Significance
The H19 imprinted gene produces a long noncoding RNA (lncRNA) exclusively expressed from the maternal allele. It is involved in the control of embryonic growth and regulates nine genes of an Imprinted Gene Network (IGN). Our goal was to decipher the molecular mechanisms that drive this control of the IGN. We show that this lncRNA represses several target genes through interaction with the methyl-CpG–binding domain protein 1 MBD1. This protein is involved in the maintenance of repressive H3K9me3 histone marks. The H19 RNA is required for the recruitment of MBD1 to some of its targets, including the adjacent insulin-like growth factor 2 gene, and acts by a fine-tuned regulation on the expression levels of these growth-controlling genes of the IGN.
Keywords: genomic imprinting, embryonic growth, long noncoding RNA partner, Dlk1, Cdkn1c
Abstract
The H19 gene controls the expression of several genes within the Imprinted Gene Network (IGN), involved in growth control of the embryo. However, the underlying mechanisms of this control remain elusive. Here, we identified the methyl-CpG–binding domain protein 1 MBD1 as a physical and functional partner of the H19 long noncoding RNA (lncRNA). The H19 lncRNA–MBD1 complex is required for the control of five genes of the IGN. For three of these genes—Igf2 (insulin-like growth factor 2), Slc38a4 (solute carrier family 38 member 4), and Peg1 (paternally expressed gene 1)—both MBD1 and H3K9me3 binding were detected on their differentially methylated regions. The H19 lncRNA–MBD1 complex, through its interaction with histone lysine methyltransferases, therefore acts by bringing repressive histone marks on the differentially methylated regions of these three direct targets of the H19 gene. Our data suggest that, besides the differential DNA methylation found on the differentially methylated regions of imprinted genes, an additional fine tuning of the expressed allele is achieved by a modulation of the H3K9me3 marks, mediated by the association of the H19 lncRNA with chromatin-modifying complexes, such as MBD1. This results in a precise control of the level of expression of growth factors in the embryo.
The imprinted H19 locus belongs to a conserved gene cluster on chromosome 7 in the mouse and 11p15.5 in human, and it plays an important role in embryonic development and growth control. The cluster contains the insulin-like growth factor 2 (Igf2) gene, located 90 kb away from the H19 gene, and both genes are coordinately regulated by an intergenic differentially methylated region (DMR) also called imprinting control region (ICR) and by downstream enhancers, with H19 being expressed from the maternal and Igf2 from the paternal allele (1, 2). The Igf2 gene is under the additional control of somatic DMRs 1 and 2 in the embryo. Both genes are strongly expressed during embryogenesis and down-regulated after birth, with H19 remaining expressed in adult skeletal muscle and heart.
The H19 gene produces a 2.3 kb spliced, capped, and polyadenylated long noncoding RNA (lncRNA) (3). The H19 locus also produces a microRNA (miR) from a highly conserved region in the first exon. This miR-675 plays a role in controlling placental growth at the end of gestation by regulating the expression of the Igf1r gene (4).
The targeted deletion of the gene (H19Δ3) induces an overgrowth phenotype (+ 8% compared with WT mice), which can be rescued by transgenic reexpression of H19 (5, 6). Expression of the Igf2 gene is affected by the deletion of the H19 gene, and it becomes biallelically expressed, with a 35% level of expression from the usually silent maternal allele. Similarly, eight other genes belonging to an Imprinted Gene Network (IGN) (7) also show an increased expression level in the absence of H19, which is restored to a normal level by transgenic reexpression. These data suggest that H19 acts in trans to regulate the expression of these genes and to control growth of the embryo (6). Whether this control is transcriptional or posttranscriptional and whether these nine targets are direct or indirect targets remain elusive.
Several lncRNAs interact with chromatin-modifying complexes and appear to exert a transcriptional control by targeting local chromatin modifications at discrete genomic regions (8, 9). In the case of imprinted clusters, the DMRs controlling the expression of imprinted genes exhibit parent-of-origin epigenetic modifications (DNA methylation and histone modifications) that govern the imprinting of the locus. In some of these clusters, lncRNAs control in cis the transcription of adjacent genes. For example, the Kcnq1ot1 lncRNA associates with the lysine methyltransferase G9a and the Polycomb Repressive Complex (PRC2) to regulate the expression of other genes of the locus in the placenta (10, 11). Similarly, in the context of X-inactivation, the Xist lncRNA associates with PRC2 and creates domains of repressive control on the inactive X chromosome (8). Alternatively, some lncRNAs, termed macroRNAs, such as Airn and Nespas, act by silencing promoters and enhancers by transcriptional overlap (12, 13). Finally, in other nonimprinted regions, several lncRNAs, such as HOTAIR, seem to exert their functional role by recruiting chromatin-modifying complexes for transregulation (14, 15).
