggest that functional arbuscules are necessary for continued fungal development. Consistent with this notion, nutrient experiments indicate that carbon and Pi exchange are linked functionally, and a reciprocal rewards model has been proposed in which Pi delivery by the fungal symbiont is rewarded with increased carbon, and vice versa (95, 96). Given these data, the extensive growth of intercellular hyphae in della1/della2 and in GA-treated roots was surprising, because the fungus proliferates in the cortex without forming arbuscules. This phenotype suggests that the symbiosis-associated link between phosphate and carbon has been uncoupled in this mutant. It is known that GA signaling can have direct effects on starch metabolism (97, 98). During AM symbiosis, starch decreases as the roots become colonized (99). It is possible that in the della1/della2 mutant, constitutive GA signaling may promote starch degradation, enabling the fungus to proliferate in the cortex in the absence of symbiotic Pi delivery.
Studies in Arabidopsis indicate that DELLA proteins influence signaling through multiple pathways and coordinate aspects of growth, development, and also responses to defense and to various abiotic stresses (51, 52). In general DELLA proteins act as repressors of GA signaling and negatively regulate plant growth and development through interactions with a myriad of transcription factors. In addition, they respond to environmental signals and mediate cross-talk with other hormone-signaling pathways, enabling the coordination of growth with adaptive responses to abiotic and biotic stresses. AM symbiosis is one of the less common examples in which DELLA proteins appear to function as positive regulators; a similar observation has been made for plant interactions with biotrophic pathogens that are promoted by DELLA proteins as a result of modulation of jasmonic acid and salicylic acid signaling (47).
There is considerable evidence that the plant coordinates the development of AM symbiosis with its nutritional requirements, carbon availability, and root growth (33, 100–103). Based on our current data and DELLA proteins’ established roles in growth restraint as well as cross-talk with other signaling pathways (43, 52, 53), we propose that DELLA proteins provide a mechanism to coordinate arbuscule formation and AM symbiosis with plant nutrient status and growth and that DELLA-mediated regulation of symbiosis occurs, in part, through regulation of the SSP. As outlined in Fig. 8, during Pi-limiting conditions, DELLA expression is high, and arbuscule formation is promoted through effects on unknown transcription factors that lie downstream of cyclops in the SSP. Once arbuscules are formed, symbiotic Pi transport leads to an increase in Pi levels which negatively regulate DELLA transcription. In addition, symbiosis-induced and/or Pi-induced expression of GA biosynthesis genes results in a rise in GA levels that leads to the proteolytic degradation of DELLA; consequently, negative feedback regulates arbuscule formation. Control of GA levels through a combination of biosynthesis and degradation provides a mechanism for regulating and fine-tuning GA levels and consequently DELLA activity in the mycorrhizal root. In addition, GA transport may further regulate cellular GA levels, although the proteins involved in this process are as yet unknown (104). The current data provide evidence that DELLA proteins control arbuscule formation, and, although the precise mechanism remains to be determined, the role of DELLA proteins as positive regulators of arbuscule formation coupled with their position at the nexus of many signaling pathways provides a mechanism for balancing symbiosis with plants’ nutritional needs, growth, and development.
Fig. 8.
A model integrating DELLAs and AM symbiosis. In Pi-deprived plants, DELLA transcript levels are high (Fig. S3) (71), and DELLA promotes arbuscule formation and hyphal growth in the cortex (Figs. 1 and 3–5) through the activation of as yet unknown transcription factors (TFs). Arbuscule development and the resulting symbiotic Pi transport leads to an increase in Pi levels in the root (94), resulting in a decrease in DELLA transcript levels (Fig. S3) (71). Genes of GA biosynthesis and GA inactivation are induced during symbiosis, indicating complex regulation of bioactive GA levels (Fig. 2D) (35). GA levels rise in the roots (40), leading to the degradation of DELLA protein. A decrease in DELLA (transcripts and protein) regulates arbuscule development through negative feedback. In plants with a high Pi status, low levels of DELLA proteins result in minimal arbuscule formation.
Materials and Methods
Plant and AM Fungal Growth Procedures.
