Significance
[NiFe] hydrogenases carry a [NiFe](CN)2CO center at the active site to catalyze the reversible oxidation of hydrogen in microorganisms. Hydrogenase pleiotropically acting protein (Hyp)E synthesizes the CN ligand for [NiFe] hydrogenases by catalyzing the dehydration of primary amide to nitrile. In the case of HypE, the primary amide is the carbamoylated C-terminal cysteine of HypE, and the nitrile is the cyanated cysteine. However, there has been no structural evidence to explain how HypE dehydrates primary amides into nitrile. We have determined the structures of these two intermediate steps of HypE, namely, carbamoylated and cyanated states. These structures reveal the detailed interactions around the carbamoylated and cyanated cysteine, providing structural basis for the biological conversion of primary amide to nitrile.
Keywords: metalloprotein maturation, intermediate states, nitrile synthesis
Abstract
Hydrogenase pleiotropically acting protein (Hyp)E plays a role in biosynthesis of the cyano groups for the NiFe(CN)2CO center of [NiFe] hydrogenases by catalyzing the ATP-dependent dehydration of the carbamoylated C-terminal cysteine of HypE to thiocyanate. Although structures of HypE proteins have been determined, until now there has been no structural evidence to explain how HypE dehydrates thiocarboxamide into thiocyanate. Here, we report the crystal structures of the carbamoylated and cyanated forms of HypE from Thermococcus kodakarensis in complex with nucleotides at 1.53- and 1.64-Å resolution, respectively. Carbamoylation of the C-terminal cysteine (Cys338) of HypE by chemical modification is clearly observed in the present structures. In the presence of ATP, the thiocarboxamide of Cys338 is successfully dehydrated into the thiocyanate. In the carbamoylated state, the thiocarboxamide nitrogen atom of Cys338 is close to a conserved glutamate residue (Glu272), but the spatial position of Glu272 is less favorable for proton abstraction. On the other hand, the thiocarboxamide oxygen atom of Cys338 interacts with a conserved lysine residue (Lys134) through a water molecule. The close contact of Lys134 with an arginine residue lowers the pKa of Lys134, suggesting that Lys134 functions as a proton acceptor. These observations suggest that the dehydration of thiocarboxamide into thiocyanate is catalyzed by a two-step deprotonation process, in which Lys134 and Glu272 function as the first and second bases, respectively.
Hydrogenases catalyze the reversible oxidation of hydrogen in microorganisms. This catalysis is essential for energy metabolism and the regulation of proton concentrations. Under anaerobic conditions, hydrogenases are involved in maintaining the redox balance in the cell. The enzymes are classified into three groups according to the metals in the reaction center, [NiFe], [FeFe], and [Fe] hydrogenases (1, 2). [NiFe] hydrogenases basically consist of one large subunit and one small subunit. The reaction center in the large subunit carries a [NiFe] cluster. The Ni atom is bound to the four conserved cysteine residues of the large subunit. Two of the four cysteine residues also bind the Fe atom, which is coordinated by two CN and one CO ligands (3, 4).
Biosynthesis of the [NiFe] cluster is catalyzed by six hydrogenase pleiotropically acting proteins (Hyp) proteins (HypA–HypF) (5). Biosynthesis of the cyano group is catalyzed by HypE and HypF. First, HypF catalyzes a transfer reaction of the carbamoyl moiety of carbamoylphosphate to the C-terminal cysteine residue of HypE. Then, HypE catalyzes an ATP-dependent dehydration to produce a thiocyanate group (6, 7). The cyano group is transferred to the HypCD complex (8), which is thought to catalyze the formation of the Fe(CN)2CO group and insert the Fe ligand into the hydrogenase large subunit (9–12). The biological origin of the CO molecule is still unclear (13). After the Fe atom insertion, HypA and HypB insert the Ni atom into the hydrogenase large subunit (14). Finally, the C terminus of the hydrogenase large subunit is cleaved by a specific protease (15).
HypE belongs to the aminoimidazole ribonucleotide synthetase (PurM) family, which catalyzes the ATP-dependent conversion of acyl compounds into various products (16). The PurM family includes PurM, formylglycinamide ribonucleotide amidotransferase (PurL), thiamine-monophosphate kinase (ThiL), and selenophosphate synthetase (SPS). HypE and other proteins of the PurM family function as a homodimer and require Mg2+ ions for their catalytic activity (17–20). The DX4GAXP motif is characteristic for the PurM family. The aspartate residue in the DX4GAXP motif is involved in holding Mg2+ ions and nucleotides.