To elucidate the mechanism of action of the H19 lncRNA on the genes of the IGN, we performed RNA immunoprecipitation (RNA-IP) with specific proteins and discovered that H19 RNA binds the methyl-CpG-binding domain protein 1 (MBD1). MBD1 belongs to the family of the methyl-CpG–binding domain proteins, such as MBD2, MBD3, MBD4, and MECP2 (methyl-CpG-binding protein 2) (16). MeCP2, MBD1, and MBD2 proteins bind to methylated DNA and recruit different histone deacetylase (HDACs)- and histone lysine methyltransferase (KMT)-containing complexes that control chromatin compaction and gene silencing (17). In particular, MBD1 associates with the KMTs SETDB1 and SUV39H1, responsible for H3K9 methylation (18, 19). MBD1 actually binds both to methylated and unmethylated DNA sequences, and its role is still unclear (20). Interestingly, it was shown that MBD1, as well as other MBD proteins, can bind RNA in vitro (21), but this was never explored in vivo.
In the current study, we show that the H19 lncRNA is one of the partners of MBD1. The H19 lncRNA–MBD1 complex participates in the control of several genes of the IGN, by modifying the repressive histone marks on DMR regions controlling their expression.
Results
The H19 RNA Controls the Imprint of Igf2 in Vivo.
Because several imprinted genes were up-regulated in the absence of a functional H19 gene (6), we investigated if this overexpression was due to loss of imprinting of these genes.
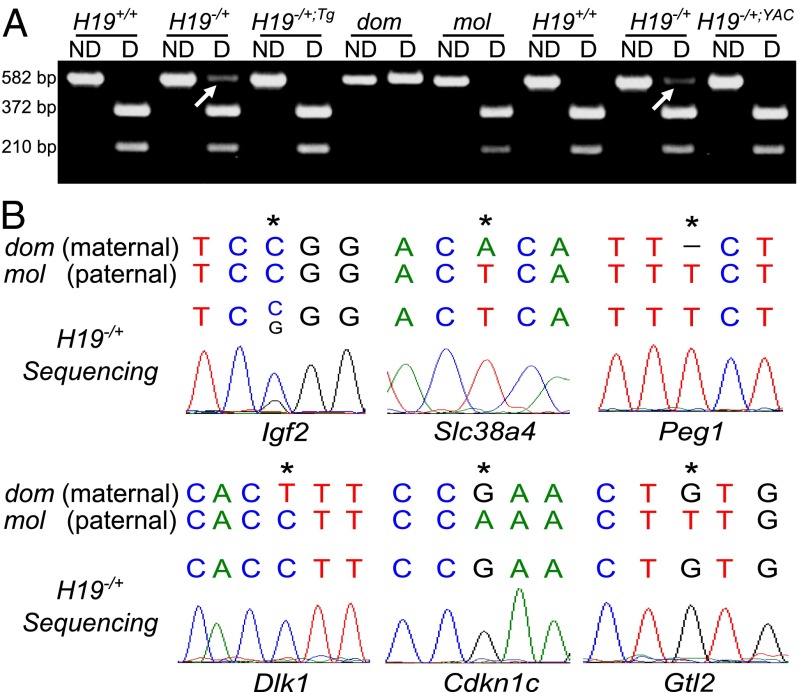
The H19Δ3 mice harbor a 3 kb deletion of the transcription unit. We established an H19 transgenic line (H19Tg) carrying the H19 transcription unit under the control of the necdin gene promoter and used the previously described YZ8 one copy YAC line (H19YAC) (22). These transgenic lines were bred onto an H19Δ3 background. H19−/+;Tg and H19−/+;YAC females were crossed with JF1 WT (Mus musculus molossinus or mol) males, to distinguish the parental origin of the alleles. Using restriction length polymorphism on cDNA samples from E14.5 muscle, we confirmed that the Igf2 gene loses its imprinted status in H19−/+ embryos (Fig. 1A). Interestingly, we observed that Igf2 imprint was rescued upon reintroduction of an H19 transgene, as Igf2 was exclusively expressed from the paternal allele in H19−/+;Tg and H19−/+;YAC embryos (Fig. 1A). These data suggest that the H19 RNA represses the maternal expression of the nearby Igf2 gene in trans and thus regulates Igf2 gene levels in embryonic limb muscle.
Fig. 1.
Allele-specific expression of the IGN. (A) Allele-specific expression analysis of the Igf2 gene detected by RT-PCR followed by MspI digestion in E14.5 limb muscle samples. Molossinus (mol, paternal) allele presents an MspI restriction site absent in the domesticus (dom, maternal) allele. Nondigested (ND) and digested (D) RT-PCR products are presented. On the left, results obtained with a H19−/+dom;Tg × JF1 mating. On the right, results obtained with a H19−/+;YAC × JF1 mating. Arrows show the maternal Igf2 allele. (B) Allele-specific expression analysis of other imprinted genes of the network detected by RT-PCR and sequencing in H19−/+ E14.5 limb muscle samples. Maternal (dom, domesticus) and paternal (mol, molossinus) sequences are indicated. Stars indicate polymorphisms between the two alleles.