Unless otherwise stated, plants were grown in a growth chamber under a 16-h light (25 °C)/8-h dark (22 °C) regime at 40% relative humidity in sterile Turface (Profile Products) inoculated with 300 surface-sterilized G. versiforme or G. intraradices spores per plant, as described (105), and fertilized once a week with modified half-strength Hoagland’s solution containing full-strength nitrogen and 20 μM potassium phosphate.
To characterize the AM phenotype in della1/della2 plants (Fig. 1 A–C, E, and F), 2-d-old seedlings were planted in a sand layer 4 cm below the top of 20.5-cm cones filled with a sterile gravel/filter sand mixture (1:1 ratio) containing 300 surface-sterilized G. versiforme spores. Seedlings were fertilized every third day with the above-mentioned fertilizer solution. Transcript analyses (Figs. 1D, 2, and 6), spore counting (Table 1), and plant phenotype evaluation (Fig. S4) were performed on plants growing in cones filled with a sterile gravel/sand mixture (1:2 ratio) inoculated with 500 surface-sterilized G. versiforme spores per cone. Plants were fertilized daily with above-mentioned fertilizer solution. Plants were harvested 21 d post inoculation.
The data shown in Fig. S5 were generated from plants grown in a double-cone system to synchronize infection (106), with the following modifications. Two seedlings were used per cone, and the cones contained sterile sand. Plants were fertilized once a week with above-mentioned fertilizer. After 2 wk, the plant cone was placed over the spore cone, which was partially filled with a sterile Turface/sand mixture (1:1 ratio) with a 1-cm sand layer at the top of the mixture. Eight hundred sterile G. versiforme spores were spread onto this sand layer. After 12 d, the nylon mesh was removed, and the plant cone was pushed down. Cones were harvested 7, 9, and 11 d after the physical contact of the roots with G. versiforme spores.
To analyze gene expression in L. japonicus Gifu wild-type and cyclops-3 roots, 7-d-old seedlings were planted in cones filled with sterile filter sand containing 500 surface-sterilized G. versiforme spores and were fertilized every third day with modified half-strength Hoagland’s solution containing full-strength nitrogen and 20 μM potassium phosphate. Plants were harvested 8 wk post planting.
Three-week-old composite L. japonicus plants were inoculated with 300 surface-sterilized G. versiforme spores and were grown in 11-cm pots filled with a sterile gravel/filter sand mixture (1:1 ratio). Plants were harvested 26 d post inoculation.
For analysis of AM symbiosis in wheat (Fig. 7 A–D), 3-d-old seedlings were transplanted to 11-cm pots containing sterile gravel/filter sand mixture (1:1 ratio) and were inoculated with 400 surface-sterilized G. versiforme spores. To monitor mycorrhiza-associated growth response (Fig. 7 E and F), seedlings were grown in a sterile gravel/filter sand mixture (1:1 ratio) with 400 surface-sterilized G. versiforme spores per seedling and were fertilized weekly with modified half-strength Hoagland’s solution containing full-strength nitrogen and 20 μM potassium phosphate. After 3 wk, plants were transferred to a sterile Lansing soil/sand mixture (1:5 ratio) and were fertilized twice a week with a modified 1/4× Hoagland’s solution containing half-strength nitrogen and without potassium phosphate. Once a week, the fertilizer was supplemented with 10 mL 0.5 mM Ca3(PO4)2 as described (107). The plants were harvested 10 wk post transfer.
To visualize fungal structures, roots were stained in 0.2 mg/mL WGA Alexafluor 488 (Molecular Probes) (94). Colonization levels specified as percent root length colonized were assessed by the modified gridline intersect method (108). Image J software was used to measure the infected root length. Infected root lengths could contain more than one infection unit. Arbuscule density was quantified by counting arbuscules within fungal colonization units (Fig. 1C) or by counting arbuscules within a defined area (Figs. S5B, S8C, and S10C).
Spore production was assessed by counting the numbers of spores in 1-cm2 areas randomly across the root system. Roots were harvested 7 wk after inoculation. Five independent samples of della1/della2 mutant and wild-type roots were analyzed.
GA Treatment.