The crystal structures of HypE have been reported from Thermococcus kodakarensis (9), Desulfovibrio vulgaris (21), Escherichia coli (22), and Caldanaerobacter subterraneus (23). The structures of all of the other Hyp proteins have also been reported (9, 24–32). The carbamoylation mechanism of the C-terminal cysteine residue of HypE was proposed based on the structure of the HypE–HypF complex (23). The N-terminal acylphosphatase-like domain of HypF catalyzes the formation of a carbamate intermediate, which can be transferred to the active site through a tunnel inside the protein. On the other hand, previous studies suggested the involvement of two basic groups in the dehydration of the carbamoyl moiety in HypE (21, 22). However, these studies provided no structural evidence to explain how HypE recognizes and dehydrates thiocarboxamide.
Here, we report the structures of the carbamoylated and cyanated forms of HypE from T. kodakarensis (TkHypE) at 1.53- and 1.64-Å resolution, respectively. These structures trap the intermediate carbamoyl/cyano steps of the reactions catalyzed by HypE and provide deeper insight into its reaction mechanism in hydrogenase maturation.
Results and Discussion
Structure Determination of the Carbamoylated and Cyanated Forms of HypE in Complex with Nucleotides.
Carbamoylation of the conserved cysteine residue of HypE was performed by chemical modification using KOCN before crystallization. KOCN carbamoylates the SH group of cysteines (33). Because TkHypE has only one cysteine residue (Cys338), KOCN was valid for selective carbamoylation. Previously, we could not obtain the structure of the nucleotide-bound states of TkHypE, despite the observation of the ATP-dependent conformational changes of its C-terminal tail (9). This seemed to be because the nucleotides were unstable under the acidic crystallization conditions used. In addition, in the previously reported structure of TkHypE, the N-terminal lobe of TkHypE was missing. Therefore, we sought new crystallization conditions for this study. The new crystal form of TkHypE belongs to the space group P43212, and contains one protein molecule in the asymmetric unit. Two diffraction datasets for adenosine 5′-(β,γ-imido)triphosphate (AMPPNP)- and ATP-bound forms were collected at 1.53- and 1.64-Å resolution, respectively. Two benzamidine molecules added in the crystallization step were found at the molecule boundary opposite the side of the dimer formation, improving the crystal packing.
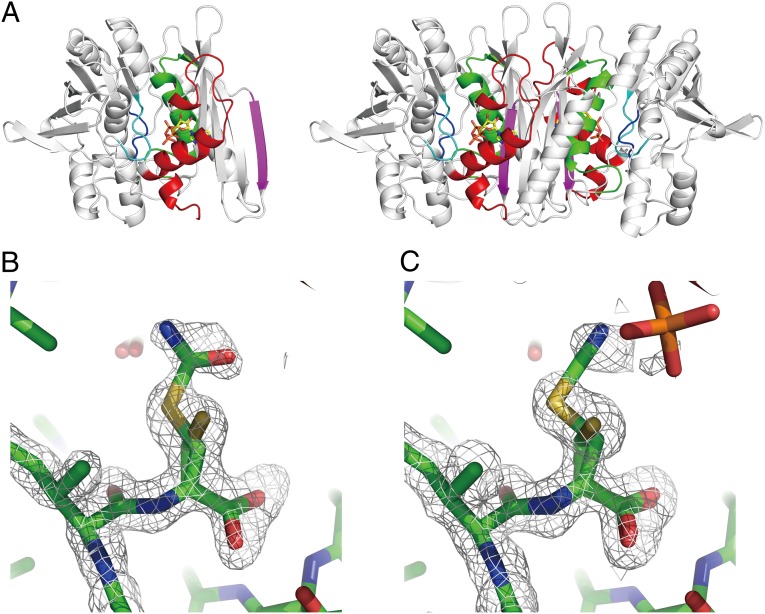
The structure of TkHypE consists of two α/β domains (domains A and B) with the C-terminal tail in the inward conformation (Fig. 1A). The electron density for the N-terminal lobe and ATP/AMPPNP are clearly visible in the present structures. The N-terminal lobe (residues 3–45) contains an α-helix and two 310 helices. As observed in the previously determined structures, the dimeric structure is formed by the interactions between the domain A and its symmetry-related counterpart. Two ATP/AMPPNP molecules are found at the boundary of the homodimer. The configuration of the N-terminal lobe of TkHypE is similar to that found in C. subterraneus HypE (CsHypE), but is different from those of D. vulgaris HypE (DvHypE) and E. coli HypE (EcHypE). A sequence alignment based on the updated structures (Fig. S1) shows that the loop length between the β2–β3 strands of TkHypE is shorter than those of EcHypE and DvHypE, indicating that the β2–β3 loop length determines the configuration of the N-terminal lobe.