We then focused on the other imprinted genes whose expression was increased in absence of the H19 gene. In contrast to the Igf2 gene, RT-PCR followed by sequencing indicated that the other H19 targets of the IGN (such as Slc38a4, Peg1, Dlk1, Cdkn1c, and Gtl2) remained monoallelicaly expressed in H19−/+ embryonic limb muscle (Fig. 1B). Dcn is biallelically expressed, however sequencing in WT embryonic limb muscle showed that this gene is actually not imprinted in this tissue (Fig. S1).
The H19 lncRNA Represses the IGN in MEFs.
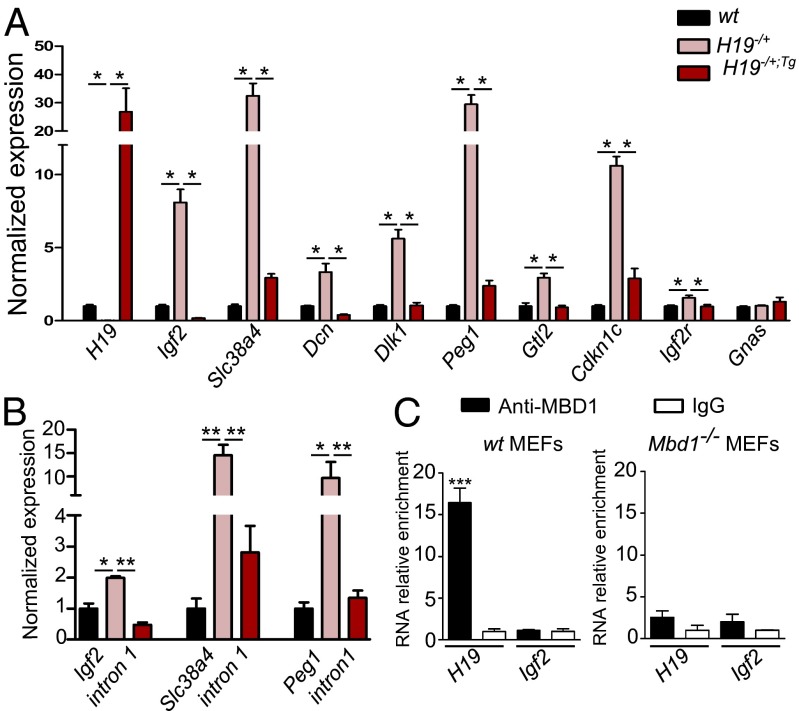
To study the molecular mechanisms that drive the H19-mediated repression of the IGN in the embryo, we chose to use primary mouse embryo fibroblasts (MEFs) as a model system. We measured IGN expression levels in WT, H19−/+, and H19−/+;Tg MEFs. We observed, as previously shown in limb muscles, that Igf2, Slc38a4, Dcn, Dlk1, Peg1, Gtl2, Cdkn1c, and Igf2r are overexpressed in cells lacking H19 RNA expression (Fig. 2A and Table S1). In MEFs that ectopically express the H19 RNA in an H19−/+ background (H19−/+;Tg), the expression of these genes is restored to WT level. Thus, both in MEFs and in embryonic limb muscle, the H19 gene negatively regulates several genes of the IGN. The level of expression of these target genes was often higher in MEFs compared with that in embryonic muscle samples. This suggests that a strict control of the expression of growth-controlling genes is exerted in vivo, whereas this control is more flexible in an in vitro culture system.
Fig. 2.
H19 modulates the IGN in MEFs and interacts with the MBD1 protein. (A) Expression levels in E14.5 primary MEF samples were detected by RT-qPCR. The expression level of WT MEFs was set at 1, and histograms show modifications relative to this level (n = 4 for each genotype). (B) Primary transcript levels were detected using primers located in introns of the genes. (C) RIP with an antibody to MBD1 indicates binding to H19 in WT MEFs. The enrichment of RNA over a random IgG is shown. Igf2 mRNA was used as a negative control. The specificity of the antibody was tested by performing the experiment in Mbd1−/− MEFs.
We also evaluated the levels of expression of the primary transcripts using primers in introns of target genes (Fig. 2B). For Igf2, Slc38a4, and Peg1 genes, an increase in the level of primary transcript expression was detected in H19−/+ compared with WT MEFs. The primary transcripts are further down-regulated in the presence of the H19 transgene, suggesting a possible transcriptional effect on the levels of expression of these three genes. However, we cannot fully exclude a posttranscriptional effect, as alternative splicing or nuclear degradation of transcripts containing introns could also occur.
H19 lncRNA Interacts with the MBD1 Protein.