A 50-mg/mL gibberellic acid stock solution (GA3; G-7645; Sigma) was created with ethanol and diluted with H2Odd to a working solution of 10−6 M GA3. Control pots received H2Odd with the equivalent volume of ethanol (solvent solution). Starting 6 d post inoculation, 50 mL GA3 working or solvent solution was applied daily to each 11-cm pot.
Cloning and Vector Construction.
pMtDELLA1:UidA was created by amplifying a 1,054-bp fragment containing the region 5′ proximal to the MtDELLA1 ATG start codon with primers that added 5′ SalI and 3′ HindIII restriction sites (Table S1). The MtDELLA1 promoter fragment was inserted between the SalI and HindIII restriction sites of a modified pCAMBIA2301 vector that lacked the CaMV 35S-promoter (109).
The dominant DELLA1 gene, della1-Δ18, was made by fusion PCR to create a gene with a 54-nt deletion, which results in a DELLA protein lacking 18 amino acids encompassing the DELLA motif, beginning at amino acid 60 (MDELLAALGYKVRSSDMA). The deletion was designed based on the Arabidopsis mutants gai-1 and rga-Δ17, which contain a 17-aa deletion of the DELLA motif (55, 110). The resulting della1-Δ18 gene was inserted between the SalI and BsrGI restriction sites of a pCAMBIA2300 vector that contains p35S:GFP (pJL33) to create p35S:della1-Δ18. pJL33 was created by inserting a HindIII-EcoRI fragment containing the CaMV 35S-promoter-sGFP-AtNOS fusion from the CaMV35S-sGFP(S65T)-Nos plasmid (111) into its multiple cloning site. pJL33 was used as vector control for the experiment.
To create pMtDELLA1:GFP-della1-Δ18, della1-Δ18 was amplified with primers that added 5′ BsrGI and 3′ PmlI/NotI restriction sites. The fragment was digested with BsrGI and NotI and was ligated into the CaMV35S-sGFP(S65T)-Nos plasmid, resulting in an in-frame 3′ fusion to GFP driven by the CaMV 35S-promoter. This plasmid was digested with PstI and PmlI and was ligated into pCAMBIA2301, resulting in p35S:GFP-della1-Δ18. The MtDELLA1 promoter was amplified with primers adding 5′ and 3′ SalI restriction sites and was inserted in 5′-to-3′ orientation between the two SalI restriction sites of p35S:GFP-della1-Δ18, replacing the CaMV 35S-promoter. pMtPT9:della1-Δ18 was created by amplifying the MtPT9 promoter with primers that added 5′ BamHI and 3′ SalI restriction sites. The MtPT9 promoter fragment (Fig. S7) was inserted between the BamHI and SalI restriction sites of p35S:della1-Δ18, replacing the CaMV 35S-promoter. To enable identification of transgenic roots, the Ubiquitin promoter:dsRED1 reporter cassette from the modified pHellsgate 8 vector was blunt end-cloned into pMtPT9:della1-Δ18 and p35S:della1-Δ18 using the BamHI restriction sites of these vectors. pJL33 was used as vector control for the experiment.
Agrobacterium rhizogenes-Mediated Transformation.
Composite M. truncatula plants were produced by Agrobacterium rhizogenes-mediated transformation (112, 113) with minor modifications as described in refs. 20 and 26. To generate composite L. japonicus cyclops-3 plants, constructs were transferred via A. rhizogenes strain AR1193 as described (114).
Genotyping of Tnt1 Insertion Lines and Generation of della1/della2.
M. truncatula R108 Tnt1 transposon insertion lines in DELLA1 (NF5155 and NF4215) and DELLA2 (NF4302) were obtained from The Noble Foundation (Fig. S3E). R1 plants were grown and genotyped by PCR using a transposon-specific primer (Tnt1-F) and DELLA1 or DELLA2 gene-specific primers (Table S1). Wild-type plants at the DELLA1 or DELLA2 loci were identified using the corresponding gene-specific primers. della1 alleles were backcrossed to R108. Backcrossed della1 alleles from NF5155 and NF4215 were crossed with della2 to create two della1/della2 double mutants. The della1/della2 double mutants show a similar plant and AM phenotype. All comparative experiments were performed with the della1/della2 mutant obtained from the NF5155 della1 allele and included wild-type segregants from the respective della1 and della2 populations as controls.