Fig. 1.
Structures of the carbamoylated and cyanated forms of HypE. (A) Overall structure of the TkHypE monomer (Left) and dimer (Right). ATP is shown in a stick model. The N-terminal lobe and the conserved motifs I, II, III, and IV are shown in red, green, magenta, cyan, and orange, respectively. (B and C). Close-up views of the carbamoylated cysteine (B) and cyanated cysteine (C). The electron density of an Fo–Fc omit map around residues 335–338 is shown at 3.0 σ in a gray mesh.
An Fo–Fc omit map clearly shows that the C-terminal cysteine (Cys338) of HypE in the AMPPNP-bound state was partially carbamoylated (Cys338–CA) (Fig. 1B). In the structure in complex with ATP, the carbamoylated cysteine was dehydrated to cyanated cysteine (Cys338–CN) (Fig. 1C). The best-fit models were a mixture of 50% of carbamoylated/cyanated cysteine and 50% unreacted cysteine. The reactivity of Cys338 to KOCN suggests that before the reaction with KOCN, 50% of the HypE proteins take the inward conformation, which seems to prevent the carbamoylation of Cys338 by KOCN.
ATP-Binding Site.
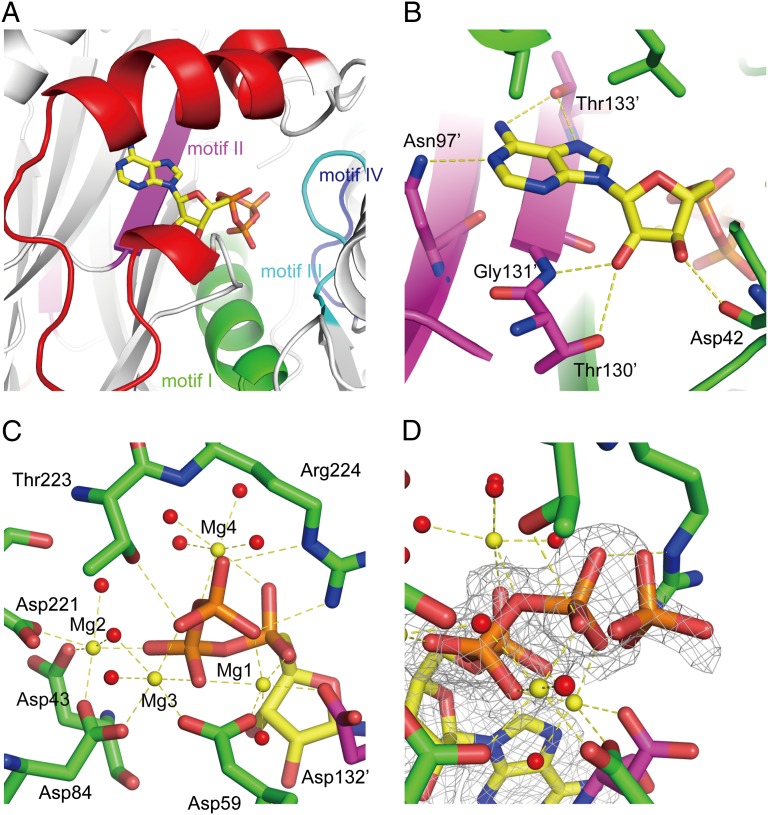
ATP/AMPPNP binding sites are formed by the N-terminal lobe and motifs I, II, and III from one protomer and the motif II from its counterpart (Fig. 2A and Fig. S1). The adenine base is sandwiched between the hydrophobic residues of the α1 helix and the main chain of motif II. The OH group of Thr133′ (primes indicate residues from the counterpart protomer) in motif II forms hydrogen bonds with the N6 and N7 atoms. The side chain NH2 of Asn97′ also forms a hydrogen bond with N1. The ribose interacts with Asp42, Thr130′, and Gly131′ (Fig. 2B).