Because the level of expression of several imprinted genes was restored to WT levels in H19−/+Tg cells, we hypothesized that the H19 lncRNA may interact with proteins involved in epigenetic modifications. RNA immunoprecipitation assays (RIPs) were performed on WT MEFs. Interestingly, we observed that the H19 RNA significantly coimmunoprecipitated with the MBD1 protein, previously shown to have a high affinity for RNA (Fig. 2C). The Igf2 mRNA was used as a control for binding specificity, and the RIP experiment was performed in Mbd1−/− MEFs to confirm the specificity of the MBD1 antibody. No interaction of the H19 RNA was detected with either EZH2 or SUZ12, components of the PRC2 complex (Fig. S2). This result led us to hypothesize that H19 could possibly control IGN expression through its interaction with the MBD1 protein.
MBD1 Is a Repressor of the IGN.
To test if the MBD1 protein was involved in the H19-mediated repression of the IGN, we first investigated the expression level of the IGN in Mbd1−/− MEFs (Fig. 3A). Five genes of the IGN (Igf2, Slc38a4, Dcn, Dlk1, and Peg1) are overexpressed in Mbd1−/− compared with WT MEFs. This indicates that the repression of these genes is not only dependent on the presence of H19 (Fig. 2A), but also on the presence of the MBD1 protein. We also performed Mbd1 siRNA-mediated knockdown experiments. In cells treated with a Mbd1 siRNA, these five genes were up-regulated compared with cells treated with a nonsilencing control siRNA (Fig. 3B). This confirms that MBD1 is necessary for the maintenance of IGN repression in MEFs. Gtl2, Cdkn1c, and Igf2r are not overexpressed in Mbd1−/− MEFs compared with WT MEFs, suggesting that their repression is dependent on H19 but does not require MBD1. Together, these results show that five out of nine genes of the IGN are common targets of both H19 and MBD1, suggesting that this protein may be involved in the function of H19 lncRNA as a repressor of this network.
Fig. 3.
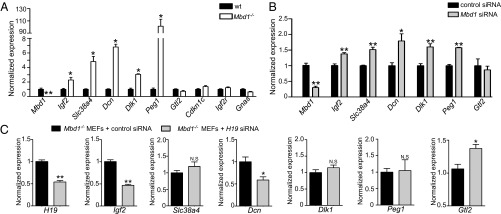
H19 represses its targets via the MBD1 protein. (A) Expression levels of the IGN in Mbd1−/− samples were detected by RT-qPCR. The expression level in WT MEFs was set at 1, and histograms show modifications relative to this level (n = 4). (B) siRNA-mediated knockdown experiments of MBD1 in WT MEFs. The expression level in MEFs treated with a nonsilencing control was set at 1, and histograms show modifications relative to this level (n = 6). (C) siRNA-mediated knockdown experiments of H19 in Mbd1−/− MEFs. The expression level in MEFs treated with a nonsilencing control was set at 1, and histograms show modifications relative to this level (n = 6).
H19 lncRNA Requires MBD1 to Repress Its Targets.
The next challenge was to test if H19 lncRNA and MBD1 act together to repress the IGN, or if they act through independent pathways. We performed siRNA-mediated knockdown of H19 lncRNA in Mbd1−/− MEFs to test if H19 requires the MBD1 protein to repress the five genes of the IGN that show H19 lncRNA and MBD1 dependence. We observed that Gtl2, a target of H19 but not of MBD1, was overexpressed in cells treated with an H19 siRNA compared with cells treated with a control siRNA, as expected (Fig. 3C). In contrast, the expression of common targets of H19 and MBD1, such as Slc38a4, Dlk1, and Peg1, was not affected by the down-regulation of H19 lncRNA in Mbd1−/− MEFs, whereas Igf2 and Dcn mRNA levels were strongly down-regulated (50% reduction) (Fig. 3C). These data indicate that H19 lncRNA requires the MBD1 protein to repress H19–MBD1 common targets. This also suggests that in the absence of MBD1, H19 lncRNA could act as an activator of Igf2 and Dcn rather than a repressor.
In summary, these data demonstrate that the H19 lncRNA represses five genes of the IGN (Igf2, Slc38a4, Dcn, Dlk1, and Peg1) in a manner dependent on the MBD1 protein.
Direct Targets of the H19 lncRNA–MBD1 Complex.
The MBD1 protein is a DNA-methylation–dependent transcriptional repressor that also binds unmethylated CpG islands via its CXXC domain (20, 23). To test if genes of the network modulated by the H19 lncRNA–MBD1 complex are direct or indirect targets, we performed MBD1 ChIP–quantitative PCR (qPCR) experiments in MEFs. MBD1 binds to the DMR1 region of Igf2 and to the DMRs of Slc38a4 and Peg1 in WT MEFs (Fig. 4A). The Slc38a4 and Peg1 paternally expressed genes harbor maternally methylated gametic DMRs. The Igf2 somatic DMR1 is an unusual DMR, as it displays DNA hypermethylation on the paternal expressed allele and is thought to bind a repressor protein on the maternal allele, which results in a silencer effect on this allele (24). Interestingly, further sequencing of the immunoprecipitated DMRs using JF1 polymorphisms showed that the MBD1 protein binds to both the paternal and the maternal alleles (Fig. 4C and Fig. S3). We next performed MBD1 ChIP experiments in H19−/+ MEFs to test if the H19 lncRNA was required for the binding of MBD1 to the Igf2 DMR1 and Slc38a4 and Peg1 DMRs observed in WT MEFs. As expected, we found that MBD1 binding was lost in the absence of H19 lncRNA (Fig. 4A). The Cdkn1c DMR, a target of H19 lncRNA but not of MBD1, was used as a control and no difference in the binding of MBD1 to this region was observed (Fig. 4A). Taken together, these results suggest that H19 lncRNA directly represses Igf2, Slc38a4, and Peg1 by recruiting the MBD1 protein to the DMRs of these three imprinted genes.