RNA Isolation, cDNA Synthesis, and Semiquantitative and Quantitative RT-PCR.
Unless otherwise stated, RNA isolation, cDNA synthesis, and PCR were carried out as described previously (35). Quantitative real-time RT-PCR was performed as described (34). Total RNA from L. japonicus roots was extracted as described in ref. 30.
Determination of Phosphorus Content, Histochemical Staining for GUS, and Laser-Scanning Confocal Microscopy.
Phosphorus content was determined by a phosphomolybdate colorimetric assay (115).
Histochemical staining for GUS activity was performed as previously described (20).
Laser-scanning confocal microscopy was carried out as described previously (73).
Statistical Analyses.
Unless otherwise stated, the Kruskal–Wallis rank sum test was applied.
Supplementary Material
Acknowledgments
We thank Dr. Armando Bravo for assistance with the phylogeny analyses, Lauren Carley for assistance with della phenotyping, Prof. Martin Parniske for the L. japonicus cyclops mutant, and Prof. Giles Oldroyd for the M. truncatula nsp1-1/nsp2-2 mutant. The M. truncatula Tnt1 insertion mutant lines were obtained from The Samuel Roberts Noble Foundation, Inc. Financial support was provided by the National Science Foundation Plant Genome Grants DBI-0421676 and IOS 1127155, and by the National Research Initiative Competitive Grant 2008-35301-19039 from the US Department of Agriculture National Institute of Food and Agriculture. D.S.F. was supported in part by Deutsche Forschungsgemeinschaft 2-y postdoctoral fellowship FL 699/1-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308973110/-/DCSupplemental.
References
- 1.Smith SE, Read DJ. Mycorrhizal Symbiosis. San Diego: Academic; 2008. [Google Scholar]
- 2.Schüßler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol Res. 2001;105(12):1414–1421. [Google Scholar]
- 3.van der Heijden MGA, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- 4.Fester T, Sawers R. Progress and Challenges in Agricultural Applications of Arbuscular Mycorrhizal Fungi. Crit Rev Plant Sci. 2011;30(5):459–470. [Google Scholar]
- 5.Harrison MJ. Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 2012;15(6):691–698. doi: 10.1016/j.pbi.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6(10):763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 7.Hodge A, Campbell CD, Fitter AH. An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature. 2001;413(6853):297–299. doi: 10.1038/35095041. [DOI] [PubMed] [Google Scholar]
- 8.Sanders FE. The effect of foliar-applied phosphate on the mycorrhizal infections of onion roots. In: Sanders FE, Mosse B, Tinker PB, editors. Endomycorrhizas. London: Academic; 1974. pp. 261–277. [Google Scholar]
- 9.Helber N, et al. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 2011;23(10):3812–3823. doi: 10.1105/tpc.111.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SE, Smith FA. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Ann Rev Plant Biol. 2011;62:227–250. doi: 10.1146/annurev-arplant-042110-103846. [DOI] [PubMed] [Google Scholar]
- 11.Oldroyd GED. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11(4):252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 12.Lévy J, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303(5662):1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 13.Banba M, et al. Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 2008;49(11):1659–1671. doi: 10.1093/pcp/pcn153. [DOI] [PubMed] [Google Scholar]
- 14.Gutjahr C, et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20(11):2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirichine L, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441(7097):1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- 16.Yano K, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA. 2008;105(51):20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messinese E, et al. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact. 2007;20(8):912–921. doi: 10.1094/MPMI-20-8-0912. [DOI] [PubMed] [Google Scholar]
- 18.Horváth B, et al. Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact. 2011;24(11):1345–1358. doi: 10.1094/MPMI-01-11-0015. [DOI] [PubMed] [Google Scholar]
- 19.Ovchinnikova E, et al. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago Spp. Mol Plant Microbe Interact. 2011;24(11):1333–1344. doi: 10.1094/MPMI-01-11-0013. [DOI] [PubMed] [Google Scholar]
- 20.Pumplin N, et al. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010;61(3):482–494. doi: 10.1111/j.1365-313X.2009.04072.x. [DOI] [PubMed] [Google Scholar]
- 21.Murray JD, et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011;65(2):244–252. doi: 10.1111/j.1365-313X.2010.04415.x. [DOI] [PubMed] [Google Scholar]
- 22.Feddermann N, et al. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 2010;64(3):470–481. doi: 10.1111/j.1365-313X.2010.04341.x. [DOI] [PubMed] [Google Scholar]