Fig. 2.
ATP-binding site of HypE. (A) Overall binding mode of ATP. The conserved motifs I, II, III, and IV are shown in green, magenta, cyan, and blue, respectively. (B) A close-up of the interactions of the adenine and ribose moieties with the conserved residues. Hydrogen bonds are shown in yellow broken lines. (C) A close-up view of the phosphates binding mode. Mg2+ ions and water molecules are shown in yellow and red spheres, respectively. (D) Partially hydrolyzed γ-phosphate. The electron density of the 2 Fo–Fc map is shown at 1.0 σ in a gray mesh.
The phosphate groups of ATP are surrounded by four Mg2+ ions, which are held by five aspartate residues (Asp43, Asp59, Asp84, Asp221, and Asp132′) (Fig. 2C). Asp84 is the characteristic residue of the PurM family, and Asp59, Asp221, and Asp132′ are highly conserved among the HypE proteins (Fig. S1). Mg1 is coordinated by the α-, β- and γ-phosphates, Asp59, Asp132′, and one water molecule. Mg1 is also observed in the other PurM family proteins (17–20). Mg2 is coordinated by the γ-phosphate, Asp221, Asp43, Asp84, and two water molecules. Mg3 binds the β- and γ-phosphates, Asp59, Asp84, and two water molecules. Mg4 binds the α- and β-phosphates and four water molecules. Mg1, Mg2, and Mg3 are also observed in the AMPPNP-bound structure, but Mg4 is missing, because the O3B of ATP is replaced with the nitrogen atom in AMPPNP. Interestingly, the Mg3 and Mg4 observed in TkHypE are not found in the structure of the DvHypE in complex with ATP (21), despite the conservation of the aspartate residues. Superposition of these structures shows that the domain B of DvHypE is slightly open compared with TkHypE and that the position of Mg2 in DvHypE is slightly distant from ATP. As a result, the conserved Asp residues of DvHypE assume different conformations, which cannot support the additional Mg ions.
The O atoms of the γ-phosphate also form hydrogen bonds with Thr223 and Arg224 in motif III. In the ATP data, extra electron density was found near the γ-phosphate, indicating that the γ-phosphate is partially hydrolyzed (Fig. 2D). The occupancies of the γ-phosphate and hydrolyzed phosphate ion were refined to 0.63 and 0.29, respectively. The phosphate ion interacts with the conserved lysine residue (Lys134′) in motif II.
The β-Phosphate Hydrolysis.
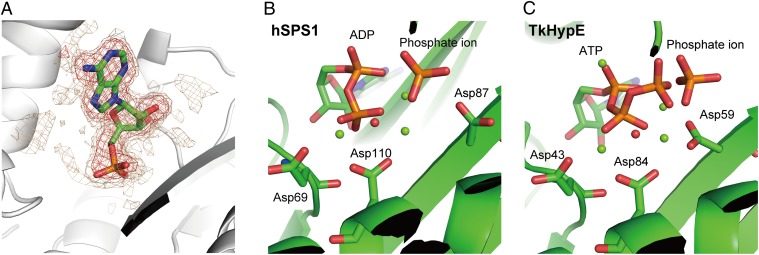
In the ATP data, the refined occupancies of the α- and β-phosphates were 0.93 and 0.80, respectively. In the case of the crystallization conditions at pH 7.0 (dataset “ATP pH7.0”), there was no electron density for either the γ-phosphate or the β-phosphate (Fig. 3A), indicating that the β-phosphate was also hydrolyzed. Among the PurM family members, human selenophosphate synthetase 1 (hSPS1) has also been reported to catalyze ATP hydrolysis to produce AMP and phosphate (20). In hSPS1, the water molecule held by two Mg2+ ions, Asp219, and Thr221 probably plays a nucleophilic role in ADP hydrolysis (Fig. 3B). Superposition of hSPS1 and TkHypE shows that the water molecule bound to Mg2, Mg3, Asp221, and Thr223 of TkHypE corresponds to the probable nucleophilic water molecule in hSPS1 (Fig. 3C). These observations indicate that HypE has an ability to hydrolyze the β-phosphate of ADP.