Fig. 4.
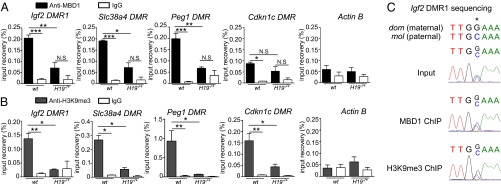
H19 is necessary for recruitment of MBD1 and H3K9me3 to the Igf2 DMR1 and Slc38a4 and Peg1 DMRs. (A) Chromatin immunoprecipitation with an antibody to MBD1 in WT and H19−/+ MEFs (n = 4). Histograms represent the ratio of immunoprecipitated DNA relative to the input sample. Igf2 DMR1, Slc38a4, Peg1, and Cdkn1c DMR regions were analyzed. Actin B was included as a negative control. (B) Chromatin immunoprecipitation with an antibody to H3K9me3 in WT and H19−/+ MEFs (n = 3). (C) Sequencing of immunoprecipitated Igf2 DMR1 in WT MEFs. Maternal (dom, domesticus) and paternal (mol, molossinus) sequences are indicated. Star shows polymorphism between the two alleles.
Decrease of H3K9me3 in the Absence of H19 lncRNA.
MBD1 is involved in the maintenance of H3K9me3 through cell division, especially by recruiting KMTs (18). We therefore hypothesized that H19 lncRNA, because it interacts with MBD1, could be involved in the establishment of this histone mark to repress its targets. To address this, we investigated H3K9me3 levels at DMRs by ChIP and qPCR. H3K9me3 was indeed present at the Igf2 DMR1 and Slc38a4 and Peg1 DMRs in WT MEFs (Fig. 4B). Further sequencing of the immunoprecipitated Igf2 DMR1 and Peg1 and Slc38a4 DMRs, using JF1 polymorphisms, also showed that this mark was present on both alleles, even if it is clearly more important on the maternal allele for the Igf2 gene (Fig. 4C and Fig. S3). In H19−/+ MEFs, we observed a loss of H3K9me3 on the Igf2, Slc38a4, and Peg1 DMRs. H3K9me3 was also lost on the Cdkn1c DMR, despite the fact that this gene is not dependent on MBD1.
This shows that H19 is important for the maintenance of the H3K9me3 transcriptional repressive mark, which is concordant with a control of Igf2, Slc38a4, and Peg1 through an interaction with MBD1 and the H3K9 KMTs.
Discussion
The H19 locus has been shown to be involved both in embryonic growth control and tumorigenesis (5, 25, 26). H19 and Igf2 belong to the IGN, first described in 2006 (7). Using loss- and gain-of-function mouse mutants, we showed that H19 itself has several targets among the IGN and is capable of repressing the expression of these genes in E14.5 embryonic muscle (6). This results in a fine-tuned regulation of embryonic growth mediated by the H19 gene. Identification of the underlying molecular mechanisms through which it controls its targets is an important issue.
Recent studies have shown that this locus could act through the production of the miR-675 to control placental growth (4) and that the full-length H19 RNA could interact with the EZH2 protein and inhibit E-cadherin expression in bladder cancer metastasis (27). Therefore, the H19 locus appears to have multiple functions by acting on specific genes through distinct molecular mechanisms, depending on the biological and spatiotemporal contexts during development and disease states.
Here we show that in primary MEFs produced from midgestation embryos, the methyl-CpG–binding domain protein MBD1 is a partner of the H19 lncRNA. This H19–MBD1 complex induces the H3K9me3 histone tail modifications on DMRs of target genes, such as Igf2, Slc38a4, and Peg1. This results in the repression of these genes in the presence of H19 lncRNA. Interestingly, the modulation of expression of the IGN is found neither in neonate or adult muscle nor in placenta (6). Therefore, we suspect that this H19 lncRNA interaction with MBD1 is specific to embryonic stages. This is probably linked with the establishment of the histone marks at a specific moment in early development. This is then maintained throughout further cell divisions by MBD1-dependent association with chromatin assembly factor CAF1 at the replication forks (18).