- 23.Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 24.Gobbato E, et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol. 2012;22(23):2236–2241. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 25.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469(7328):58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, et al. Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell. 2003;15(9):2106–2123. doi: 10.1105/tpc.014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Güimil S, et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA. 2005;102(22):8066–8070. doi: 10.1073/pnas.0502999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogekamp C, et al. Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol. 2011;157(4):2023–2043. doi: 10.1104/pp.111.186635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 2012;69(3):510–528. doi: 10.1111/j.1365-313X.2011.04810.x. [DOI] [PubMed] [Google Scholar]
- 30.Kistner C, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17(8):2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce A, Smith SE, Tester M. The development of mycorrhizal infection in cucumber - effects of P-supply on root growth, formation of entry points and growth of infection units. New Phytol. 1994;127(3):507–514. [Google Scholar]
- 32.Pearson JN, Smith SE, Smith FA. Effect of photon irradiance on the development and activity of VA mycorrhizal infection in Allium porrum. Mycol Res. 1991;95(6):741–746. [Google Scholar]
- 33.Breuillin F, et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010;64(6):1002–1017. doi: 10.1111/j.1365-313X.2010.04385.x. [DOI] [PubMed] [Google Scholar]
- 34.Javot H, et al. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 2011;68(6):954–965. doi: 10.1111/j.1365-313X.2011.04746.x. [DOI] [PubMed] [Google Scholar]
- 35.Gomez SK, et al. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009;9(10):10. doi: 10.1186/1471-2229-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guether M, et al. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009;182(1):200–212. doi: 10.1111/j.1469-8137.2008.02725.x. [DOI] [PubMed] [Google Scholar]
- 37.Manthey K, et al. Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact. 2004;17(10):1063–1077. doi: 10.1094/MPMI.2004.17.10.1063. [DOI] [PubMed] [Google Scholar]
- 38.Ortu G, et al. Plant genes related to gibberellin biosynthesis and signaling are differentially regulated during the early stages of AM fungal interactions. Mol Plant. 2012;5(4):951–954. doi: 10.1093/mp/sss027. [DOI] [PubMed] [Google Scholar]
- 39.Garrido JMG, Morcillo RJL, Rodríguez JAM, Bote JA. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant Microbe Interact. 2010;23(5):651–664. doi: 10.1094/MPMI-23-5-0651. [DOI] [PubMed] [Google Scholar]
- 40.Shaul-Keinan O, et al. Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomu intraradices. New Phytol. 2002;154(2):501–507. doi: 10.1046/j.1469-8137.2002.00388.x. [DOI] [PubMed] [Google Scholar]
- 41.El Ghachtouli N, Martin-Tanguy J, Paynot M, Gianinazzi S. First report of the inhibition of arbuscular mycorrhizal infection of Pisum sativum by specific and irreversible inhibition of polyamine biosynthesis or by gibberellic acid treatment. FEBS Lett. 1996;385(3):189–192. doi: 10.1016/0014-5793(96)00379-1. [DOI] [PubMed] [Google Scholar]
- 42.Brian PW. EFFECTS OF GIBBERELLINS ON PLANT GROWTH AND DEVELOPMENT. Biol Rev Camb Philos Soc. 1959;34(1):37–84. [Google Scholar]
- 43.Harberd NP, et al. Gibberellin: Inhibitor of an inhibitor of...? Bioessays. 1998;20(12):1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 44.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311(5757):91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 45.Achard P, et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143(3):1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18(9):656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 47.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18(9):650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 48.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444(1):11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 50.Alvey L, Harberd NP. DELLA proteins: Integrators of multiple plant growth regulatory inputs? Physiol Plant. 2005;123(2):153–160. [Google Scholar]
- 51.Sun TP. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154(2):567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davière JM, Achard P. Gibberellin signaling in plants. Development. 2013;140(6):1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 53.Harberd NP. Botany. Relieving DELLA restraint. Science. 2003;299(5614):1853–1854. doi: 10.1126/science.1083217. [DOI] [PubMed] [Google Scholar]