Fig. 3.
The β-phosphate hydrolysis. (A) A close-up view of AMP in the “ATP pH 7.0” data. The electron density of the Fo–Fc map at 3.0 and 2.0 σ is shown in red and gold mesh, respectively. (B and C) Comparison of hSPS1 and TkHypE. The two structures are superimposed and also shown independently. The structure of hSPS1 is shown in B, and that of TkHypE is shown in C. Three conserved aspartate residues are shown in stick models. The putative nucleophilic water and two Mg2+ ions are shown as red and green spheres, respectively.
Interactions Around the Carbamoylated C-Terminal Cysteine.
The present structure reveals the detailed interactions around the carbamoyl group of Cys338–CA (Fig. 4 A and B). Due to the partial carbamoylation of Cys338, two conformations of residues surrounding Cys338 are observed in the present structure (Fig. S2). Therefore, the structure analysis described below is based on the structure of the carbamoylated state.
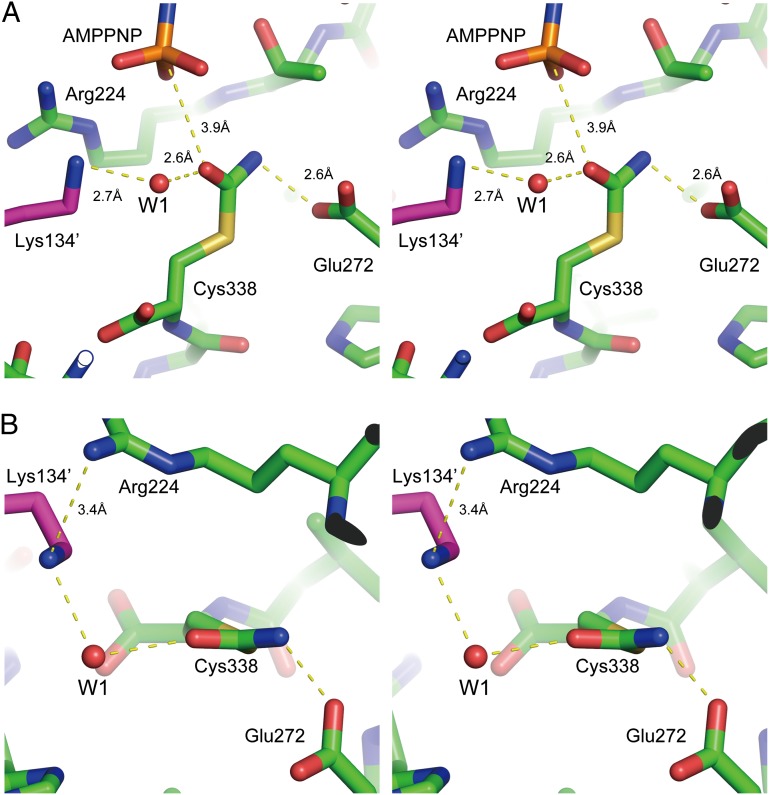
Fig. 4.
Interactions around carbamoylated cysteine. (A and B) Stereoview of a close-up of interactions around carbamoylated cysteine (Cys338–CA) from a side view (A) or top view (B). The remarkable interactions surrounding Cys338–CA are shown in yellow broken lines (see text).
Compared with the unreacted cysteine, the chi 1 angle of Cys330–CA and Cys338–CN is rotated about 180°. The thiocarboxamide oxygen atom of Cys338–CA is located within van der Waals distance of the γ–phosphate, with a distance of 3.9 Å between the Oδ and PG (Fig. 4A). The angle of N3B–PG–Oδ is ∼153°, and therefore the thiocarboxamide oxygen atom is in a proper position for an in-line attack on the γ-phosphate of ATP. The thiocarboxamide nitrogen atom of Cys338–CA is close to the Oδ atom of the conserved glutamate residue (Glu272) in motif IV (2.6 Å). However, the NH–O angle is ∼90° (Fig. 4B), suggesting that the position of Glu272 is less favorable for the proton abstraction from the nitrogen atom of the thiocarboxamide of Cys338–CA. On the other hand, a water molecule (W1) is located near the thiocarboxamide oxygen atom (Fig. 4B). W1 forms hydrogen bonds with the thiocarboxamide oxygen atom, Nζ of the conserved lysine residue (Lys134′) in motif II, the backbone oxygen atom, and a water molecule. Interestingly, Nζ of Lys134 is very close to the guanidium group of Arg222 at a distance of 3.4 Å. This close contact would be expected to decrease the pKa of this lysine residue. Indeed, the calculated pKa value of Lys134 by PROPKA (34) is 5.1. These results suggest that Lys134 is deprotonated in the ATP-binding state and functions as a proton acceptor.