Interestingly, MBD1 has been described as having affinity for RNA by in vitro experiments (21). Here we identify H19 as one of the RNA partners of MBD1. The H19 gene was thought to be essentially found in the cytoplasm, as it associates with polysomes (28). Our experiments now also provide evidence for a nuclear role for this lncRNA.
Finally, MBD1 has been described as having affinity both for methylated and unmethylated DNA sequences (23, 29). Recent data have suggested that a mutant of MBD1 lacking the MBD domain but conserving the CXXC region shows preferential binding for unmethylated sequences (20). It is possible that the binding of H19 RNA to the MBD1 protein modifies the binding capacities of MBD1 and directs it to both methylated and unmethylated DMRs.
In our study, we identified five genes as being targets of both H19 lncRNA and MBD1. The double deletion experiment using siRNA against H19 in Mbd1−/− MEFs confirmed that MBD1 is required for the function of H19 lncRNA to repress these genes. Interestingly, these five common targets of H19 and MBD1 are all paternally expressed genes (except for Dcn, which is not imprinted at this stage of development). This observation could reinforce the parental conflict theory, in the sense that the maternally expressed H19 gene controls growth by repressing paternally expressed genes.
In contrast, four other genes (Gtl2, Cdkn1c, Igf2r, and Gnas) are regulated by H19 lncRNA but are not affected by the absence of MBD1. For example, Cdkn1c is a target of H19 but not of MBD1, and H3K9 methylation is reduced on its DMR. This therefore suggests that H19 could repress these other genes through other chromatin-modifying partners. The control of expression of the IGN genes can therefore be mediated by an epigenetic effect via the H19 lncRNA. Alternatively, these genes could be controlled by posttranscriptional effects, although an effect of the miR-675 can be excluded, as it is not expressed in the embryo (4).
In addition, an indirect effect of H19 could be mediated by direct targets acting on the other IGN genes. For example, Igf2r is a negative regulator of the insulin and IGF signaling pathway. Therefore, in H19−/+ mice, increased Igf2 expression may be compensated by Igf2r overexpression as a response to maintain overall homeostasis.
Because the IGN genes seem to play an essential role in controlling the growth of the embryo, it would seem likely that a complex regulation involving several levels of control, including transcriptional, posttranscriptional, and indirect mechanisms, would be required for this crucial process. Our experiments show that H19 lncRNA is required for direct binding of MBD1 to regulatory regions of Igf2, Slc38a4, and Peg1 genes. Therefore, we postulate that H19 could recruit the MBD1 protein to its DNA targets. How H19 lncRNA recognizes the correct DNA regions remains unknown. The H19 RNA could possibly produce a triplex structure with target DNA sequences, similar to what has been described for rDNA genes (30).
Our detailed study of the Igf2 gene and the binding of MBD1 to the Igf2 DMR1 shows that MBD1 targets both alleles of the Igf2 gene. The H3K9me3 modification is therefore brought to both alleles (Fig. 5). The reduction of H3K9me3 on the Igf2 DMR in the absence of H19 is quite straightforward, as this reduction will lead to expression of the normally silent maternal Igf2 allele. Although it is difficult to evaluate if the expression of the paternal allele is also increased, this is reminiscent of a previous observation in which the DNA methylation profiles of the Igf2 locus were studied in H19Δ3 and H19Δ13 mice (31). DNA methylation was acquired on the maternal Igf2 allele (and resulted in loss of imprint) but was also lost on the paternal allele. The authors suggested a cross-talk between the two alleles with exchange of methylation from one allele to the other. Thus, this DMR1 region appears to be a flexible region with respect to epigenetic marks, such as DNA and histone methylation. This may reflect a specific property of the somatic DMRs.
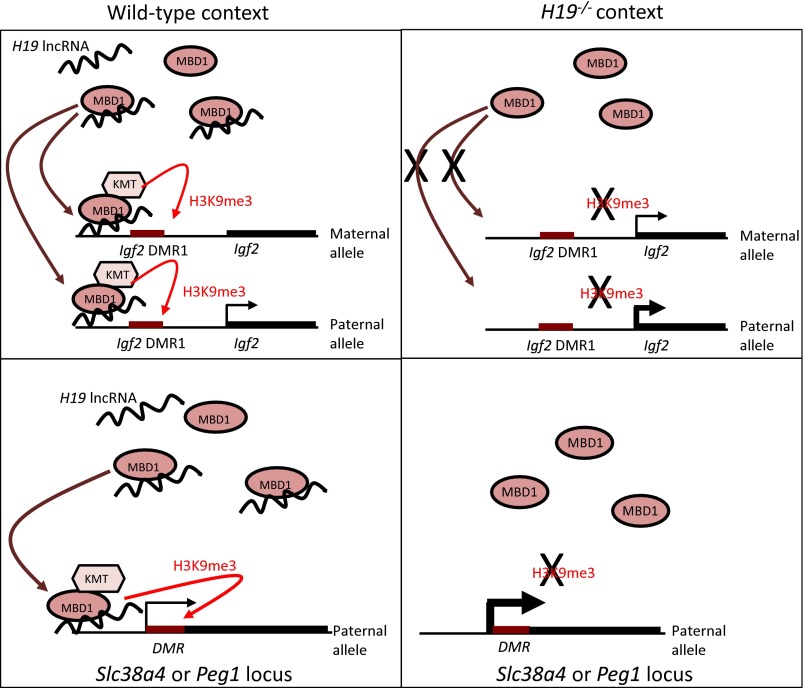
Fig. 5.