- 54.Sasaki A, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299(5614):1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 55.Peng JR, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11(23):3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SC, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16(5):646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dill A, Sun TP. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159(2):777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverstone AL, Ciampaglio CN, Sun TP. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10(2):155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen CK, Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell. 2002;14(1):87–100. doi: 10.1105/tpc.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weston DE, et al. The Pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol. 2008;147(1):199–205. doi: 10.1104/pp.108.115808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda A, et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13(5):999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218(5):683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 63.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21(5):1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 65.Feng SH, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451(7177):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallego-Bartolomé J, et al. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(33):13446–13451. doi: 10.1073/pnas.1119992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirano K, et al. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012;71(3):443–453. doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- 68.Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24(6):2635–2648. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou XL, Lee LYC, Xia KF, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19(6):884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 70.Park J, Nguyen KT, Park E, Jeon J-S, Choi G. DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell. 2013;25(3):927–943. doi: 10.1105/tpc.112.108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang CF, Gao XH, Liao L, Harberd NP, Fu XD. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145(4):1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foo E, Ross JJ, Jones WT, Reid JB. Plant hormones in arbuscular mycorrhizal symbioses: An emerging role for gibberellins. Ann Bot (Lond) 2013;111(5):769–779. doi: 10.1093/aob/mct041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison MJ, Dewbre GR, Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 2002;14(10):2413–2429. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frenzel A, et al. Combined transcriptome profiling reveals a novel family of arbuscular mycorrhizal-specific Medicago truncatula lectin genes. Mol Plant Microbe Interact. 2005;18(8):771–782. doi: 10.1094/MPMI-18-0771. [DOI] [PubMed] [Google Scholar]
- 75.Isayenkov S, Fester T, Hause B. Rapid determination of fungal colonization and arbuscule formation in roots of Medicago truncatula using real-time (RT) PCR. J Plant Physiol. 2004;161(12):1379–1383. doi: 10.1016/j.jplph.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 76.Zentella R, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19(10):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silverstone AL, et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13(7):1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGinnis KM, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15(5):1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ubeda-Tomás S, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10(5):625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 80.Takeda N, Maekawa T, Hayashi M. Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell. 2012;24(2):810–822. doi: 10.1105/tpc.111.091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maekawa T, et al. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009;58(2):183–194. doi: 10.1111/j.1365-313X.2008.03774.x. [DOI] [PubMed] [Google Scholar]
- 82.Lauressergues D, et al. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012;72(3):512–522. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]
- 83.Delaux PM, Bécard G, Combier JP. NSP1 is a component of the Myc signaling pathway. New Phytol. 2013;199(1):59–65. doi: 10.1111/nph.12340. [DOI] [PubMed] [Google Scholar]
- 84.Liu W, et al. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell. 2011;23(10):3853–3865. doi: 10.1105/tpc.111.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng JR, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400(6741):256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 86.Evans L. Feeding the Ten Billion: Plants and Population Growth. Cambridge, UK: Cambridge Univ Press; 1998. [Google Scholar]
- 87.Allan RE. Influence of Semidwarfism and Genetic Background on Stand Establishment of Wheat. Crop Sci. 1980;20(5):634–638. [Google Scholar]
- 88.Allan RE. Harvest indexes of backcross-derived wheat lines differening in culm height. Crop Sci. 1983;23(6):1029–1032. [Google Scholar]
- 89.Allan RE. Registration of 16 lines of soft white spring wheat germplasm. Crop Sci. 1989;29(4):1098–1099. [Google Scholar]