Proposed Reaction Mechanism.
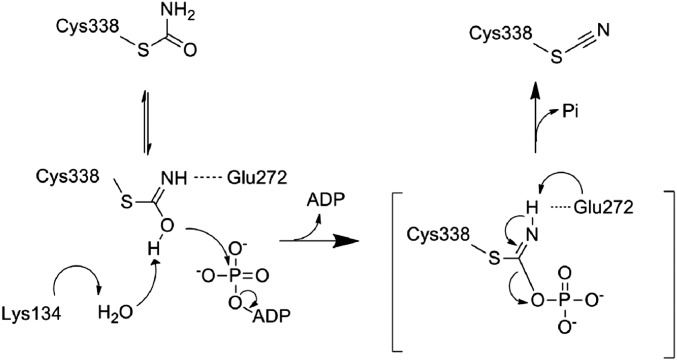
PurM family members including HypE are assumed to catalyze a common reaction scheme via an iminophosphate intermediate (35). In the case of HypE, two bases have been proposed to be involved in abstracting protons from thiocarboxamide (21, 22). Previous studies have proposed that the conserved glutamate residue in motif IV (Glu272 in TkHypE) functions as the first base that deprotonates the nitrogen atom of thiocarboxamide. However, the second base has not been clarified. The present structures of the carbamoylated HypE reveal that Glu272 in motif IV does not take an effective configuration for proton abstraction (Fig. 4 A and B). On the other hand, Lys134 in motif II is shown to be stable in the deprotonated form in the ATP-binding state.
Based on our structural findings as well as synthetic studies of nitriles from primary amides (36, 37), we propose a reaction mechanism for the ATP-dependent dehydration of thiocarboxamide by the conserved lysine and glutamate residues of HypE (Fig. 5). In the proposed mechanism, the conserved lysine residue in motif II (Lys134′) functions as the first base. The close contact with the conserved arginine residue (Arg222) lowers the pKa of Lys134′. The decrease in the pKa value allows Lys134′ to abstract the proton from the imidic acid of Cys338–CA through the water molecule W1. The oxygen atom of Cys338–CA attacks the γ-phosphate, resulting in formation of an iminophosphate intermediate. Finally, the intrinsic glutamate (Glu272) in motif IV functions as the second base, which deprotonates the imino nitrogen and promotes formation of the cyano group.
Fig. 5.
Proposed reaction mechanism of the carbamoyl cysteine dehydration.
The intrinsic ATPase activity of HypE (7) can also be explained by W1 and Lys134′. W1 in the unreacted state is also 3.9 Å distant from the γ-phosphate. The W1–PG–PO angle is about 157°. Therefore, the W1 activated by Lys134′ probably performs an in-line attack on the γ-phosphate of ATP. In fact, in the ATP data, the partially hydrolyzed phosphate occupies the position of W1.
In this study, we present structural evidence of the ATP dependent-dehydration of HypE. The conserved lysine of motif II and glutamate of motif IV probably function as essential bases in the conversion of thiocarboxamide into thiocyanate or ATP hydrolysis of HypE. These results shed light on the general mechanism via iminophosphate intermediates.