Model of H19-mediated regulation of Igf2, Slc38a4, and Peg1 genes. In WT cells, the H19 lncRNA interacts with the MBD1 protein and recruits it to the Igf2 DMR1, both on the maternal and paternal allele (Upper Left). This recruitment induces H3K9me3 on both alleles, probably via interaction with an H3K9 KMT. In H19−/+ cells, MBD1 cannot be recruited to the Igf2 DMR1, leading to a loss of H3K9me3 (Upper Right). This results in an increase of Igf2 transcription, concomitant with a loss of Igf2 imprinting. On the Slc38a4 and Peg1 paternal DMRs, the H19 lncRNA recruits MBD1 and induces H3K9me3 (Lower Left). In absence of H19, the lack of binding of MBD1 results in a loss of H3K9me3 and in overexpression of the paternal allele (Lower Right). Therefore, H19 exerts a fine-tuned regulation of these genes, by modulating the presence of the repressive H3K9me3 histone mark on the active alleles.
Finally, we have provided evidence that Slc38a4 and Peg1 overexpression in the absence of H19 lncRNA is linked to a decrease in H3K9me3 at their gametic DMR. In the WT samples, this repressive histone mark is present on both alleles, a situation that has also been previously described for the Rasgrf1 gene, another paternally expressed gene (32). Other marks independent of H19, such as H3K27me3 marks and/or DNA methylation, must act as an additional lock to prevent expression from the silent allele (33). The importance of controlling the level of expression of the Slc38a4 gene (a system A amino acid transporter) was recently illustrated by the observation that human placentas from low birth weight children (with fetal macrosomia) showed an increase in Slc38a4 expression (34). The control of H19 on the DMR of this gene could be involved in maintaining low expression of Slc38a4 in the embryo to obtain normal development.
In conclusion, we have identified the MBD1 protein as a partner of the H19 full-length lncRNA. This complex brings H3K9me3 modifications to chromatin at DMRs of certain imprinted gene targets of H19 (Fig. 5). The DMR regions could therefore represent areas in which repressive H3K9me3 histone marks are present on both alleles, even though they are clearly defined by differential DNA methylation. The effect of the H19 RNA is to tether the MBD1 protein to these regions and to provide H3K9me3 modifications to finely control the level of expression of the normally expressed allele. This is an interesting observation, as it was thought up to now that imprinted genes were “on” or “off” depending on their parental origin. Our data suggest that even the expressed alleles are under a precise control to avoid overexpression of these genes controlling growth. Mediating this control through lncRNAs associated with chromatin-modifying complexes brings an additional level of fine tuning of embryonic growth.
Materials and Methods
Mouse Strains.
All experimental designs and procedures were in agreement with the guidelines of the animal ethics commitee of the Ministère de l’Agriculture (France). The H19Δ3 (H19−/−), Tg, and YAC line used in this work were previously described (6). H19Δ3/+;Tg or H19Δ3/+;YAC females were mated with WT JF1 males. Wt, H19−/+ and H19−/+;Tg, or H19−/+;YAC embryos (E14.5) were then dissected to collect limb muscle or to produce primary MEFs.
Allele-Specific Expression Analysis.
Polymorphims between Mus musculus molossinus (JF1) and Mus musculus domesticus were extracted from previous studies (35), or from the National Institute of Genetics (NIG) mouse genome database (http://molossinus.lab.nig.ac.jp/msmdb/index.jsp). Reverse transcription was performed using PrimeScript Reverse Transcriptase (Takara Bio Inc.) followed by PCR with GoTaq polymerase (Promega) according to the manufacturer’s guidelines, with oligos that span an exon region that harbors a single defined base difference or a restriction fragment length polymorphism between the two mouse strains. Allele-specific expression was determined either by sequencing or digestion by MspI for the Igf2 gene. Primers are listed in Table S2.
Gene Expression Analysis.
Total RNAs were extracted using miRNeasy kit (Qiagen), and reverse transcription was performed as above. RT-qPCR was performed on 10 ng cDNA in 10 mL final volume with SYBR qPCR Premix Ex Taq (Takara Bio Inc.) in a LightCycler 2.0 apparatus (Roche). Gene expression levels were normalized to the geometric mean of the expression levels of Sdha, Tfrc, and ActB housekeeping genes with geNorm software (v3.4) (Table S1) (36).
RIP Experiments.