- 90.Sisaphaithong T, Kondo D, Matsunaga H, Kobae Y, Hata S. Expression of plant genes for arbuscular mycorrhiza-inducible phosphate transporters and fungal vesicle formation in sorghum, barley, and wheat roots. Biosci Biotechnol Biochem. 2012;76(12):2364–2367. doi: 10.1271/bbb.120782. [DOI] [PubMed] [Google Scholar]
- 91.Winkler RG, Helentjaris T. Dominant dwarfs. Maize Genet. Coop. Newsl. 1993;67:110–111. [Google Scholar]
- 92.Hans J, Hause B, Strack D, Walter MH. Cloning, characterization, and immunolocalization of a mycorrhiza-inducible 1-deoxy-d-xylulose 5-phosphate reductoisomerase in arbuscule-containing cells of maize. Plant Physiol. 2004;134(2):614–624. doi: 10.1104/pp.103.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wild M, et al. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 2012;24(8):3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2007;104(5):1720–1725. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333(6044):880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 96.Hammer EC, Pallon J, Wallander H, Olsson PA. Tit for tat? A mycorrhizal fungus accumulates phosphorus under low plant carbon availability. FEMS Microbiol Ecol. 2011;76(2):236–244. doi: 10.1111/j.1574-6941.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 97.Ritchie S, Gilroy S. Tansley Review No. 100 - Gibberellins: Regulating genes and germination. New Phytol. 1998;140(3):363–383. doi: 10.1111/j.1469-8137.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- 98.Chen PW, Chiang CM, Tseng TH, Yu SM. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell. 2006;18(9):2326–2340. doi: 10.1105/tpc.105.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gutjahr C, et al. Presymbiotic factors released by the arbuscular mycorrhizal fungus Gigaspora margarita induce starch accumulation in Lotus japonicus roots. New Phytol. 2009;183(1):53–61. doi: 10.1111/j.1469-8137.2009.02871.x. [DOI] [PubMed] [Google Scholar]
- 100.Koide RT, Schreiner RP. Regulation of the vesicular-arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:557–581. [Google Scholar]
- 101.Amijee F, Stribley DP, Tinker PB. The development of endomycorrhizal root systems. VIII. Effects of soil phosphorus and fungal colonization on the concentration of soluble carbohydrates in roots. New Phytol. 1993;123(2):297–306. [Google Scholar]
- 102.Amijee F, Tinker PB, Stribley DP. The development of endomycorrhizal root systems. New Phytol. 1989;111(3):435–446. doi: 10.1111/j.1469-8137.1989.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 103.Tester M, Smith SE, Smith FA, Walker NA. Effects of photon irradiance on the growth of shoots and roots, on the rate of initiation of mycorrhizal infection and on the growth of infection units in Trifolium subterraneum L. New Phytol. 1986;103(2):375–390. [Google Scholar]
- 104.Shani E, et al. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA. 2013;110(12):4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu J, Blaylock L, Harrison MJ. cDNA arrays as tools to identify mycorrhiza-regulated genes: Identification of mycorrhiza-induced genes that encode or generate signaling molecules implicated in the control of root growth. Can J Bot. 2004;82(8):1177–1185. [Google Scholar]
- 106.Lopez-Meyer M, Harrison MJ. An experimental system to synchronize the early events of development of the arbuscular mycorrhizal symbiosis. In: Sánchez F, Quinto C, López-Lara IM, Geiger O, editors. Biology of Molecular Plant-Microbe Interactions. Vol 5. St. Paul: International Society for Molecular Plant-Microbe Interactions; 2006. pp. 546–551. [Google Scholar]
- 107.Hong JJ, et al. Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta. 2012;236(3):851–865. doi: 10.1007/s00425-012-1677-z. [DOI] [PubMed] [Google Scholar]
- 108.McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method that gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990;115(3):495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 109.Liu JY, et al. Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J Biol Chem. 2008;283(36):24673–24681. doi: 10.1074/jbc.M802695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dill A, Jung H-S, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA. 2001;98(24):14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiu W, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6(3):325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 112.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14(6):695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 113.Limpens E, et al. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot. 2004;55(399):983–992. doi: 10.1093/jxb/erh122. [DOI] [PubMed] [Google Scholar]
- 114.Charpentier M, et al. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 2008;20(12):3467–3479. doi: 10.1105/tpc.108.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 116.Hartweck LM. Gibberellin signaling. Planta. 2008;229(1):1–13. doi: 10.1007/s00425-008-0830-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.