Materials and Methods
The detailed methods are described in SI Materials and Methods. The overexpression and purification of TkHypE were performed as described previously (38). The protein sample for crystallization was concentrated to 20 mg mL−1 in 20 mM Mes–NaOH pH 6.0, 5 mM MgCl2, 1 mM tris (2-carboxyethyl) phosphine (TCEP). The sample was mixed with 10 mM KOCN and incubated for 4 h at room temperature or for 20 h at 277 K, and 5 mM AMPPNP or 10 mM ATP was added. Crystals suitable for data collection were obtained using reservoir solution containing 0.1 M Mes–NaOH pH 6.0, 1.4–1.55 M ammonium sulfate, 1.7% (vol/vol) polyethylene glycol 400, and 6% (vol/vol) glycerol. Drops were prepared by mixing 1.0 μL protein, 0.8 μL reservoir solution, and 0.2 μL Additive Screen reagent [20% (wt/vol) benzamidine hydrochloride]. The dataset “ATP pH7.0” was collected from a crystal grown in the drop made by mixing the protein sample in 20 mM Hepes pH 7.0, 5 mM MgCl2, 1 mM TCEP, 10 mM ATP, and the above described reservoir solution. Diffraction datasets were collected at the beamlines BL-1A, BL-5A, and AR-NW12A (Photon Factory). The structures were determined by the molecular replacement method with MOLREP (39), using the previously determined HypE structure in its ternary complex (PDB entry 3VYT). Statistics for the crystallographic data are summarized in Table S1.
Supplementary Material
Acknowledgments
We thank the beamline scientists of the Photon Factory for their help with the X-ray data collection. This work was supported by Grants-in-Aid for Scientific Research (A) (20247009 and 23247014 to K.M.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors of HypE in the carbamoylated state, the cyanated state, and the AMP-bound state have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3WJP, 3WJQ, and 3WJR, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313620110/-/DCSupplemental.
References
- 1.Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem Rev. 2007;107(10):4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 2.Thauer RK, et al. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem. 2010;79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 3.Volbeda A, Fontecilla-Camps JC. The active site and catalytic mechanism of NiFe hydrogenases. Dalton Trans. 2003;21:4030–4038. [Google Scholar]
- 4.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem Rev. 2007;107(10):4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 5.Böck A, King PW, Blokesch M, Posewitz MC. Maturation of hydrogenases. Adv Microb Physiol. 2006;51:1–71. doi: 10.1016/s0065-2911(06)51001-x. [DOI] [PubMed] [Google Scholar]
- 6.Reissmann S, et al. Taming of a poison: Biosynthesis of the NiFe-hydrogenase cyanide ligands. Science. 2003;299(5609):1067–1070. doi: 10.1126/science.1080972. [DOI] [PubMed] [Google Scholar]
- 7.Blokesch M, et al. Analysis of the transcarbamoylation-dehydration reaction catalyzed by the hydrogenase maturation proteins HypF and HypE. Eur J Biochem. 2004;271(16):3428–3436. doi: 10.1111/j.1432-1033.2004.04280.x. [DOI] [PubMed] [Google Scholar]
- 8.Blokesch M, et al. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J Mol Biol. 2004;344(1):155–167. doi: 10.1016/j.jmb.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe S, et al. Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: Insights into cyanation reaction by thiol redox signaling. Mol Cell. 2007;27(1):29–40. doi: 10.1016/j.molcel.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe S, Matsumi R, Atomi H, Imanaka T, Miki K. Crystal structures of the HypCD complex and the HypCDE ternary complex: Transient intermediate complexes during [NiFe] hydrogenase maturation. Structure. 2012;20(12):2124–2137. doi: 10.1016/j.str.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Bürstel I, et al. A universal scaffold for synthesis of the Fe(CN)2(CO) moiety of [NiFe] hydrogenase. J Biol Chem. 2012;287(46):38845–38853. doi: 10.1074/jbc.M112.376947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soboh B, et al. [NiFe]-hydrogenase maturation: Isolation of a HypC-HypD complex carrying diatomic CO and CN- ligands. FEBS Lett. 2012;586(21):3882–3887. doi: 10.1016/j.febslet.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Bürstel I, et al. Probing the origin of the metabolic precursor of the CO ligand in the catalytic center of [NiFe] hydrogenase. J Biol Chem. 2011;286(52):44937–44944. doi: 10.1074/jbc.M111.309351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaluarachchi H, Chan Chung KC, Zamble DB. Microbial nickel proteins. Nat Prod Rep. 2010;27(5):681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- 15.Theodoratou E, Huber R, Böck A. [NiFe]-Hydrogenase maturation endopeptidase: Structure and function. Biochem Soc Trans. 2005;33(Pt 1):108–111. doi: 10.1042/BST0330108. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Kappock TJ, Stubbe J, Weaver TM, Ealick SE. X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution. Structure. 1999;7(9):1155–1166. doi: 10.1016/s0969-2126(99)80182-8. [DOI] [PubMed] [Google Scholar]