RNA ChIP Kit (Active Motif) was used to perform RIP experiments, according to the manufacturer’s instructions. Two million cells were used per IP. Cells were fixed in 1% formaldehyde solution (Sigma). Total chromatin and RNAs were sonicated into 200–800 bp fragments, using a Bioruptor (Diagenode). We used 1% of the chromatin/RNA to purify the input RNA. The remaining chromatin/RNA was immunoprecipitated with 2 µg of MBD1 antibody (Diagenode pAb-078-050), EZH2 antibody (Diagenode pAb-039-050), SUZ12 antibody (Diagenode pAb-029-050), or 2 µg of nonspecific rabbit IgG (negative control, Diagenode kch-504-250). RNA was purified using TRIzol LS reagent (Life Technologies), reverse-transcribed as above, and analyzed by RT-qPCR.
SiRNA-Mediated Knockdown Experiments.
RNAi-mediated knockdown was performed with Stealth RNAi siRNA against Mbd1 (Life Technologies, mss206539) and by Silencer Select siRNA against H19 (Life technologies, 4390771) or a nontargeting control (12935–400 and 4390843), at a final concentration of 50 pmol/mL. Transfection of oligos into cell lines was achieved using Lipofectamine RNAimax (Life Technologies). RNA was extracted 48 h later.
Chromatin Immunoprecipitation Experiments.
ChIP experiments were performed using the HighCell ChIP kit (Diagenode), according to the manufacturer’s instructions. One million cells were used per IP. Cells were grown to 80–90% confluency and then fixed with 1% formaldehyde solution (Sigma). Chromatin was sonicated into 200–800 bp fragments, and 1% of the chromatin was used to purify the input DNA. Chromatin was immunoprecipitated with 2 µg of MBD1 antibody, H3K9me3 antibody (Diagenode CS-056-100), or nonspecific rabbit IgG. DNA was purified and analyzed by qPCR.
Statistical Analysis.
Data are presented as the mean ± SEM. Statistical significance of the different experiments was determined using a Kruskal–Wallis test followed by post hoc paired comparisons, or a Mann–Withney test, using Prism software (v5.0a). Results were considered statistically significant when P < 0.05 compared with WT *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We are grateful to J. F.-X. Ainscough and M. A. Surani for providing the YZ8 YAC transgenic line. We are grateful to Y. Styczen for additional experiments and to T. Forné for helpful advice. We are very thankful to E. Heard for constant encouragement. This work was supported by funding from the Ligue contre le Cancer, the Association Française contre les Myopathies, the Agence Nationale de la Recherche Epinet Project (to L.D.), and the Association de la Recherche contre le Cancer and Grants ANR-11-IDEX-0005-02 and ANR-11-LABX-0071 (to P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310201110/-/DCSupplemental.
References
- 1.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12(23):3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 3.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 6.Gabory A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 7.Varrault A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Wutz A. RNA-mediated silencing mechanisms in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:351–376. doi: 10.1016/B978-0-12-387685-0.00011-1. [DOI] [PubMed] [Google Scholar]
- 9.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Wagschal A, et al. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol. 2008;28(3):1104–1113. doi: 10.1128/MCB.01111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338(6113):1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 13.Williamson CM, et al. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 2011;7(3):e1001347. doi: 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18(11):6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defossez PA, Stancheva I. Biological functions of methyl-CpG-binding proteins. Prog Mol Biol Transl Sci. 2011;101:377–398. doi: 10.1016/B978-0-12-387685-0.00012-3. [DOI] [PubMed] [Google Scholar]
- 18.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15(4):595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 19.Fujita N, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003;278(26):24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 20.Baubec T, Ivánek R, Lienert F, Schübeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153(2):480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279(47):49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 22.Ainscough JFX, Koide T, Tada M, Barton S, Surani MA. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124(18):3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol. 2004;24(8):3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constância M, et al. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26(2):203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 25.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimizu T, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105(34):12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo M, et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 28.Li YM, et al. The H19 transcript is associated with polysomes and may regulate IGF2 expression in trans. J Biol Chem. 1998;273(43):28247–28252. doi: 10.1074/jbc.273.43.28247. [DOI] [PubMed] [Google Scholar]
- 29.Clouaire T, de Las Heras JI, Merusi C, Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res. 2010;38(14):4620–4634. doi: 10.1093/nar/gkq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24(20):2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forné T, et al. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc Natl Acad Sci USA. 1997;94(19):10243–10248. doi: 10.1073/pnas.94.19.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindroth AM, et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 2008;4(8):e1000145. doi: 10.1371/journal.pgen.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh P, et al. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol Cell Biol. 2011;31(8):1757–1770. doi: 10.1128/MCB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, et al. Association of SLC38A4 and system A with abnormal fetal birth weight. Exp Ther Med. 2012;3(2):309–313. doi: 10.3892/etm.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue K, et al. Faithful expression of imprinted genes in cloned mice. Science. 2002;295(5553):297. doi: 10.1126/science.295.5553.297. [DOI] [PubMed] [Google Scholar]
- 36.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):H0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.