- 17.Morar M, Anand R, Hoskins AA, Stubbe J, Ealick SE. Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima describe a novel ATP binding protein superfamily. Biochemistry. 2006;45(50):14880–14895. doi: 10.1021/bi061591u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCulloch KM, Kinsland C, Begley TP, Ealick SE. Structural studies of thiamin monophosphate kinase in complex with substrates and products. Biochemistry. 2008;47(12):3810–3821. doi: 10.1021/bi800041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh Y, et al. Structure of selenophosphate synthetase essential for selenium incorporation into proteins and RNAs. J Mol Biol. 2009;385(5):1456–1469. doi: 10.1016/j.jmb.2008.08.042. [DOI] [PubMed] [Google Scholar]
- 20.Wang KT, Wang J, Li LF, Su XD. Crystal structures of catalytic intermediates of human selenophosphate synthetase 1. J Mol Biol. 2009;390(4):747–759. doi: 10.1016/j.jmb.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 21.Shomura Y, et al. Crystal structures of hydrogenase maturation protein HypE in the Apo and ATP-bound forms. J Mol Biol. 2007;372(4):1045–1054. doi: 10.1016/j.jmb.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Rangarajan ES, et al. Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF. J Bacteriol. 2008;190(4):1447–1458. doi: 10.1128/JB.01610-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shomura Y, Higuchi Y. Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases. J Biol Chem. 2012;287(34):28409–28419. doi: 10.1074/jbc.M112.387134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia W, Li H, Sze KH, Sun H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J Am Chem Soc. 2009;131(29):10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe S, et al. Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. J Mol Biol. 2009;394(3):448–459. doi: 10.1016/j.jmb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki D, et al. Identification and structure of a novel archaeal HypB for [NiFe] hydrogenase maturation. J Mol Biol. 2013;425(10):1627–1640. doi: 10.1016/j.jmb.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Chan KH, Li T, Wong CO, Wong KB. Structural basis for GTP-dependent dimerization of hydrogenase maturation factor HypB. PLoS ONE. 2012;7(1):e30547. doi: 10.1371/journal.pone.0030547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasper R, Scrima A, Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem. 2006;281(37):27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Xia B, Jin C. Solution structure of Escherichia coli HypC. Biochem Biophys Res Commun. 2007;361(3):665–669. doi: 10.1016/j.bbrc.2007.07.094. [DOI] [PubMed] [Google Scholar]
- 30.Rosano C, et al. Crystal structure and anion binding in the prokaryotic hydrogenase maturation factor HypF acylphosphatase-like domain. J Mol Biol. 2002;321(5):785–796. doi: 10.1016/s0022-2836(02)00713-1. [DOI] [PubMed] [Google Scholar]
- 31.Petkun S, et al. Structure of hydrogenase maturation protein HypF with reaction intermediates shows two active sites. Structure. 2011;19(12):1773–1783. doi: 10.1016/j.str.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Tominaga T, et al. Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 10):1153–1157. doi: 10.1107/S1744309112036421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark GR, Stein WH, Moore S. Reactions of cyanate present in aqueous urea with amino acids and proteins. J Biol Chem. 1960;235(11):3177–3181. [Google Scholar]
- 34.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 2008;65(23):3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahsavari-Fard Z, Sardarian AR. Diethyl chlorophosphate: A new alternative reagent for dehydration of primary amides to nitriles in solvent and solvent-free conditions. J Iran Chem Soc. 2011;8(1):204–208. [Google Scholar]
- 37.Kuo C-W, et al. A convenient new procedure for converting primary amides into nitriles. Chem Commun (Camb) 2007;3:301–303. doi: 10.1039/b614061k. [DOI] [PubMed] [Google Scholar]
- 38.Arai T, et al. Crystallization and preliminary X-ray crystallographic study of [NiFe]-hydrogenase maturation factor HypE from Thermococcus kodakaraensis KOD1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63(Pt 9):765–767. doi: 10.1107/S1744309107038833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.